Abstract
Bloom's syndrome (BS) is a cancer predisposition disorder caused by mutation of the BLM gene, encoding a member of the RecQ helicase family. Although the phenotype of BS cells is suggestive of a role for BLM in repair of stalled or damaged replication forks, thus far there has been no direct evidence that BLM associates with any of the three human replicative DNA polymerases. Here, we show that BLM interacts specifically in vitro and in vivo with p12, the smallest subunit of human POL δ (hPOL δ). The hPOL δ enzyme, as well as the isolated p12 subunit, stimulates the DNA helicase activity of BLM. Conversely, BLM stimulates hPOL δ strand displacement activity. Our results provide the first functional link between BLM and the replicative machinery in human cells, and suggest that BLM might be recruited to sites of disrupted replication through an interaction with hPOL δ. Finally, our data also define a novel role for the poorly characterized p12 subunit of hPOL δ.
INTRODUCTION
The faithful completion of chromosomal DNA replication is of crucial importance in determining the fidelity with which genetic information is passed from mother to daughter cells. Incomplete replication or the erroneous copying of a damaged DNA template can give rise to genome instability, accumulation of mutations and, in multicellular organisms, to neoplastic transformation (1). Chromosomal DNA replication in eukaryotic cells requires three distinct DNA polymerases named DNA polymerase α (POL α), ε (POL ε) and δ (POL δ). POL δ and POL ε are required for replication of the leading strand and for completion of lagging strand DNA synthesis. Their respective roles in the replication of leading and lagging strands are still uncertain, though it has been proposed that POL δ and POL ε function specifically at the lagging and leading strands of the replication fork, respectively. POL δ is also involved in different DNA repair pathways as a gap filling enzyme (2). The mammalian POL δ has been studied extensively as a core enzyme consisting of four subunits named p125, p66, p50 and p12 (3). Two of the subunits form a tightly-associated catalytic heterodimer consisting of the catalytic p125 subunit, which has both 5′ to 3′ DNA polymerase and 3′ to 5′ exonuclease activities, and p50. The role of the p66 subunit is to bind PCNA, the homotrimeric sliding clamp that functions as a processivity factor for POL δ during DNA replication (4). A specific role for the p12 subunit has not been identified thus far, although it has been shown to interact with the p125 and p50 subunits of POL δ and Proliferating Cell Antigen (PCNA) (5), and data from in vitro DNA replication assays indicate that addition of p12 enhances the DNA polymerizing activity of the enzyme (6). The levels of p12 are regulated by the proteasome through the mechanism that is not dependent upon p12 ubiquitination (7). Apart from PCNA, no other interacting protein has been characterized that specifically associates with p12.
The RecQ family of DNA helicases represents a group of evolutionarily conserved enzymes that are involved in the maintenance of genome stability (8,9). There are five members of this family known in humans called RECQL1, BLM, WRN, RECQL4 and RECQL5. Defects in three of these give rise to defined clinical disorders associated with cancer predisposition and various aspects of premature aging: mutations in the WRN and RECQL4 genes result in Werner's syndrome (WS) and Rothmund–Thomson syndrome (RTS), respectively, both of which feature genome instability, predisposition to some types of cancer and the early onset of several aging features. Mutations in the BLM gene cause Bloom's syndrome (BS), which is also associated with excessive chromosomal instability and a high incidence of cancers of all types. In contrast to WS and RTS, no obvious premature aging has been observed in BS patients (10). Cells derived from BS patients show a 10-fold higher frequency of reciprocal exchanges between sister chromatids (SCEs), as well as excessive chromosome breakage (11). The BLM protein is a DNA structure-specific helicase that unwinds DNA in 3′ to 5′ direction (12), and shows an apparent preference for unwinding of synthetic Holliday junctions, G-quadruplex (G4) DNA and D-loop DNA substrates (13,14). These substrates represent different DNA structures that can be formed in vivo during DNA replication and homologous recombination (HR) processes. Cell biological studies have shown that BLM is localized in the nucleus of human cells within discrete foci termed promyelocytic leukemia (PML) nuclear bodies (15). BLM also localizes to nucleoli in S-phase cells (16), and to telomeres in cells lacking telomerase (17). On the basis of the aforementioned reports, it has been proposed that BLM functions at the interface of DNA replication and recombination, and facilitates the repair of damaged DNA replication forks (9,18).
A large body of evidence implicates BLM in DNA replication. First, DNA replication defects, such as a retarded rate of nascent DNA chain elongation (19) and accumulation of abnormal replication intermediates (20), have been described in BS cells. Second, BLM interacts physically and functionally with several proteins that play important roles during DNA replication, such as replication protein A (RPA) (21), FEN-1 (22) and chromatin assembly factor 1 (CAF-1) (23). Third, BLM is localized to replication foci, particularly during late S phase, and this co-localization increases in the presence of agents that inhibit DNA replication (23). Fourth, BLM expression is activated at the G1/S boundary and peaks in late S-phase/G2 (15,16,24,25). Fifth, BS cells are hypersensitive to agents that perturb DNA replication, such as hydroxyurea (HU) (26).
In this work, we report the physical and functional interaction of BLM with p12, the smallest subunit of human POL δ (hPOL δ). Consistent with this interaction playing an important biological function, we show that the presence of the hPOL δ enzyme, as well as the p12 subunit alone, can specifically stimulate the DNA helicase activity of BLM. We also find that BLM specifically promotes hPOL δ strand displacement activity. Furthermore, we show that the co-localization of BLM and hPOL δ in nuclear foci is activated during replicative stress. Our data are consistent with a role for hPOL δ in the recruitment of BLM to sites of arrested or disrupted DNA replication forks, in order for it to effect its role in fork repair and/or stabilization.
MATERIALS AND METHODS
Purification of the hPOL δ enzyme
Four-subunit hPOL δ was expressed by infection of insect cells with four recombinant baculoviruses, each encoding a subunit of hPOL δ. Recombinant baculoviruses encoding the hPOL δ subunits were a kind gift from Dr Valdimir Podust. In order to produce an exonuclease deficient hPOL δ mutant, a D402A substitution mutation was introduced into the p125/wt cDNA by PCR-based site-directed mutagenesis of the transfer vector pVL1393/p125. Primer sequences are available upon request. Baculovirus-mediated expression of p125 D402A in insect cells using the BacPAK6 system was conducted in accordance with the manufacturer's instructions (Clontech Laboratories, Mountain View, California, USA). hPOL δ enzymes, wt and the exonuclease deficient mutant, as well as a three-subunit exonuclease-deficient mutant lacking the p12 subunit, were purified from insect cells as described previously (6).
