Abstract
Many experiments involving nucleic acids require the hybridization and ligation of multiple DNA or RNA molecules to form a compound molecule. When one of the constituents is single stranded, however, the efficiency of ligation can be very low and requires significant individually tailored optimization. Also, when the molecules involved are very long (>10 kb), the reaction efficiency typically reduces dramatically. Here, we present a simple procedure to efficiently and specifically end-join two different nucleic acids using the well-known biotin–streptavidin linkage. We introduce a two-step approach, in which we initially bind only one molecule to streptavidin (STV). The second molecule is added only after complete removal of the unbound STV. This primarily forms heterodimers and nearly completely suppresses formation of unwanted homodimers. We demonstrate that the joining efficiency is 50 ± 25% and is insensitive to molecule length (up to at least 20 kb). Furthermore, our method eliminates the requirement for specific complementary overhangs and can therefore be applied to both DNA and RNA. Demonstrated examples of the method include the efficient end-joining of DNA to single-stranded and double-stranded RNA, and the joining of two double-stranded RNA molecules. End-joining of long nucleic acids using this procedure may find applications in bionanotechnology and in single-molecule experiments.
INTRODUCTION
Many experiments involving the manipulation of nucleic acids require the synthesis of complicated molecular constructs from different constituting molecules of DNA or RNA. In single-molecule experiments, for example, constructs typically contain molecular handles (1–3) or specific sequences such as DNA hairpins (4,5), DNA–RNA hybrids (6,7), or promoter sequences (8). For certain applications the molecules involved are very long, either because the experimental setup requires a certain minimum length (9–11) or because the studied phenomena take place over long distances (12). An example of an experiment requiring very long molecules involves the translocation of single DNA molecules through solid-state nanopores (13–15). Here, molecules are passed through a nanometer-sized hole in a thin membrane by an applied electrical field and can be detected by a commensurate change in the ionic current through the pore. As this process is typically very fast (13), individual molecules have to be very long (>>1 kb) in order to be resolved. Furthermore, increased sophistication of these experiments, for instance via integration with optical tweezers (9,16), will permit the interrogation of more complex lengthy molecular constructs.
In such cases, the synthesis of molecular constructs can pose a serious challenge to the experimentalist, because standard biochemical reactions can become much less efficient for long molecules (17,18). The standard way to construct these molecules is to create complementary overhangs, which hybridize together and can then be ligated to form a stable construct. Although this typically works well for relatively short molecules, the yield of these reactions can drop off dramatically when the molecules involved reach lengths over 10 kb. Similar techniques using a complementary ‘splint’ molecule to join single-stranded molecules of DNA or RNA have been shown to be useful for joining short oligonucleotides (19,20), but they require the careful selection of the optimal splint molecule and have not demonstrably led to high yields for molecules longer than several hundred bases. To resolve some of these difficulties, we here present a straightforward procedure to efficiently end-join two arbitrary molecules in a manner that is independent of the molecule length (up to at least 20 kb).
Our method relies on the use of streptavidin (STV) to facilitate efficient end-coupling between two arbitrary nucleic acids A and B, where both A and B can be either DNA or RNA and of arbitrary length. In this scheme the STV acts as a linker between the molecule ends, which can be biotinylated chemically or enzymatically (e.g. using terminal transferase, Klenow polymerase, or poly(A) polymerase) using standard available techniques (21–26). Although a considerable amount of previous work using STV to crosslink DNA molecules has been performed, this has primarily focused on the coupling of oligonucleotides or relatively short DNA molecules (27–31) up to 2 kb (32). Also, such research typically had the aim of either forming multimers of DNA–STV complexes or long strings of bis-biotinylated molecules (biotin on both ends) for the purpose of scaffolding in nanoengineering (30,31,33) or as useful aids in detection assays (29,34). Methods that specifically aim to optimize the formation of a unique end-product, such as a heterodimer A+B, are lacking.
STV is an excellent choice as a coupling agent, because it binds biotin with an extremely high binding affinity (Kd ∼ 10−15 M) and has multiple biotin binding sites (35,36). Indeed, wild-type STV contains four binding sites for biotin, so it could in principle join up to four biotin-labeled molecules. By simply adding STV to a collection of different biotinylated molecules, one would therefore expect to form a variety of dimers, trimers and tetramers. To specifically promote the formation of the heterodimer A+B, we here introduce a two step approach, in which the STV is first bound only to molecule A. Molecule B is then added only after the remaining unbound STV is removed, so that B must bind to the STV attached to molecule A. This not only avoids the formation of homodimers A+A and B+B, but, as we show below, also significantly suppresses the formation of trimers and tetramers.
MATERIALS AND METHODS
Synthesis of DNA and RNA
DNA molecules were synthesized by PCR derived from phage lambda DNA (Promega). For all DNA molecules used to bind STV, only one of the primers was 5′-biotinylated, so that only one end of each molecule can bind STV. The following forward and reverse primers (Biolegio, Leiden, The Netherlands) were used for the fragments: 2.2 kb (2250 bp) DNA: 5′-CCAGAAAAATGCATTCCGTG-3′ (forward) and 5′-biotin-GGATATTAATACTGAAACTGAGATCAAGC-3′ (reverse). 3.5 kb (3524 bp) DNA: 5′-AAAAGAATTCAGCCTCAGCTGACCAGCCAGAAAACGACC-3′ (forward) and 5′-biotin- CTTGTTGGGCTTGTTTAATCCAGTAACTGC-3′ (reverse). 4.2 kb (4215 bp) DNA: 5′-TGATATTGCCAAAACAGAGCTG (forward) and 5′-GGAAAGGGCCCGTAAAGTGATAATGATTATCATC-3′ (reverse). The underlined sequence corresponds to a restriction site for ApaI. 9.6 kb (9571 bp) DNA: 5′-AAAAAAGCTTGCGAGAATTTTTAGCCCAAGC-3′ (forward) and 5′-biotin-GGATATTAATACTGAAACTGAGATCAAGC (reverse). 12.7 kb (12668 bp) DNA: 5′-biotin-GAGGCCGGGTTATTCTTGTTCTCTGG (forward) and 5′-GGAAAGGGCCCGTAAAGTGATAATGATTATCATC-3′ (reverse). 20 kb (19945 bp) DNA: 5′-biotin-GACGCAGGGGACCTGCAG (forward) and 5′-AAAAGGTCTCTTCATGCGTTCAGTCTTAAAAGCAATT (reverse).