Purification of 6xHis-p12 protein
The p12 cDNA was cloned into the pRSETb vector. The resulting pRSETb-p12 construct was verified by DNA sequencing. p12 was expressed in Escherichia coli BL21(DE3) (Novagen, Merck KGaA, Darmstadt, Germany). Expression of p12 was induced by addition of 1 mM IPTG to cultures grown at 37°C to an A600 of 0.4. After incubation at 37°C for 3 h, the cells were harvested by centrifugation. The E. coli pellet was resuspended in 30 ml of buffer A (30 mM phosphate buffer, 10 mM Tris–HCl, pH 8.0, 500 mM NaCl, 10 mM imidazole, 1 mM PMSF, 1 μM benzamidine, 5 μg/ml leupeptin and 2 μg/ml pepstatin A). The cells were disrupted with a French Press (twice) and the lysate was sonicated on ice for 1 min. After centrifugation (20 000 r.p.m. for 30 min at 4°C in a SS-34 rotor), the soluble fraction was loaded onto a 1 ml HiTrap Chelating (Ni+) column pre-equilibrated with buffer A. The column was washed with 50 ml buffer A, and then with 20 ml buffer A containing 50 mM imidazole. The bound protein was eluted with 300 mM imidazole in buffer A. After desalting to buffer B (40 mM Tris–HCl pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM 2-mercaptoethanol, 15% (v/v) glycerol, 1 mM PMSF, 1 μM benzamidine, 5 μg/ml leupeptin and 2 μg/ml pepstatin A) using a HiTrap Desalting column, the eluate was loaded onto a 1 ml HiTrap Heparin column pre-equilibrated with buffer B. The column was washed with 20 ml buffer B, and the p12 protein was eluted with a 20 ml linear NaCl gradient (50–1000 mM). p12 was eluted at 300 mM NaCl as tested by SDS–PAGE and western blotting using an antibody against p12. The pool of p12 protein was diluted to 50 mM NaCl and finally loaded onto a Mono S column pre-equilibrated with buffer B. Chromatography was performed as for the HiTrap Heparin column. The yield from a 1 l culture was ∼0.3 mg of p12 protein with a purity of over 99.5%, as judged by Coomassie Blue staining. Protein concentrations were determined by the Bradford method (27) by using Bovine Serum Albumin (BSA) as a calibration standard.
Production and purification of the p12 antibody
To produce the p12 antibody, the 6xHis-p12 protein was expressed in E. coli from the pRSETb-p12 plasmid, and purified from E. coli using conventional chromatography (see above). Rabbits were immunized three times with 300 µg of 6xHis-p12 protein being used for each immunization. After the third immunization, the rabbit was sacrificed to obtain the serum. The p12 serum was first purified over a Protein G sepharose column, to isolate total IgG, and then purified over a column coupled to the p12 protein, to obtain the IgGp12. For all subsequent studies, the anti-IgG p12 was used at concentration of 0.2 μg/ml in dilution 1:100 in TBS, 0.05% Tween-20. Control IgG was purified from preimune serum using Protein G sepharose column.
Far-western analysis
Far-western assays were performed as described previously (28). Human WRN was a kind gift from Dr Pavel Janscak, University of Zürich. Briefly, total extracts of Sf21 insect cells (0.825 µg), 0.8 µg of hPOL δ, 0.3 µg of hPOL λ and 0.2 μg of p12 were subjected to 12% SDS–PAGE and transferred to a nitrocellulose filter. After blocking in 10% milk, 0.3% Tween-20 in TBS, for 1 h at RT, the filter was incubated for 2 h at 4°C with BLM (0.5 µg/ml) in TBS supplemented with 0.25% milk, 0.3% Tween-20, 1 mM DTT and 1 mM PMSF (hybridization solution). After the washing step (4 × 15 min, 0.25% milk, 0.3% Tween-20 in TBS), western blot was performed using the anti-BLM IHIC33 antibody (29) to detect the presence of BLM. For the experiment presented in Figure 1, BLM (1.2 µg), WRN (0.8 µg) and BSA (0.4 µg) were separated on a 7.5% SDS gel and transferred to a nitrocellulose membrane. The membrane was incubated with 6xHis-p12 in hybridization solution, with a final concentration 6xHis-p12 of 0.1 µg/ml. After washing, the membrane was probed with an anti-p12 antibody (this study) to detect the presence of p12. The input samples were visualized with antibodies against BLM [IHIC33, (29)] and WRN (ab-200, Abcam, Cambridge, UK). The membrane was incubated with the BLM protein in hybridization solution, with a final concentration of BLM of 0.5 µg/ml. After washing, western blot using an anti-BLM IHIC33 antibody was performed to detect the presence of BLM. The input samples were visualized with antibodies against hPOL λ (ab5954, Abcam) and p12 (this study).
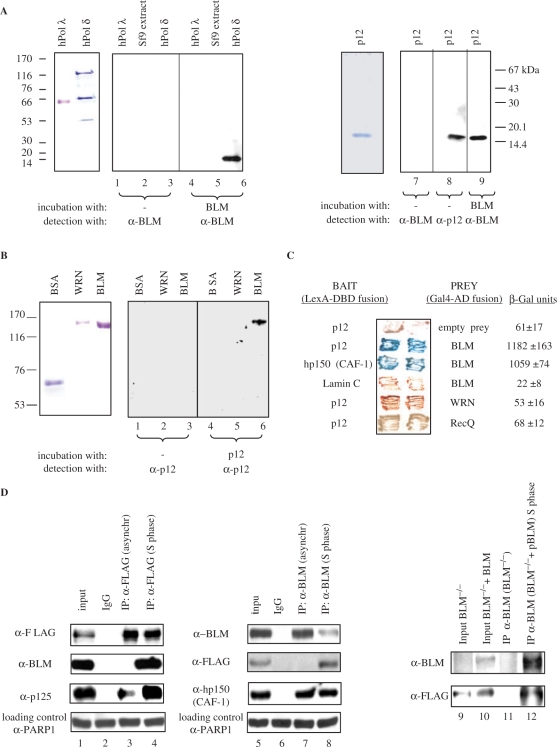
Figure 1.
BLM and p12 of hPOL δ interact in vitro and in vivo. (A) Left panel, far-western analysis. hPOL λ, total protein extract from Sf9 insect cells, and hPOL δ enzyme were subjected to SDS–PAGE, transferred to a nitrocellulose membrane, and were incubated with purified recombinant BLM. Anti-BLM antibodies were used to permit the detection of p12 as a novel BLM-interacting protein (lane 6). Molecular weight markers are also indicated on the left. Right panel, purified p12 subunit of hPOL δ was subjected to SDS–PAGE, transferred to nitrocellulose membrane, incubated with BLM, and subsequently probed with the anti-BLM antibody (lane 9). (B) Reciprocal far-western analysis. BSA, WRN and BLM (left panel) were hybridized with the purified p12 and probed with the anti-p12 antibody (right panel). Anti-p12 antibodies were used to confirm that p12 specifically binds to BLM (lane 6). (C) BLM and p12 of hPOL δ interact in the YTH assay. The L40 yeast reporter strain was co-transformed with plasmids encoding the indicated full-length ‘bait’ (LexA-DBD) and ‘prey’ (Gal4-AD) fusions. Two independent colonies were grown on SD agar plates lacking tryptophan and leucine, but containing X-gal, prior to assessment of β-galactosidase activity. Also shown are two negative controls, p12 co-transformed with an empty prey vector and BLM co-transformed with lamin C protein. The previously described BLM/hp150 (CAF-1) interaction (23) was used as a positive control. (D) BLM and hPOL δ form a complex in human cells. 293T cells were transiently transfected with FLAG-p12, and were synchronized in S phase using 1 mM HU. Nuclear extracts derived from either unsynchronized (lane 3) or S-phase synchronized cells (lane 4) were immunoprecipitated with the anti-FLAG antibody or control IgG, and were analysed by SDS–PAGE. One-tenth (50 µg) of the same nuclear extract was used as input control (lane 1). Immunoprecipitated FLAG-p12 and BLM were detected by western blotting using the anti-FLAG and anti-BLM antibody, respectively (lane 4). p125, the largest subunit of hPOL δ, was also efficiently co-immunoprecipitated using the same anti-FLAG antibody (lanes 3 and 4). Reciprocal co-IP is shown in the middle panel: lane 5, input; lane 6, IP with the control IgG; lane 7, IP with an anti-BLM antibody (C-18) from nuclear extracts derived from unsynchronized 293T cells; lane 8, IP with an anti-BLM antibody (C-18) from nuclear extracts derived from the S-phase synchronized 293T cells. The known BLM interacting protein, hp150 (CAF-1) was also efficiently co-immunoprecipitated using the same anti-BLM antibody (lanes 7 and 8). As a loading control for lanes 1–8, 50 µg of the corresponding nuclear extract was probed with an anti-PARP1 antibody. Right panel shows co-IP with anti-BLM antibody (C-18) from BS cell nuclear extracts (BS) and BS cells containing the BLM cDNA (BS + pBLM). p12 could be co-immunoprecipitated in the presence of BLM from the S-phase synchronized nuclear extracts (lane 12) but not in the absence of BLM (lane 11). Lanes 9 and 10 are the inputs of the two different nuclear extracts.
Yeast two-hybrid assay
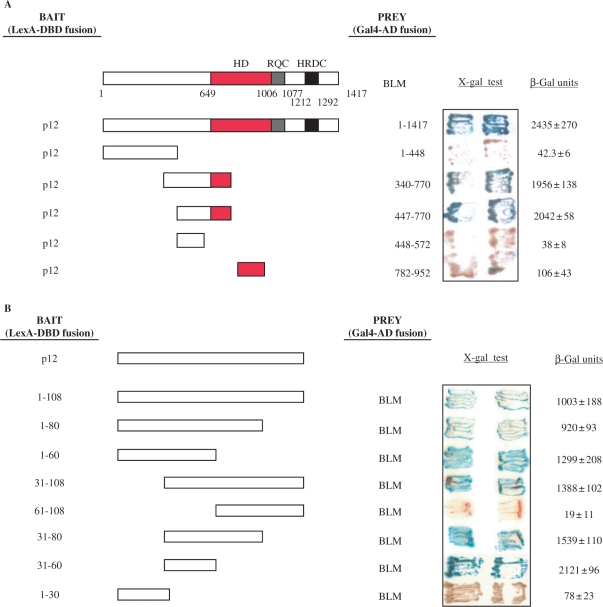
Yeast two-hybrid (YTH) assays were performed as described previously (23). The activity of the reporter gene (β-galactosidase) was assessed using a liquid culture assay with O-nitrophenyl-d-galactopyranoside as a substrate. The constructs used to map the region of BLM that interacts with p12 have been described previously (30). The different p12 constructs were generated by PCR using pMALc2e-p12 as a template, and were cloned into the pBTM116 + 2 (MBN) vector. Sequences of all plasmids, primers used and construction schemes are available upon request.
Transfections and immunoprecipitation assay
The p12 cDNA was cloned into the p3xFLAG-myc-CMV-23 (Sigma-Aldrich, Buchs, SG, Switzerland) vector using plasmid pRSETb-p12 as a template and primers 5′-ggaagatctcataggggatagagatgccag -3′ (BglII) and 5′- cccaagcttatgggccggaagcggctc - 3′ (HindIII). The resulting construct was sequenced. 293T cells were transiently transfected with p3xFLAG-myc-CMV-23-p12 by the calcium phosphate precipitation method. Thirty-six hours after transfection, cells were treated with 1 mM HU for 24 h, and were then harvested. Nuclear extracts were prepared from 293T cells as described previously (31). Aliquots (500 µg) of the nuclear extracts were incubated with 3 µg of anti-FLAG antibody coupled to magnetic, tosyl-activated Dynabeads (Dynal Biotech, Invitrogen, Paisley, UK, M-280) according to manufacturer's instructions in immunoprecipitation (IP) buffer (20 mM HEPES pH 7.5, 150 mM NaCl, 5 mM MgCl2, 0.1% Nonidet P 40, protease inhibitor) at 4°C for 3 h. As a control, nuclear extracts were incubated with a control rabbit IgG. The beads were washed three times in IP buffer, before any protein complexes bound to beads were eluted and analysed by SDS–PAGE. A 50 µg portion of nuclear extract was used as input control. Subsequently, western blot analysis was performed using the anti-FLAG (Sigma M2) and anti-BLM ab 476 antibody (Abcam). C-18 (anti-BLM, Santa Cruz) was used for the reciproical co-IP in the above-mentioned IP buffer containing 150 mM KCl. Nuclear extracts from BS cells were used as a negative control for the C-18 reciprocal co-IP.
DNA polymerase primer extension assay
A 18-nt primer was 5′-end labeled with 32P using T4 polynucleotidekinase and purified on a Sephadex G25 microcolumn. The X-poly template was generated by annealing an 18-nt primer to four complementary oligonucleotides. Three of these oligonucleotides are 50-nt long; the fourth oligonucleotide has an extended arm that is complementary to the 18-mer primer at its 3′ end. Twenty-five nucleotides of each four oligonucleotides are fully complementary to two out of three other oligonucleotides, so that the result of annealing is a cruciform structure. Annealing and purification of the X-poly substrate was carried out as described previously (32). Sequences of the primers used are available upon request. Reactions (10 μl final volume) were carried out in buffer containing 40 mM Tris–HCl buffer (pH 8.0), 3 Mm MgCl2, 1 mM ATP, 50 mM NaCl, 2 mM DTT, 0.1 mg/ml BSA, 10% glycerol, 0.15 pmol of 32P-18-nt-X-poly template, 100 μM each of dATP, dGTP, dCTP and dTTP, 10 ng of hPOL δ exo, and the indicated amounts of BLM, human PCNA or E. coli RecQ. Reactions were incubated at 37°C for 30 min, and were terminated by rapid cooling on ice and addition of an equal volume of denaturing loading buffer. The samples were boiled, and 10 μl of sample were electrophoresed through 12.5% polyacrylamide–8 M urea gel in 0.5 × TBE buffer, and the extension products were visualized by autoradiography.
DNA helicase assays
Recombinant BLM protein was purified from yeast cells as described previously (12). The splayed arm DNA substrate that mimics a replication fork was generated by annealing two partially complementary oligonucleotides, consisting of 25 nt of fully complementary and 25 nt of non-complementary sequences, and was purified as described previously (13,32). The helicase reactions were carried out under presumed single-turnover conditions; that is with helicase concentration in excess over substrate concentration. The 10 μl reactions contained 1 × helicase buffer [33 mM Tris–acetate (pH 7.8); 1 mM MgCl2; 66 mM sodium acetate; 0.1 mg/ml BSA; 1 mM DTT; 1 mM ATP], 100 pM substrate, various concentrations of BLM and other proteins as stated in figure legends. The reaction was allowed to progress for 15 min at 37°C unless otherwise stated. Analysis of reaction products was carried out as described previously (32).