The 4.2 kb and 12.7 kb DNA molecules were used in the ligation reaction in Figure 2b. The reverse primer is identical for both molecules and includes an ApaI site (underlined). The molecules were digested overnight with an excess of ApaI at 25°C. After digestion, both molecules contain the same overhang for ligation. The ligation reactions were performed in equimolar concentrations in a 20 µl volume with T4 DNA ligase (New England Biolabs, Ipswich, MA, USA) in T4 ligase buffer. All PCR fragments and digested molecules were purified with the nucleospin extract II kit (Machery-Nagel, Düren, Germany) and concentrations were determined using the commercial instrument Nanodrop (Isogen, IJsselstein, The Netherlands).
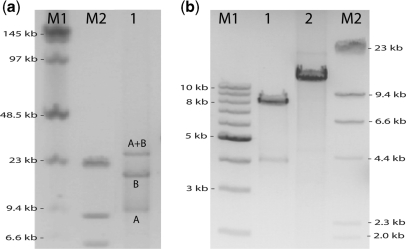
Figure 2.
End-joining very long DNA. (a) Pulsed field electrophoresis gel image illustrating the efficient end-joining of two very long DNA molecules. Lane 1 contains the product after joining a 20 kb (A) DNA molecule to a 9.6 kb (B) DNA molecule using our two step protocol. Using markers M1 and M2 as a reference, we can see bands corresponding to the molecules A, B, and an additional band corresponding to DNA longer than 23 kb, which is likely the A + B dimer of ∼30 kb DNA. The efficiency of dimer formation (∼50%) is similar to that of the shorter molecules in Figure 1. (b) Pulsed field electrophoresis image illustrating that the ligation efficiency to join two molecules is strongly reduced for longer molecules: In lane 1 we show the ligation of a 4.2 kb long DNA molecule to itself (the molecules have a small self-complementary overhang), which clearly has a very high efficiency of about 90%, judging from the relative intensity of the band at about 8.5 kb DNA. However, the ligation of a 12.7 kb molecule with exactly the same overhang is much less efficient: a band at ∼25 kb is not observed.
The 4.2 kb single-stranded RNA (ssRNA) and double-stranded RNA (dsRNA) molecules used in Figure 3a were synthesized via run-off transcription of DNA PCR fragments derived from pBADb10 (a recombinant pBAD vector) using T7 RNA polymerase. The PCR primers (Biolegio, The Netherlands) for the template DNA were designed to incorporate a T7 RNA promoter sequence at one end of the PCR fragment. The template DNA for the ssRNA molecule contained the following forward and reverse primers (the T7 promoter is underlined): 5′-TAATACGACTCACTATAGGAAGATTAGCGGATCCTACCTGAC-3′ (forward) and 5′-CGCAGCCAGCCATCGGAACCGGGGTTAACCTCAACTTCC-3′ (reverse).
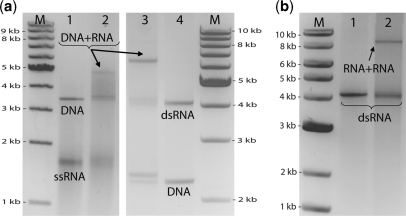
Figure 3.
End-joining DNA to RNA and RNA to RNA. (a) Joining DNA to RNA. Lane 1 contains the result following our two step protocol without addition of STV, using a 3.5 kb DNA (A) molecule and a 4.2 kb ssRNA molecule (B). The ssRNA is seen to migrate at the approximately at the same speed as 1.5 kb DNA and –as expected- we see only bands corresponding to molecules A and B. In lane 2, where the protocol was followed in the presence of STV, a band appears at ∼5 kb DNA, corresponding to the A + B dimer. In lanes 3 and 4 we illustrate the same procedure with two different molecules: a 2.2 kb DNA molecule (A) and a 4.2 kb dsRNA molecule (B). Again, in the control experiment without STV no dimers are formed (lane 4). Conversely, in lane 3, after following our regular protocol in the presence of STV, an additional band is observed at ∼6.5 kb. As the bare dsRNA molecule migrates at ∼4.2 kb DNA, the band at 6.5 kb most likely corresponds to the A + B dimer. The efficiencies in both cases are similar to or even better than DNA–DNA binding shown in Figure 1. (b) Joining RNA to RNA. The same 4.2 kb dsRNA molecule used above was used with our protocol to create an 8.4 kb dsRNA molecule. Lane 1 contains the result following our two-step protocol without addition of STV, using the 4.2 kb RNA molecule as both A and B: no dimers are formed. However, in the presence of STV (lane 2) an additional band is formed at ∼8.5 kb DNA corresponding to the dsRNA–dsRNA dimer.
A small biotinylated oligonucleotide, 5′-bio-CGCAGCCAGCCAUCGGAACC-3′, was hybridized to the 5′-end of the 4.2 kb ssRNA molecule to facilitate the binding to STV.