Indirect immunofluorescence analysis
GM00637 transformed normal human fibroblasts were grown on coverslips and were either treated with 2.5 mM HU for 18 h or cultured untreated, and were then pulse labeled with 25 μM BrdU for 5 min. The coverslips were then rinsed with ice-cold PBS. Soluble proteins were removed by incubating the slides in pre-extraction buffer [10 mM PIPES, 300 mM sucrose, 3 mM MgCl2, 20 mM NaCl, 0.5% Triton X-100 (pH 6.8)] for 5 min on ice. The cells were then fixed in 4% paraformaldehyde for 20 min on ice. The immunostaining was performed as described earlier (33) using the IHIC34 rabbit polyclonal antibody (29) and the AlexaFluor 488 conjugated donkey anti-rabbit secondary antibody (Molecular Probes, Invitrogen, Paisley, UK) to detect BLM, at 1 : 200 and 1 : 800 dilutions, respectively. The A-9 mouse monoclonal antibody (Santa Cruz Biotechnology Inc., Santa Cruz, California, USA) against the catalytic subunit of hPOL δ and the CY3 conjugated sheep anti-mouse secondary antibody (Sigma-Aldrich, Gillingham, UK) were used to detect hPOL δ, at 1 : 400 and 1 : 1000 dilutions, respectively. BrdU incorporation was detected after repeated paraformaldehyde fixation and HCl denaturation with rat anti-BrdU primary antibody (Abcam) and AlexaFluor 350 conjugated goat anti-rat secondary antibody (Molecular Probes), each at 1 : 300 dilution. Epifluorescence microscopy, image acquisition and analysis were carried out on a Nikon Eclipse 80i microscope with the Lucia G software (Laboratory Imaging s.r.o., Prague, Czech Republic). Grabbed images were scored manually using the Adobe Photoshop program. Foci obtained following staining with either antibody (green or red) were marked and counted. Foci were counted as co-localizing if more than 50% of the green and red signal was overlapping. Co-localization was expressed as percentage of the total number of BLM (green) or POL δ (red) foci. The total number of cells scored in each treatment was 100. Two independent experiments were conducted with nearly identical results, of which only one is presented.
RESULTS
BLM and hPOL δ interact in vitro and in vivo
To analyse the possible functional interaction of BLM and hPOL δ, we first purified both BLM and the four subunit hPOL δ enzyme (Supplementary Figure. 1). A far-western assay was then used to test for a specific interaction between BLM and one or more of the hPOL δ subunits. As shown in Figure 1A, the BLM protein specifically interacted with a protein of apparent molecular mass of 14 kDa (lane 6), which corresponds to p12, the smallest subunit of hPOL δ. In contrast, BLM did not interact with any of the other three hPOL δ subunits, with an unrelated human DNA polymerase, hPOL λ (lane 4), or with any protein from the extract of Sf9 insect cells from which the recombinant hPOL δ enzyme was purified (lane 5). In order to confirm that BLM specifically binds to p12, the far-western analysis was repeated using full-length BLM and purified recombinant p12 immobilized on nitrocellulose. Clear evidence of binding was obtained (Figure 1A, lane 9). Moreover, in a reverse far-western, purified p12 specifically bound to full-length BLM (Figure 1B, lane 6), but not to BSA (Figure 1B, lane 4) or to another human RecQ helicase, WRN (Figure 1B, lane 5). No cross-reactivity of either the anti-BLM antibody with p12, or the anti-p12 antibody with BLM was detected (Figure 1A, lanes 1–3 and 7 and Figure 1B, lanes 1–3). Taken together, these data indicate that BLM and p12 interact specifically in vitro.
To validate the results obtained from far-western assays, we next performed YTH analysis. As shown in Figure 1C, p12 interacted with full-length BLM, but not with full-length WRN or E. coli RecQ, indicating that the physical interaction between p12 and BLM is specific. Furthermore, no YTH interaction could be detected between the full-length BLM fused to Gal4-AD and any of the other three hPOL δ subunits (p125, p66 and p50) fused to LexA-DBD (data not shown).
To gain insight into the possible association of BLM and p12 in human cells, we performed co-IP assays (Figure 1D). Because the endogenous levels of p12 are very low and our newly generated anti-p12 antisera did not work in IP assays, we were forced to use ectopically expressed p12. For this, FLAG-p12 was transiently transfected into 293T cells and the cells were subsequently synchronized in S phase by treatment with 1 mM HU for 24 h. Synchronization of the 293T cells was confirmed by fluorescence activated cell sorter (FACS) analysis (data not shown). Using an anti-FLAG antibody, we were able to specifically co-immunoprecipitate BLM from HU-treated cells (Figure 1D, lane 4), but not from unsynchronized cells (Figure 1D, lane 3). As expected, the anti-FLAG antibody efficiently precipitated p125, the largest subunit of hPOL δ, from both HU-treated and unsynchronized cells (Figure 1D, lanes 3 and 4, lower panel). No BLM was present in the precipitate when a control antibody was used (Figure 1D, lane 2). A reciprocal co-IP experiment was also carried out, in which an anti-BLM polyclonal antibody was used to immunoprecipitate FLAG-p12 from 293T nuclear extracts. As shown in Figure 1D, p12 could be specifically co-immunoprecipitated with endogenous BLM from the S phase-synchronized cells (lane 8), but not from unsynchronized 293T cells (lane 7). Furthermore, in addition to p12, the anti-BLM antibody efficiently co-immunoprecipitated hp150, the largest subunit of CAF-1 (Figure 1D, lanes 7 and 8, lower panel), a protein shown previously to interact with BLM (23). Similar co-IP results were obtained when 293T-derived nuclear cell extracts were incubated with ethidium bromide, indicating that the in vivo interaction of p12 and BLM is unlikely to be mediated by DNA (data not shown). Finally, the specificity of co-IPs with the anti-BLM antibody was demonstrated in a control experiment using nuclear extracts from BS (BLM−/−) cells: in this case, the anti-BLM antibody could not co-immunoprecipitate p12 from these cells (Figure 1D, lane 11, lower panel), whereas it could efficiently co-immunoprecipitate p12 from S phase-synchronized BS cells containing the BLM cDNA (BLM−/− + pBLM) (Figure 1D, lane 12, lower panel). Collectively, these data indicate that BLM directly associates with the p12 subunit of hPOL δ in vitro and in vivo, and that the BLM/p12 interaction in human cells is exclusively or predominantly seen in cells in which DNA replication is arrested.
Mapping of the interacting regions on BLM and p12
BLM contains several important domains; a conserved helicase domain (aa 649–1006), an RecQ familiy C-terminal (RQC) domain (aa 1006–1077) and an Helicase, RNAse D C terminal (HRDC) domain (aa 1212–1292), all of which are involved in mediating protein–DNA interactions, and possibly also protein–protein interactions (Figure 2A). To identify the region of BLM that mediates the interaction with p12, we generated a series of BLM deletion mutants and tested them for their ability to interact with full-length p12 in the YTH assay (Figure 2A). The results indicated that p12 binds BLM in the region between amino acids 447 and 770. This fragment of BLM comprises the helicase-proximal region of the N-terminal domain and part of the helicase domain. A similar YTH approach was used to map the region of p12 that interacts with BLM. A single short region of p12, comprising amino acids 31–60, was found to be necessary and sufficient for interaction with BLM (Figure 2B).
Figure 2.
Interaction region mapping of BLM and p12. (A) Mapping of the BLM interaction region. The L40 yeast strain was co-transformed with plasmids encoding the indicated BLM fragments fused to Gal4-AD and the full-length p12 fused to LexA-DBD. Two independent colonies were grown on SD agar plates lacking tryptophan and leucine, but containing X-gal, prior to assessment of β-galactosidase activity. Blue coloration of colonies is a marker of interaction. Full length BLM is also shown, with a red bar indicating its conserved helicase domain, a blue bar indicating RQC and a black bar indicating the HRDC domain. Interactions between a given bait/prey pair were quantified by measurements of β-galactosidase activity. Values represent means ± SD of three independent experiments. (B) Mapping of the p12 interaction region. The L40 yeast strain was co-transformed with plasmids encoding the indicated p12 fragments fused to LexA-DBD and full length BLM fused to Gal4-AD. In both (A) and (B) the sequence boundaries of deletion mutants tested are shown with the corresponding amino acid positions indicated on the right. Values obtained from liquid β-galactosidase assay are shown on the right and represent means ± SD of three independent experiments.
The hPOL δ enzyme stimulates the BLM-mediated unwinding of a model replication fork substrate
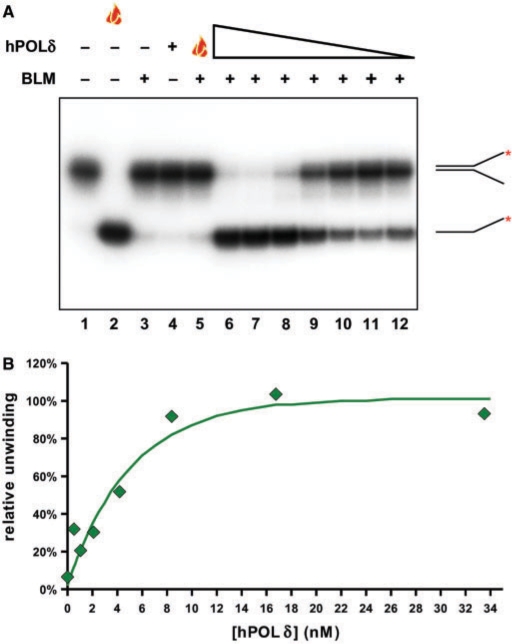
The physical interaction between BLM and hPOL δ suggested the possibility that the two proteins might functionally regulate each other's activities during DNA replication, recombination or repair. To test this hypothesis, we first determined whether the hPOL δ enzyme influences BLM helicase activity. In order to see a potential effect of hPOL δ, either positive or negative, the BLM concentration used in the helicase assays was sufficient only for partial unwinding of the substrate during a 15-min reaction period. To prevent the potential degradation of the helicase substrate or product by the exonuclease activity of hPOL δ, the hPOL δ enzyme used carried a mutation in the exonuclease domain of p125 (D402A) and was, therefore, exonuclease defective. When hPOL δ was added to the reaction in concentrations giving molar ratios of 25 to 0.4 times that of the BLM concentration, we saw a significant, concentration-dependent stimulation of helicase activity (Figure 3A, lanes 6–12). BLM alone (Figure 3A, lane 3) and BLM incubated with heat-denatured hPOL δ (Figure 3A, lane 5) showed the expected low level of helicase activity, and hPOL δ alone displayed no DNA helicase activity (Figure 3A, lane 4). We were also able to demonstrate the stimulation of BLM helicase activity by hPOL δ in time-course experiments; hPOL δ reproducibly increased the initial velocity of the unwinding reaction (Figure 4C center panel and D; and data not shown).
Figure 3.
The hPOL δ enzyme specifically stimulates the BLM-mediated unwinding of the replication fork substrate in a concentration-dependent manner. (A) A total of 1.3 nM BLM was pre-incubated with the exonuclease-defective hPOL δ enzyme in various concentrations (33.5, 16.8, 8.4, 4.2, 2.1, 1 and 0.5 nM; lanes 6–12, respectively) on ice for 3 min, and the samples were then warmed to 37°C. The unwinding reaction was initiated immediately by the addition of substrate and ATP. Flame symbol depicts heat-denatured substrate (lane 2), or BLM incubated with heat-denatured hPOL δ enzyme at the highest concentration of the titration range (33.5 nM; lane 5), as described earlier. (B) Quantification of data presented in (A). Data were normalized to the non-treated (lane 1, taken as 0%) and boiled (lane 2, taken as 100%) samples. Maximal stimulation (at 6 times above BLM basal helicase activity) was achieved with a 13 × molar excess of hPOL δ.
Figure 4.
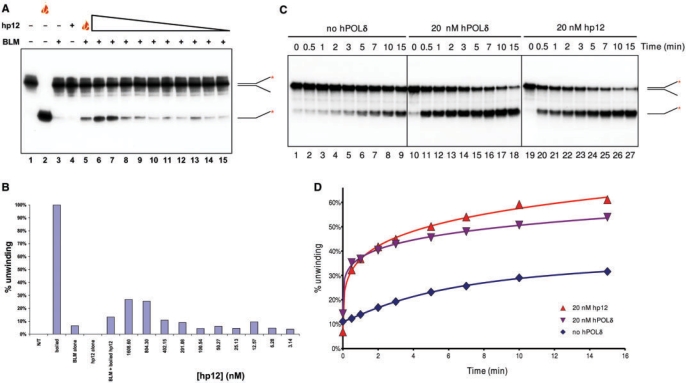
The small subunit of the hPOL δ enzyme, p12, is sufficient to stimulate BLM helicase activity. (A) A total of 1.3 nM BLM was pre-incubated with various concentrations of p12 (1609, 804.5, 402.25, 201.13, 100.56, 50.28, 25.14, 12.57, 6.29 and 3.14 nM; lanes 6–15, respectively) on ice for 3 min, and the samples were then warmed to 37°C. The unwinding reaction was initiated immediately by the addition of substrate and ATP. Controls and symbols are as in Figure 3. (B) Quantification of data from (A). (C) A total of 0.85 nM BLM was incubated at 37°C for 5 min alone (lanes 1–9), or with 20 nM hPOL δ (lanes 10–18), or 20 nM p12 (lanes 19–27). The unwinding reaction was then initiated by the addition of ATP and substrate. Samples were withdrawn at the time points indicated above the lanes. (D) Quantification of data from (C).
Next, we analysed the specificity of this apparent functional interaction. We found that this stimulatory effect was specific to the BLM/hPOL δ complex, as hPOL δ had no effect on another RecQ helicase, E. coli RecQ, in the same concentration range (Supplementary Figure 2A, lanes 7–17, and B). Moreover, we showed that another human DNA polymerase, hPOL λ, which we had shown did not bind to BLM in far-western analysis (Figure 1A), did not influence BLM helicase activity (Supplementary Figure 2C, lanes 6–17, and D). Taken together, these data indicate that hPOL δ shows a specific, functional interaction with BLM.
The far-western and YTH analyses localized the BLM/hPOL δ interaction to the smallest p12 subunit of hPOL δ between residues 31 and 60 (Figures 1 and 2). We tested, therefore, the effect of recombinant p12 subunit on the helicase activity of BLM. p12 showed a concentration-dependent stimulatory effect on BLM similar to that of the hPOL δ enzyme; however, the effective concentration range was such that there was a large molar excess of p12 over BLM (Figure 4A, lanes 6–15). Interestingly, heat-denatured p12 was also able to stimulate BLM helicase activity at this high concentration (1.609 μM; Figure 4A, lane 5), which, in the light of the results showing that a small peptide is also able to stimulate the helicase activity of BLM, is not inexplicable (see below). As described earlier, we saw concentration-dependent stimulation of BLM helicase activity by p12 only at high p12 concentration. Therefore, we designed a different system to study this effect and monitored the progress of the unwinding reaction over time in the presence or absence of p12. This revealed that p12 and the hPOL δ enzyme increased the kinetics of the BLM unwinding reaction to a similar extent (Figure 4C and D). The effect was concentration-dependent and reproducible using various molar ratios of BLM and hPOLδ or p12 (1 : 0.8–1 : 3.9, Supplementary Figure 3 and data not shown; 1 : 23.5, Figures 4C, D and 5). The initial linear part of the progress curves permitted us to approximate the initial velocity of the reaction. Depending on the molar ratio of hPOL δ or hp12 and BLM, we saw a 2–15 times increase in the initial velocity of unwinding (Supplementary Figure 3, and data not shown).