For the dsRNA molecule, the complementary strand of the same DNA template was also transcribed by introducing the T7 promoter in the reverse primer. In this case, the PCR primers used for the DNA template were: 5′-AAGATTAGCGGATCCTACCTGAC-3′ (forward) and 5′-TAATACGACTCACTATAGGGGTTAACCTCAACTTCCATTTCC-3′ (reverse).
Run-off transcription of this DNA template yields an ssRNA molecule that is complementary to and slightly shorter than the 4.2 kb ssRNA molecule described above. After hybridization of these two RNA molecules, the double-stranded product includes a 20 nt 3′ overhang, to which the same biotinylated oligonucleotide was hybridized as used above. All hybridizations were performed following a procedure described previously (37).
DNA–STV and RNA–STV binding
All incubations were performed in buffer 1 (0.25 × TBE at pH 8.3, 2 mM MgCl2). Unless stated otherwise, the first incubation step was always performed at 37°C for a duration of 30 min, and the second incubation step was performed at 4°C for 30 min up to several hours.
Removal of STV
We used two different methods to remove unbound STV following the first incubation step: column purification using the Microcon YM-100 (Millipore, Amsterdam, The Netherlands), and gel purification. The column purification was carried out by loading the sample onto the column and adding ddH2O up to a total volume of 200 µl. After ∼1 min of centrifugation at 10 000 rpm, 200 µl ddH2O was added, and the sample was again centrifuged for ∼1–3 min at 10 000 rpm. The sample was then eluted in 20 µl ddH2O and used for further experiments. Removal of the free STV by gel purification was done by loading the sample onto an agarose gel, excising the A+STV fragment and cutting the gel slice into small pieces, which were then centrifuged for 15 min at 14 000 rpm (38). The supernatant containing the A+STV fragments was afterwards collected and used in the second incubation step. Although this technique for gel extraction has a relatively low recovery for long molecules, it was chosen because the nucleic acid–STV complex does not survive the buffers used in most commercial gel extraction kits. Other techniques either require high temperatures and/or ethanol precipitation, which destabilize the nucleic acid–STV complex. We note that extraction of the nucleic acid–STV complex using electro-elution might also be a viable alternative, with possibly higher recovery.
Gel analysis
Samples containing molecules up to ∼10 kb in length were all analyzed by gel electrophoresis in nondenaturing 0.8% agarose gels in TAE buffer at 5 V/cm. DNA samples of longer molecules were analyzed by pulsed field electrophoresis (CHEF Mapper, Bio-RAD technologies, Hercules, CA, USA) on 1.0% agarose gels in 0.5 × TBE buffer using a program optimized for separating DNA molecules between 10 and 80 kb. All agarose gels were stained with ethidium bromide (EtBr).
Atomic Force Microscope (AFM) images
The gel-extracted heterodimers of 2.2 kb+3.5 kb DNA were imaged with tapping-mode AFM (National Instruments, Austin, TX, USA, Nanoscope IV) in air by depositing 5 µl of 1 ng/ml of DNA solution on freshly cleaved mica. After 2 min, the sample was rinsed for 10 s with ddH2O water, blown dry with N2, and directly imaged in the AFM. The AFM image in Figure 1c was analyzed using the freeware WsXM (39).
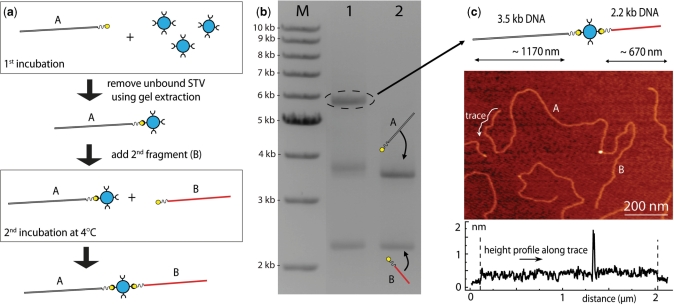
Figure 1.
Protocol and formation of DNA–STV–DNA dimers. (a) Schematic showing the two step incubation to optimize the efficiency of the dimer A+B, where A and B can be any nucleic acid with a biotin at one end. First, we bind molecule A to STV by 30 min incubation with a 30× molar excess of STV. We then remove all the unbound STV using gel purification and incubate the STV-bound molecule A together with molecule B at 4°C in a 1: 1 ratio for 30 min up to 15 h depending on the length of the molecules. (b) 0.8% agarose gel image showing the result of this protocol with 3.5 kb DNA (A) and 2.2 kb DNA (B) molecules (lane 1), and the control experiment without STV (lane 2). There is a very clear band at 5.7 kb (lane 1), corresponding to the dimer A + B. The efficiency of this reaction is ∼60%. (c) AFM image of the A+B heterodimer, and a height profile traced along the molecule. The STV can be seen as a clear peak in the height profile, and joins two molecules of about 1200 ± 50 nm and 750 ± 50 nm in length, corresponding well to the expected lengths for molecule A and B.
Determination of binding efficiency
The efficiency of the joining reactions is determined as follows: the intensities of the bands corresponding to the biotinylated molecules A+STV, B+STV, A+B, A+A and B+B were determined using the program LabImage 1D (Kapelan GmbH, Germany) and each was divided by the respective molecular weight to obtain a relative molar amount NA+STV, NB+STV, NA+B, NA+A and NB+B. From this calculation, we determined which of the reactants A or B was limiting (i.e. which reactant was present in the lowest amount before the reaction). The efficiency of the reaction was then calculated as follows: efficiency (A+B) = NA+B/(NA+STV+NA+A+NA+B) in the case of limiting reactant A, and efficiency (A+B) = NA+B/(NB+STV+NB+B+NA+B) in the case of limiting reactant B. We do not take into account the A and B molecules that did not bind STV, because these represent molecules that were not biotinylated and therefore do not participate in the reaction. Depending on the application, the final product itself may require further purification, which, depending on the protocol employed, could additionally affect the overall efficiency [e.g. the gel purification method described above has a typical recovery of 50–80% (38)].