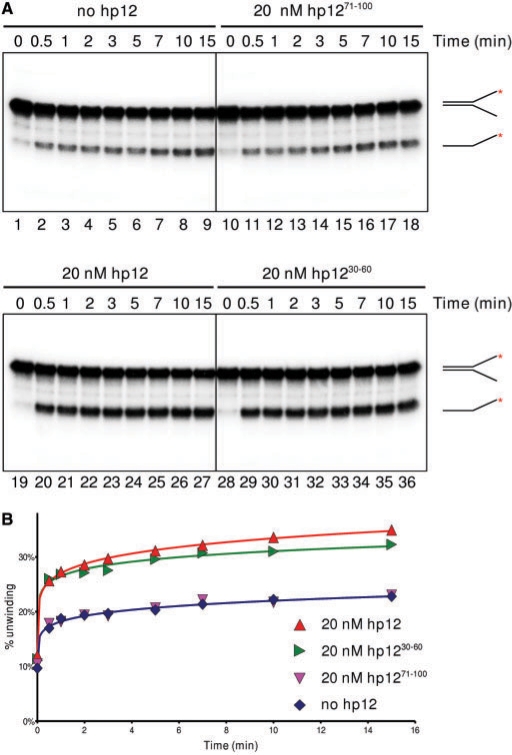
This result prompted us to test the effect on BLM activity of the tightly-defined interacting region of p12. For this, two peptides were chemically synthesized: p1230–60 covers the BLM-binding region, while p1271–100 maps to a region of p12 that shows no BLM-binding, as revealed in the YTH mapping studies (Figure 2B). Full-length p12 and p1230–60 showed a similar degree of stimulation of BLM activity in a time-course experiment. In contrast, p1271–100 had no effect on BLM helicase activity (Figure 5A and B). Taken together, these results confirm that the helicase activity of BLM is stimulated by hPOL δ, and that this stimulation is dependent on the binding between the two proteins through amino acid residues 30–60 in the p12 subunit of hPOL δ.
Figure 5.
The stimulatory effect of p12 can be localized to a small peptide, spanning the region that binds BLM. (A) A total of 0.85 nM BLM was incubated at 37°C for 5 min alone (lanes 1–9), or with 20 nM p1271–100 (a peptide that shows no binding to BLM; lanes 10–18), full length p12 (lanes 19–27) or p1230–60 (a peptide that spans the binding region to BLM; lanes 28–36). Reactions were run as in Figure 4B. (B) Quantification of data from (A).
BLM increases the strand displacement activity of hPOL δ
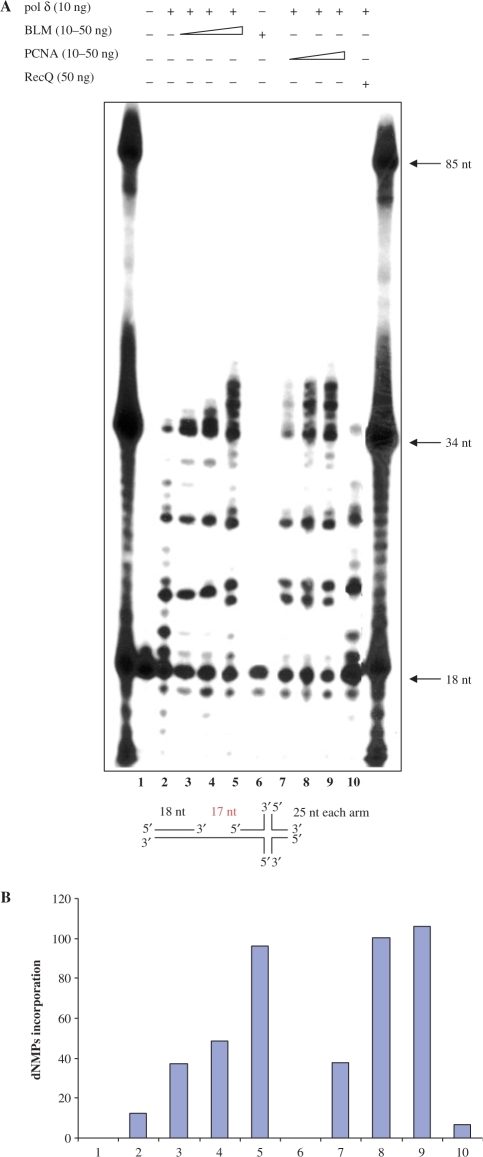
Given the data described earlier indicating that hPOL δ has a significant stimulatory effect on BLM helicase activity, it was important to examine whether BLM affects the polymerase activity of hPOL δ. To that end, we performed primer extension assays (Figure 6). hPOL δ-specific polymerization activity was monitored by visualizing extension of a 5′-end labeled 18-mer primer on an 85-mer DNA template, which contains an X-junction at the 3′end. This template was chosen because we wanted to examine whether BLM can help hPOL δ to traverse X-junction at the end of template. We observed that hPOL δ alone was able to incorporate dNTPs up to the start of the X-junction structure (Figure 6, lane 2). The inability of hPOL δ to extend the primer beyond the pause site indicates that the X-junction structure effectively blocked progression of hPOL δ along the template strand.
Figure 6.
BLM stimulates hPOL δ strand displacement activity. (A) Ten nanogram of hPOL δ alone (lane 2) or in the presence of an increasing amount (10, 20 or 50 ng) of BLM (lanes 3–5), PCNA (lanes 7–9) or 50 ng of E. coli RecQ (lane 10) were tested in primer extension assays using the X-poly DNA template as described in Materials and methods section. Eighteen-nucleotide primer was 5′ end labeled. Lane 6 contains 50 ng of BLM alone and shows that BLM does not have DNA polymerase activity. Lane 1: substrate alone; positions of oligonucleotide size-markers are indicated. Schematic representation of the X-poly DNA template is shown. The limit of extension is indicated with the arrow. (B) Quantification of products longer than 34 nt from (A).
When BLM was added to the reaction in concentrations giving molar ratios of 1.5–7.5 times that of the hPOL δ concentration, we observed an increase in both the amount of primer extended and the maximal length of product (Figure 6, lanes 3–5) such that the longest products represented polymerization extending 5 nt beyond the position of the X-junction. This effect was similar to that produced by PCNA, although the degree of stimulation of hPOL δ by PCNA was more pronounced (Figure 6, lanes 7–9). Importantly, BLM alone showed no polymerase activity (Figure 6, lane 6). The stimulatory effect of BLM on hPOL δ was found to be specific, as another member of the RecQ helicase family, E. coli RecQ, had no effect on hPOL δ (Figure 6, lane 10). In order to test whether BLM had a stimulatory effect on hPOL δ polymerase activity per se we performed primer extension assay using a more conventional DNA substrate comprising a 72-nt long template and a 17-nt primer. As shown in Supplementary Figure 4, we found that BLM does not stimulate hPOL δ polymerase activity on such a DNA template.
Altogether, these results indicate that BLM does not stimulate hPOL δ polymerase activity but, similar to PCNA, BLM promotes strand displacement by hPOL δ.