RESULTS
Formation of DNA–STV–DNA heterodimers
In our experiments, we aim to optimize the specific formation of the dimer A+B starting from unlinked molecules A and B. To achieve this, we perform a two-step reaction, shown schematically in Figure 1a. In the first step, molecule A is incubated in buffer 1 for 30 min together with a large excess of STV (>30 × molar excess). Due to the excess STV, all biotinylated molecules will rapidly be bound to single STV molecules. Because these A+STV complexes cannot bind to each other (they can only bind to those A molecules in which the biotin remains unbound), this has an additional advantage that it almost completely suppresses the formation of homodimers A+A. After this first step, the free STV is removed by gel purification and the second nucleic acid molecule B is added in approximately a 1 : 1 molar ratio to molecule A. This is incubated at 4°C for a duration of 30 min up to several hours. Provided that all free STV has been successfully removed after the first incubation, the B molecules exclusively bind to the STV-tagged A molecules, resulting in the formation of heterodimers A+B.
The result of this protocol with two monobiotinylated DNA molecules of lengths 3.5 kb (molecule A) and 2.2 kb (molecule B) is shown in Figure 1b. For reference, Lane M contains a DNA marker. In lane 2 we show the result after following the protocol without STV, which as expected shows clear bands corresponding to 2.2 kb DNA and 3.5 kb DNA. In lane 1, molecules A and B were incubated according to the protocol described above. The clear band at ∼5.7 kb (circled) corresponds to the dimer A+B.
An AFM image of the resulting A+B dimer deposited on mica is shown in Figure 1c: here, we can clearly distinguish two DNA molecules joined at the ends via a slightly higher intermediary. The intermediary is likely the STV, as STV is significantly larger (4–6 nm diameter) than the DNA (2.2 nm diameter) and should therefore appear higher in the AFM (40) (note that the apparent height of both the DNA and the STV in the AFM is lower, because the AFM tip deforms the DNA and STV as is commonly observed in AFM imaging). The height profile of a trace along the contour of the construct is taken to confirm the lengths: we find the distances along the DNA to the STV are about 1200 ± 50 nm and 750 ± 50 nm. This matches well with the expected length values for molecule A (3524 bp × 0.34 nm/bp = 1192 nm) and molecule B (2250 bp × 0.34 nm/bp = 765 nm).
We now investigate the bands in Figure 1b, lane 1, that do not correspond to the desired end-product dimer A+B. These bands migrate at approximately the same speed as the unbound molecules A and B in lane 2. However, closer inspection reveals a slight shift in the band corresponding to molecule A. Because this shift only appears in the presence of STV, it can be attributed to molecules A that are bound to STV, and is observable due to the fact that the bound STV reduces the electrophoretic mobility of DNA molecules up to at least 3.5 kb in length (Supplementary material). In the context of the second step of our reaction, the appearance of this band corresponding to A+STV may nonetheless appear somewhat surprising: for if STV is bound, why does dimer formation not follow and remove this band? To address this question, we have investigated whether the presence of this band could be decreased by longer incubation times or by varying the relative concentrations of A and B; however, no observable effect was found (Supplementary material). Consequently, we speculate that it is an imperfection in the STV that causes this band to remain: it is consistent with the existence of a fraction of STV molecules that can only bind single, but not multiple, biotins.
Using the intensities of the bands at ∼3.7 kb (A+STV) and 5.7 kb (A+B), we can estimate the efficiency of formation of the dimer A+B and find that to be ∼60% (see Materials and methods section). Note that in order to achieve such a high efficiency, it is necessary to minimize the formation of homodimers A+A and B+B (Supplementary material). We have employed two technical steps to do so: first, the gel extraction following the first incubation ensures that all free STV is removed. Unfortunately, this is not the case for many other purification methods, and small amounts of free STV can remain and will lead to the formation of B+B homodimers (Supplementary material). Second, the low temperature in the second step prevents dissociation of the STV from the A+STV complex, which would otherwise also allow formation of homodimers (Supplementary material).
Applications of the method
End-joining of very long molecules
Having demonstrated that we can specifically form the heterodimers A+B with high efficiency, we now demonstrate the utility of this method in three applications: the end-joining of two very long DNA molecules, the end-joining of DNA to RNA, and the end-joining of RNA to RNA. In Figure 2a, lane 1, we demonstrate that we can efficiently end-join two very long molecules of DNA of 20 kb (A) and 9.6 kb (B). This is shown by the appearance of a band that migrates more slowly than the 23 kb marker in lane M2. This likely corresponds to the dimer A+B, which should migrate at approximately the same speed as 30 kb DNA. From the intensities of the bands, we deduce an efficiency of formation of the A+B dimer of ∼50%, similar to the case of the much shorter molecules in Figure 1. We therefore conclude that the efficiency does not significantly decrease when joining substantially longer molecules.
This is in sharp contrast to ligation, where the efficiency can decrease dramatically for such long molecules. This is illustrated in Figure 2b: Lane 1 shows the self-ligation of a 4.2 kb DNA molecule, resulting in an 8.4 kb DNA molecule. This 4.2 kb DNA molecule was digested with a restriction enzyme at a site in one of the PCR primers, leaving a 4 base pair self-complementary overhang (see Materials and methods section). After overnight incubation of this molecule with T4 DNA ligase, almost all molecules are ligated (lane 1). However, when the same reaction was carried out with a 12.7 kb molecule containing exactly the same self-complementary overhang, the efficiency of the ligated product was dramatically reduced, to almost zero (lane 2). Note that in these two ligation reactions the molar concentrations of both molecules were kept the same for the long and the short molecules. Efforts to improve the efficiency of the ligation reaction by performing the same reaction in the presence of poly-ethylene glycol (41), did not have noticeable effects (results not shown). Further optimization may yield a higher efficiency, but this may be a time-consuming task. Conveniently, our biotin–STV linkage procedure offers a way to join such molecules without the need for further optimization.
End-joining of DNA to RNA
A second advantage of using STV as a linker is its versatility: for instance, it allows us to very straightforwardly join RNA molecules to DNA molecules. Normally, this would require the synthesis of complementary overhangs on the DNA and RNA molecules, and possibly the subsequent ligation of the RNA–DNA hybrid. This can be challenging, because secondary structure in the single-stranded DNA parts and RNA can prevent efficient hybridization. In addition, DNA–RNA hybrid ligation reactions can be very inefficient (42,43). In contrast, our biotin–STV linkage procedure only requires that both molecules are biotinylated at one end.
In Figure 3a we show two examples in which we bind DNA molecules to single-stranded and double-stranded RNA molecules, respectively. The DNA molecules are the 3.5 kb and 2.2 kb monobiotinylated PCR products used above, and the RNA molecules were synthesized by in vitro transcription from a 4.2 kb long piece of DNA. A short 5′-biotinylated RNA oligonucleotide was hybridized to one of the ends of both RNA molecules to provide the biotin group for STV binding. The result of our protocol in the absence of STV with the 3.5 kb DNA piece (A) and the single-stranded 4.2 kb RNA (B) molecule is shown in lane 1. The ssRNA migrates approximately at the same speed as 1.5 kb DNA and clearly no dimer formation occurs. The result of our protocol in the presence of STV is shown in lane 2: an additional band appears at ∼5 kb DNA, which is where the ssRNA–DNA dimer is expected. The intensity of this band compared with the others implies an efficiency of ∼40% for the formation of A+B dimers, demonstrating that the efficiency of DNA–ssRNA dimers is quite similar to that obtained for the DNA–DNA dimers above. We also demonstrate the use of our protocol in binding a 2.2 kb DNA (A) molecule to a 4.2 kb dsRNA (B) molecule. Lane 4 shows the result after following the protocol without STV: the dsRNA migrates at the same speed at 4.2 kb DNA, and no dimer formation occurs. In lane 3, on the other hand, we observe an additional band at about 6.5 kb, corresponding to the heterodimer A+B. From the intensities of the bands we calculate an efficiency of ∼75% for the formation of A+B dimers, and indeed almost all of the RNA is bound to DNA. Achieving a similar efficiency in binding long RNAs to DNA using enzymatic ligation would be very challenging because the efficiency of these reactions may vary depending on the molecules involved.
End-joining of RNA to RNA
As a final application, we demonstrate that our method can also be applied to efficiently end-join two RNA molecules (Figure 3b). Here, we used our protocol to end-join two 4.2 kb dsRNA molecules (A) to create dimers A+A. In lane 2, two distinct bands can be seen, one migrating at ∼4.2 kb DNA (corresponding to molecule A) and one migrating at ∼8.5 kb DNA, which must correspond to the dimers A+A. We find that ∼20% of the final product consists of dimers A+A. As before, we have also performed a control in the absence of STV (Figure 3b, lane 1), and here no dimer formation occurs, as expected.
DISCUSSION
Having demonstrated several applications of our method, we now analyze in more detail a number of aspects that influence the overall efficiency of dimer formation. Specifically, we will address the importance of temperature and the role of STV quality in dimer formation, and we explain why our protocol disfavors the formation of multimers.
Influence of temperature on homodimer formation
We have mentioned above that a low temperature during the second incubation step is important in preventing homodimer formation (see also Supplementary material). The reason for this is that the rate of dissociation of STV is much reduced at a lower temperature, so that the STV dissociates less frequently from the A+STV complex and renders formation of A+A dimers (and also B+B dimers) less likely. Assuming that the dissociation rate of STV at 37°C is approximately koff = 10−4 s−1 [estimated from ref. (44)], we expect that after 1 hr about 30% of the STV has dissociated once and reassociated again to form a new complex (either a DNA–STV–DNA dimer or a single DNA–STV complex). At 4°C, however, the rate of dissociation is approximately three orders of magnitude lower (44), meaning that only 0.03% of the STV molecules would have had time to dissociate once. The formation of unwanted dimers such as A+A and B+B due to STV dissociation is dramatically suppressed, therefore. One could argue that the lower temperature may also require longer incubation times to compensate for slower diffusion. A longer incubation time would in turn allow for more dissociation to occur, which could cancel the benefit gained from the lower dissociation rate. However, the rate of diffusion is reduced by only about 10% by lowering temperature from 37°C to 4°C, which is hardly significant compared with the three orders of magnitude gained in the stability of the A+STV complex.