BLM and hPOL δ partially co-localize in vivo in response to perturbation of DNA replication
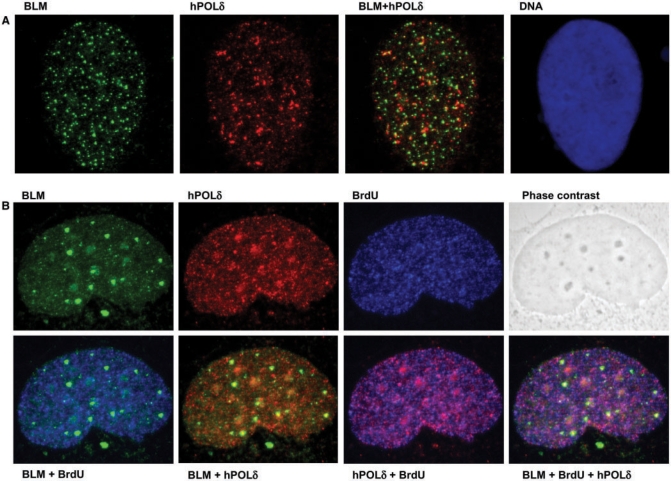
The in vitro studies described earlier prompted us to analyse whether hPOL δ and BLM might interact in vivo. As indicated earlier (Figure 1D), BLM and hPOL δ could be co-immunoprecipitated from 293T cells only after HU treatment, which suggested that the interaction is either S-phase specific or induced specifically in response to perturbation of replication. To differentiate between these possibilities, we addressed whether BLM and hPOL δ co-localize either in unperturbed, cycling cells or in cells blocked with HU. For this, GM00637 (BLM proficient) human fibroblast cells were plated on coverslips and treated with 2.5 mM HU for 18 h, sufficient to block replication, as evidenced by the depression of BrdU incorporation into the nuclei (not shown). A control culture was incubated in parallel without HU treatment. In both sets of cells, BrdU was added to a final concentration of 25 μM 5 min before the cells were fixed. The fixed samples were stained for the presence of hPOL δ, BLM and BrdU, as described in Materials and methods section. To detect hPOL δ were restricted in our use of antibodies, because our antibody to p12 did not work in immunofluorescence analysis. Hence, we chose an antibody against the catalytic subunit p125, which directly interacts with p12 (Figure 1D, lanes 3 and 4). As expected from previous analyses, BLM and hPOL δ showed a punctate nuclear pattern of localization (15,25,34). Representative images of the staining patterns are depicted in Figure 7A and B. The degree of co-localization of nuclear foci was then scored. This analysis indicated a low degree of co-localization in an untreated asynchronous cell population, which increased in response to HU treatment (Supplementary Figure 5). This increase in the extent of co-localization might be restricted to S phase or might be an effect of the HU-induced replication perturbation. To address this, we took advantage of the BrdU labeling of the actively replicating cells. When only the BrdU-positive subpopulation was scored in the untreated cultures, the degree of co-localization was similar to that of the whole asynchronous population. These results suggest that BLM and hPOL δ co-localize only to a limited extent in vivo, even during an unperturbed S phase, but that perturbation of DNA replication causes an increase in their co-localization. Nevertheless, even after treatment of cells with HU, most BLM and hPOL δ foci still did not co-localize, indicating that a significant fraction of BLM remains distant from sites of stalled replication forks.
Figure 7.
Dual staining for BLM and hPOL δ suggests co-localization in vivo, which is stimulated during replicative stress. (A) GM00637 cells were arrested with 2.5 mM HU, and were stained as described in Materials and methods section. A representative image showing punctate BLM (green, left) and hPOL δ (red, second from left) staining. Coincidence of the green and red signals (yellow signal) suggests co-localization of the two proteins. The right panel shows the nucleus of the same cell stained with the DNA dye Hoechst 33258. (B) GM00637 cells grown on coverslips without HU treatment and were pulse-labeled with 25 μM BrdU for 5 min, and then stained as described in Materials and methods section. Representative images show staining of the same nucleus for BLM in green (top left), for hPOL δ in red (top, second from left), for BrdU in blue (top, second from right). Phase contrast image of the same nucleus is depicted on the top right. In the bottom row, combined images of the individual stainings from the top row are shown. As expected, the BrdU signal correlates well with the signal for hPOLδ (magenta-color pattern second panel from bottom right). The coincidence of BLM signal with either BrdU (cyan signal in bottom left) or hPOLδ (yellow signal in second panel from bottom left) is less pronounced.
DISCUSSION
We have shown that the BS helicase, BLM and the major replicative DNA polymerase, hPOL δ, interact specifically in vitro and in vivo. This interaction is direct and is mediated via the thus far poorly characterized p12 subunit of hPOL δ. We mapped the site of interaction of BLM with p12 to a region representing amino acids 447–770. This fragment includes the N-terminal part of the BLM helicase domain and it has been shown previously that this is involved in interaction with the WRN helicase (35). In addition, we mapped the region of p12 that interacts with BLM to a short fragment comprising amino acids 30–60. Database searches revealed that this p12 fragment does not represent any known conserved protein domain and that it shows no sequence homology to any known BLM interacting partner. Of most importance, we have also shown that BLM and hPOL δ can functionally associate in that their interaction leads to stimulation of the BLM helicase activity and hPOL δ strand displacement activity.
There are a number of possible scenarios in DNA replication/repair where the productive co-operation of BLM and hPOL δ might be advantageous. Perhaps the most plausible is during the process of fork regression under circumstances where a replication fork is arrested by a DNA adduct or another blockade, particularly those that block leading strand synthesis. We have shown recently that BLM can promote the regression of a model replication fork in vitro (36). This may be a reaction that is beneficial for DNA repair simply because, through promoting fork back-tracking, it allows access to the blocking ‘lesion’. However, there is also potential for fork regression to be important in a lesion bypass pathway of DNA damage tolerance. If the lesion blocks leading strand synthesis, it appears that the synthesis of the lagging strand may continue for some distance ahead of the site of the blocked leading strand. This apparently futile uncoupling of leading and lagging strand synthesis has the potential, after fork regression, to permit template switching with the shorter leading strand being extended by copying of the longer lagging strand template. In this way, once the regressed fork is reset, the leading strand would be extended beyond the site of the lesion and normal DNA replication could commence. hPOL δ might both recruit BLM to stalled forks and then stimulate its catalytic activity once there. Conversely, BLM might play a role in assisting hPOL δ to access the regressed ‘4th arm’ and catalyse extension of the leading strand.
It has been suggested that RecQ helicases together with the type 1A topoisomerase, Top3, act at damaged replication forks to resolve recombination structures likely resulting from template switching (37). We suggest that BLM plays a dual role in rescuing arrested replication forks. One function is in assisting POL δ to bypass DNA lesions via fork regression and template switching, and the second function of BLM is to resolve the resulting pseudo double Holliday junction with a help of Top3. These two functions of BLM may be independent of each other.
A striking feature of the ability of hPOL δ to stimulate the helicase activity of BLM was our finding that a short peptide (aa 30–60) representing the minimal binding region of p12 is as efficient in this stimulatory role as is the hPOL δ enzyme. This would seem to rule out many possible mechanisms for the stimulation, including recruitment of BLM to the DNA substrate. Instead, these results argue for hPOL δ acting to alter the conformation of BLM in such a way as to enhance its helicase function. Such a mechanism is further supported by the unusual reaction progress curves: after an initial burst of activity the reaction very quickly reaches its maximum without attaining 100% unwinding. Unwinding by helicases is believed to be a multi-step process: (i) changes of the quaternary structure of the enzyme alter its binding specificity; (ii) the enzyme binds to the substrate with an increased affinity; (iii) DNA binding initiates ATP hydrolysis; and (iv) unwinding commences. Under our single turnover unwinding conditions, we postulate that the rate-limiting step is the activation of the helicase. We postulate that hPOL δ facilitates these structural changes, and when the enzyme is mixed with its substrates (DNA and ATP) it is already in its active conformation ready to initiate unwinding immediately upon binding. Furthermore, hPOL δ also increases the extent of unwinding by BLM. This increase might be a consequence of increased processivity of the helicase, which in turn might also be the result of conformational changes initiated by hPOL δ. A major goal for the future will be to characterize in molecular detail how exactly this stimulation occurs, and how this short peptide can have such a marked effect on the BLM helicase.