STV quality and the efficiency of dimer formation
We have found that the number of molecules that binds STV but does not form dimers (in other words, A+STV and B+STV complexes) is unaffected by either the incubation time or the relative concentration of molecules A and B (Supplementary material). Our speculation is that the STV molecules in these single DNA–STV complexes do not have multiple binding sites available for biotin, but only a single one. Presumably, partial degradation of the STV molecules may be the cause of this. We have indeed observed degradation over time in individual stocks of STV: in all cases, the efficiency of forming single DNA–STV complexes was not diminished (judged from the observable electrophoretic shift due to STV binding), but the relative amount of (homo- or hetero-) dimers would dramatically reduce over the course of several weeks to months, to the point where dimer formation was no longer observed. Switching to a fresh STV stock would always immediately result again in high efficiencies of dimer formation. This observation also rules out any possible influence of other factors, such as the buffers or DNA molecules used. The STV used for the experiments presented here (purchased from Roche Applied Sciences, Almere, The Netherlands) showed the best results: stocks were found to be stable for at least 2 weeks up to several months when stored in ddH2O at 4°C.
Formation of multimers
We have observed almost no formation of DNA trimers and tetramers in our experiments, even though wild-type STV has four binding sites for biotin. This is in accordance with earlier reports (29,32), where it was also found that direct incubation of biotinylated DNA with STV would not result in multimer formation, except at certain specific ratios of STV:DNA. Indeed, in some cases we have also observed the formation of trimers and tetramers when both molecules were incubated directly with STV (in a single incubation rather than our preferred two step approach). Formation of trimers and tetramers becomes more likely when the STV concentration is less than twice that of the DNA molecules, although it probably remains much slower than dimer formation because of steric hindrance (29). Indeed, Monte-Carlo simulations (45) imply that the probability of forming trimers and tetramers under optimized molar concentrations is still orders of magnitude lower than the probability of forming dimers. It is likely that our two-step protocol similarly disfavors the formation of trimers and tetramers. The likelihood of multimer formation in the first step of our protocol is reduced significantly, due to the very large excess of STV used. This makes it much more likely that all the biotin-groups on the molecules A are rapidly saturated by binding to single STV molecules rather than forming dimers. In the second step, when molecule B is added, it can only bind to A+STV complexes. Due to steric constraints, the probability that two molecules B bind to the same A+STV complex is lower than for two molecules B to bind two different A+STV complexes. Also, the concentrations of A and B are typically matched so that all B molecules can bind to A+STV molecules. Dimer formation is thus much more likely then formation of trimers and tetramers.
CONCLUSIONS
We have presented a simple procedure to specifically end-join two biotinylated nucleic acid molecules A and B using STV as an intermediary. The method avoids the time-consuming optimization usually required for ligation reactions when long molecules are involved or when joining DNA to RNA or RNA to RNA. The method presented here appears to be insensitive to the length of the molecules (up to at least 20 kb long DNA) or to whether the molecules are DNA or RNA. The motivation to use STV to end-join two nucleic acids was based on the fact that the STV–biotin linkage is the strongest noncovalent intermolecular bond known, but also because it is relatively straightforward to enzymatically biotinylate DNA and RNA molecules. We note that this procedure could in principle make use of any other pair of strongly binding antigens, such as digoxygenin and antidigoxygenin or fluorescein and antifluorescein (21,24–26). The use of such antigen-pairs could also be useful in typical single molecule applications, such as magnetic or optical tweezers, where also the other ends of the molecules A and B will be attached to either a bead or a surface (46–48). Conveniently, such an approach will automatically filter out the A+B product (the A+STV and B+STV alone will not tether between both surfaces), which will increase the overall yield of the protocol even more. The binding protocol presented here can be performed easily and rapidly in a standard biochemical buffer, and no special chemicals and subsequent purifications are required. It is therefore an ideal choice to create long nucleic acid constructs that may find many applications in bionanotechnology and single-molecule experiments.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Martin van den Heuvel and Gary Skinner for useful discussions and Serge Donkers for help with the final experiments. This work was financially supported by the Netherlands Organization for Scientific Research (Vidi grant to N.H.D). Funding to pay the Open Access publication charges for this article was provided by the Netherlands Organization for Scientific Research.
Conflict of interest statement. None declared.
REFERENCES
- 1.Marko JF, Siggia ED. Stretching DNA. Macromolecules. 1995;28:8759–8770. [Google Scholar]
- 2.Seol Y, Skinner GM, Visscher K, Buhot A, Halperin A. Stretching of homopolymeric RNA reveals single-stranded helices and base-stacking. Phys. Rev. Lett. 2007;98:158103-1–158103-4. doi: 10.1103/PhysRevLett.98.158103. [DOI] [PubMed] [Google Scholar]
- 3.Seol Y, Skinner GM, Visscher K. Elastic properties of a single-stranded charged homopolymeric ribonucleotide. Phys. Rev. Lett. 2004;93:118102-1–118102-4. doi: 10.1103/PhysRevLett.93.118102. [DOI] [PubMed] [Google Scholar]
- 4.Woodside MT, Behnke-Parks WM, Larizadeh K, Travers K, Herschlag D, Block SM. Nanomechanical measurements of the sequence-dependent folding landscapes of single nucleic acid hairpins. Proc. Natl. Acad. Sci. USA. 2006;103:6190–6195. doi: 10.1073/pnas.0511048103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Woodside MT, Anthony PC, Behnke-Parks WM, Larizadeh K, Herschlag D, Block SM. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liphardt J, Onoa B, Smith SB, Tinoco I, Bustamante C. Reversible unfolding of single RNA molecules by mechanical force. Science. 2001;292:733–737. doi: 10.1126/science.1058498. [DOI] [PubMed] [Google Scholar]
- 7.Onoa B, Dumont S, Liphardt J, Smith SB, Tinoco I, Bustamante C. Identifying kinetic barriers to mechanical unfolding of the T-thermophila ribozyme. Science. 2003;299:1892–1895. doi: 10.1126/science.1081338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Skinner GM, Baumann CG, Quinn DM, Molloy JE, Hoggett JG. Promoter binding, initiation, and elongation by bacteriophage T7 RNA polymerase: A single-molecule view of the transcription cycle. J. Biol. Chem. 2004;279:3239–3244. doi: 10.1074/jbc.M310471200. [DOI] [PubMed] [Google Scholar]
- 9.Keyser UF, Koeleman BN, Van Dorp S, Krapf D, Smeets RMM, Lemay SG, Dekker NH, Dekker C. Direct force measurements on DNA in a solid-state nanopore. Nat. Phys. 2006;2:473–477. [Google Scholar]
- 10.Storm AJ, Chen JH, Zandbergen HW, Dekker C. Translocations of double-strand DNA through a silicon oxide nanopore. Phys Rev E. 2005;71 doi: 10.1103/PhysRevE.71.051903. 051903-1–051903-10. [DOI] [PubMed] [Google Scholar]
- 11.Li JL, McMullan C, Stein D, Branton D, Golovckenko J. Solid state nanopores for single molecule detection. Biophys. J. 2001;80:339A. [Google Scholar]
- 12.Bianco PR, Brewer LR, Corzett M, Balhorn R, Yeh Y, Kowalczykowski SC, Baskin RJ. Processive translocation and DNA unwinding by individual RecBCD enzyme molecules. Nature. 2001;409:374–378. doi: 10.1038/35053131. [DOI] [PubMed] [Google Scholar]
- 13.Dekker C. Solid-state nanopores. Nat. Nanotechnol. 2007;2:209–215. doi: 10.1038/nnano.2007.27. [DOI] [PubMed] [Google Scholar]
- 14.Storm AJ, Storm C, Chen JH, Zandbergen H, Joanny JF, Dekker C. Fast DNA translocation through a solid-state nanopore. Nano Lett. 2005;5:1193–1197. doi: 10.1021/nl048030d. [DOI] [PubMed] [Google Scholar]
- 15.Li JL, Gershow M, Stein D, Brandin E, Golovchenko JA. DNA molecules and configurations in a solid-state nanopore microscope. Nat. Materials. 2003;2:611–615. doi: 10.1038/nmat965. [DOI] [PubMed] [Google Scholar]
- 16.Keyser UF, van der Does J, Dekker C, Dekker NH. Optical tweezers for force measurements on DNA in nanopores. Rev. Sci. Instrum. 2006;77:105105-1–105105-9. [Google Scholar]
- 17.Pheiffer BH, Zimmerman SB. Polymer-Stimulated Ligation: Enhanced Blunt-End or Cohesive-End Ligation of DNA or Deoxyribooligonucleotides by T4-DNA Ligase in Polymer-Solutions. Nucleic Acids Res. 1983;11:7853–7871. doi: 10.1093/nar/11.22.7853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rusche JR, Howardflanders P. Hexamine Cobalt Chloride Promotes Intermolecular Ligation of Blunt End DNA Fragments by T4 DNA-Ligase. Nucleic Acids Res. 1985;13:1997–2008. doi: 10.1093/nar/13.6.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kurschat WC, Muller J, Wombacher R, Helm M. Optimizing splinted ligation of highly structured small RNAs. Rna Publ. Rna Soc. 2005;11:1909–1914. doi: 10.1261/rna.2170705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stark MR, Pleiss JA, Deras M, Scaringe SA, Rader SD. An RNA ligase-method for the efficient creation of large, synthetic RNAs. Rna Publ. Rna Soc. 2006;12:2014–2019. doi: 10.1261/rna.93506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmitz GG, Walter T, Seibl R, Kessler C. Nonradioactive Labeling of Oligonucleotides Invitro with the Hapten Digoxigenin by Tailing with Terminal Transferase. Anal. Biochem. 1991;192:222–231. doi: 10.1016/0003-2697(91)90212-c. [DOI] [PubMed] [Google Scholar]
- 22.Yehudai-Resheff S, Schuster G. Characterization of the E. coli poly(A) polymerase: nucleotide specificity, RNA-binding affinities and RNA structure dependence. Nucleic Acids Res. 2000;28:1139–1144. doi: 10.1093/nar/28.5.1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Feinberg AP, Vogelstein B. A Technique for Radiolabeling DNA Restriction Endonuclease Fragments to High Specific Activity. Anal. Biochem. 1983;132:6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- 24.Huang Z, Szostak JW. A simple method for 3′-labeling of RNA. Nucleic Acids Res. 1996;24:4360–4361. doi: 10.1093/nar/24.21.4360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Proudnikov D, Mirzabekov A. Chemical methods of DNA and RNA fluorescent labeling. Nucleic Acids Res. 1996;24:4535–4542. doi: 10.1093/nar/24.22.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lingner J, Keller W. 3′-End Labeling of Rna with Recombinant Yeast Poly(a) Polymerase. Nucleic Acids Res. 1993;21:2917–2920. doi: 10.1093/nar/21.12.2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Niemeyer CM, Adler M, Gao S, Chi LF. Nanostructured DNA-protein aggregates consisting of covalent oligonucleotide-streptavidin conjugates. Bioconjug. Chem. 2001;12:364–371. doi: 10.1021/bc000090x. [DOI] [PubMed] [Google Scholar]
- 28.Niemeyer CM, Ceyhan B, Blohm D. Functionalization of covalent DNA-streptavidin conjugates by means of biotinylated modulator components. Bioconjug. Chem. 1999;10:708–719. doi: 10.1021/bc980150n. [DOI] [PubMed] [Google Scholar]
- 29.Niemeyer CM, Adler M, Pignataro B, Lenhert S, Gao S, Chi LF, Fuchs H, Blohm D. Self-assembly of DNA-streptavidin nanostructures and their use as reagents in immuno-PCR. Nucleic Acids Res. 1999;27:4553–4561. doi: 10.1093/nar/27.23.4553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Niemeyer CM, Burger W, Peplies J. Covalent DNA-Streptavidin conjugates as building blocks for novel biometallic nanostructures. Angew. Chem. Int. Ed. 1998;37:2265–2268. doi: 10.1002/(SICI)1521-3773(19980904)37:16<2265::AID-ANIE2265>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- 31.Niemeyer CM, Sano T, Smith CL, Cantor CR. Oligonucleotide-Directed Self-Assembly of Proteins: Semisynthetic DNA Streptavidin Hybrid Molecules as Connectors for the Generation of Macroscopic Arrays and the Construction of Supramolecular Bioconjugates. Nucleic Acids Res. 1994;22:5530–5539. doi: 10.1093/nar/22.25.5530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tian Y, He Y, Ribbe AE, Mao CD. Preparation of branched structures with long DNA duplex arms. Org. Biomol. Chem. 2006;4:3404–3405. doi: 10.1039/b605464a. [DOI] [PubMed] [Google Scholar]
- 33.Niemeyer CM, Boldt L, Ceyhan B, Blohm D. DNA-directed immobilization: Efficient, reversible, and site-selective surface binding of proteins by means of covalent DNA-streptavidin conjugates. Anal. Biochem. 1999;268:54–63. doi: 10.1006/abio.1998.3017. [DOI] [PubMed] [Google Scholar]
- 34.Niemeyer CM, Wacker R, Adler M. Hapten-functionalized DNA-streptavidin nanocircles as supramolecular reagents in a competitive immuno-PCR assay. Angew. Chem. Int. Ed. 2001;40:3169–3172. doi: 10.1002/1521-3773(20010903)40:17<3169::AID-ANIE3169>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 35.Green NM. Avidin and Streptavidin. Methods Enzymol. 1990;184:51–67. doi: 10.1016/0076-6879(90)84259-j. [DOI] [PubMed] [Google Scholar]
- 36.Jones ML, Kurzban GP. Noncooperativity of Biotin Binding to Tetrameric Streptavidin. Biochemistry. 1995;34:11750–11756. doi: 10.1021/bi00037a012. [DOI] [PubMed] [Google Scholar]
- 37.Vilfan ID, Kamping W, van den Hout M, Candelli A, Hage S, Dekker NH. An RNA toolbox for single-molecule force spectroscopy studies. Nucleic Acids Res. 2007;35:6625–6639. doi: 10.1093/nar/gkm585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wu W, Welsh MJ. A Method for Purification of DNA Species by High-Speed Centrifugation of Agarose-Gel Slices. Anal. Biochem. 1995;229:350–352. doi: 10.1006/abio.1995.1425. [DOI] [PubMed] [Google Scholar]
- 39.Horcas I, Fernandez R, Gomez-Rodriguez JM, Colchero J, Gomez-Herrero J, Baro AM. WSXM: A software for scanning probe microscopy and a tool for nanotechnology. Rev. Sci. Instrum. 2007;78:013705-1–013705-8. doi: 10.1063/1.2432410. [DOI] [PubMed] [Google Scholar]
- 40.Pignataro B, Chi L, Gao S, Anczykowski B, Niemeyer C, Adler M, Fuchs H. Dynamic scanning force microscopy study of self-assembled DNA-protein nanostructures. Appl. Phys. Mater. Sci. Process. 2002;74:447–452. [Google Scholar]
- 41.Zimmerman SB, Minton AP. Macromolecular Crowding: Biochemical, Biophysical, and Physiological Consequences. Annu. Rev. Biophys. Biomol. Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 42.Sugino A, Goodman HM, Heyneker HL, Shine J, Boyer HW, Cozzarelli NR. Interaction of Bacteriophage-T4 Rna and DNA Ligases in Joining of Duplex DNA at Base-Paired Ends. J. Biol. Chem. 1977;252:3987–3994. [PubMed] [Google Scholar]
- 43.Persson T, Willkomm DK, Hartmann RK. In: Handbook of RNA Biochemistry. Hartmann RK, Bindereif A, Schön A, Westhof E, editors. Vol. 1. Wiley-VCH GmbH & Co; 2005. pp. 53–74. [Google Scholar]
- 44.Chilkoti A, Stayton PS. Molecular-Origins of the Slow Streptavidin-Biotin Dissociation Kinetics. J. Am. Chem. Soc. 1995;117:10622–10628. [Google Scholar]
- 45.Richter J, Adler M, Niemeyer CM. Monte Carlo simulation of the assembly of bis-biotinylated DNA and streptavidin. Chemphyschem. 2003;4:79–83. doi: 10.1002/cphc.200390012. [DOI] [PubMed] [Google Scholar]
- 46.Smith SB, Finzi L, Bustamante C. Direct mechanical measurements of the elasticity of single DNA-molecules by using magnetic beads. Science. 1992;258:1122–1126. doi: 10.1126/science.1439819. [DOI] [PubMed] [Google Scholar]
- 47.Lionnet T, Allemand J-F, Revyakin A, Strick TR, Saleh OA, Bensimon D, Croquette V. In: Single Molecule Techniques. Selvin PR, Ha T, editors. Vol. 1. 2008. pp. 347–369. [Google Scholar]
- 48.Gore J, Bryant Z, Nollmann M, Le MU, Cozzarelli NR, Bustamante C. DNA overwinds when stretched. Nature. 2006;442:836–839. doi: 10.1038/nature04974. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.