Our data indicate that BLM and hPOL δ do not significantly co-localize in the nucleus of human cells under normal growth conditions, including within an unperturbed S phase. This is consistent with the known localization of BLM to PML bodies and not to sites of ongoing replication. However, we were able to detect a consistent increase in the percentage of nuclear BLM foci that co-localize with hPOL δ when cells were treated with HU. Taken together, these data indicate that this association at replication foci is driven by replication perturbation and not by cell-cycle phase per se. To support these results, we could show direct association of BLM with the p12 subunit of hPOL δ in vivo in cells which were synchronized in S phase by HU treatment. No interaction between BLM and p12 could be observed in unsynchronized cells. Our data showing that BLM and p12 co-immunoprecipitate from the nuclear extracts of the 293T cells after treatment with 1 mM HU seemingly contrast with the recent data of Zhang and colleagues (38), who reported that, in HeLa and 293T cells, p12 undergoes ubiquitination and degradation 20 h after the treatment with 2 mM HU. These differences are possibly the result of the different HU concentrations that have been employed in these two studies. Alternatively, it may well be that binding of BLM to p12 prevents p12 ubiquitination and degradation, which is the reason why we did not observe degradation of p12 in our experiments.
We have also shown that BLM, similar to PCNA (39,40), promotes the strand displacement activity of hPOL δ. BLM might help hPOL δ to traverse short regions of DNA secondary structure during the process of DNA replication, but we have no direct evidence for this and such a possibility still needs to be investigated. However, we could not observe any effect of BLM on hPOL δ polymerase activity on a simple primer/template substrate. It has been shown previously that WRN has a stimulatory effect on the polymerization activity of Saccharomyces cerevisiae POL δ in the absence of PCNA (41). However, WRN does not apparently have an effect on DNA synthesis catalysed by the POL δ-PCNA complex. These results suggest that WRN may not function in processive DNA synthesis reactions during normal DNA replication; instead, WRN may function in replication restart of stalled or collapsed replication forks from which the replication machinery has dissociated (41).
Our current hypothesis is that BLM may, in a similar manner to WRN, be involved in replication restart of stalled replication forks blocked by DNA damage or unusual secondary structures in DNA template (42). One such scenario is during the replication of ribosomal DNA in the nucleolus. Similar to telomeres, ribosomal DNA is GC rich and can adopt alternative DNA structures such as hairpins and G-quadruplexes. These structures are efficiently resolved in vitro by BLM and WRN (13,43–45). When overexpressed in HeLa cells, WRN can recruit the p50 and p125 subunit of hPOL δ from nucleoplasm to the nucleolus (46). In an unperturbed cell cycle, BLM is found primarily in PML nuclear bodies, except during late S phase when it co-localizes with WRN in the nucleolus (16). Additionally, BLM binds to ribosomal DNA and primarily at the non-transcribed spacer region where replication forks initiate (47). These findings suggest that BLM may be directly coupled to replication fork initiation and/or progression at particular sites of DNA synthesis. The fact that BLM and hPOL δ co-localize more prominently after blockade of replication, but that not all replication foci contain BLM under these conditions, suggests that BLM may be recruited to sites of DNA synthesis only in response to the formation of a particular DNA structure that needs BLM helicase function for its resolution. It will be interesting to address in the future whether BLM is recruited to hPOL δ foci only following posttranslational modification of one or both of these factors, or whether the association is mediated by other replication factors to which BLM binds, such as RPA (21).
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We thank Pavel Janscak for the gift of purified recombinant WRN, Vladimir Podust for the gift of recombinant baculoviruses encoding the hPOL δ subunits, and Grant Brown for critical reading of the article and helpful discussions. C.Z.B. and I.D.H. are supported by Cancer Research UK. N.S. was supported by a Swiss National Foundation Grant (Nr. 3100A0-100256/1) to I.S. The Stagljar lab is supported by grants from the Canadian Foundation for Innovation (CFI), the Canadian Institute for Health Research (CIHR), the National Cancer Institute of Canada (NCIC), Gebert Rüf Foundation, Genentech, and Novartis. Funding to pay the Open Access publication charges for this article was provided by the National Cancer Institute of Canada (NCIC).
Conflict of interest statement. None declared.
REFERENCES
- 1.Bell SP, Dutta A. DNA replication in eukaryotic cells. Annu. Rev. Biochem. 2002;71:333–374. doi: 10.1146/annurev.biochem.71.110601.135425. [DOI] [PubMed] [Google Scholar]
- 2.Hubscher U, Maga G, Spadari S. Eukaryotic DNA polymerases. Annu. Rev. Biochem. 2002;71:133–163. doi: 10.1146/annurev.biochem.71.090501.150041. [DOI] [PubMed] [Google Scholar]
- 3.Liu L, Mo J, Rodriguez-Belmonte EM, Lee MY. Identification of a fourth subunit of mammalian DNA polymerase delta. J. Biol. Chem. 2000;275:18739–18744. doi: 10.1074/jbc.M001217200. [DOI] [PubMed] [Google Scholar]
- 4.Maga G, Hubscher U. Proliferating cell nuclear antigen (PCNA): a dancer with many partners. J. Cell Sci. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- 5.Li H, Xie B, Zhou Y, Rahmeh A, Trusa S, Zhang S, Gao Y, Lee EY, Lee MY. Functional roles of p12, the fourth subunit of human DNA polymerase delta. J. Biol. Chem. 2006;281:14748–14755. doi: 10.1074/jbc.M600322200. [DOI] [PubMed] [Google Scholar]
- 6.Podust VN, Chang LS, Ott R, Dianov GL, Fanning E. Reconstitution of human DNA polymerase delta using recombinant baculoviruses: the p12 subunit potentiates DNA polymerizing activity of the four-subunit enzyme. J. Biol. Chem. 2002;277:3894–3901. doi: 10.1074/jbc.M109684200. [DOI] [PubMed] [Google Scholar]
- 7.Liu G, Warbrick E. The p66 and p12 subunits of DNA polymerase delta are modified by ubiquitin and ubiquitin-like proteins. Biochem. Biophys. Res. Commun. 2006;349:360–366. doi: 10.1016/j.bbrc.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 8.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem. J. 2003;374:577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Opresko PL, Cheng WH, Bohr VA. At the junction of RecQ Helicase biochemistry and human disease. J. Biol. Chem. 2004;279:18099–18102. doi: 10.1074/jbc.R300034200. [DOI] [PubMed] [Google Scholar]
- 10.Hickson ID. RecQ helicases: caretakers of the genome. Nat. Rev. Cancer. 2003;3:169–178. doi: 10.1038/nrc1012. [DOI] [PubMed] [Google Scholar]
- 11.Ray JH, German J. The cytogenetics of the chromosome-breakage syndromes. In: German J, editor. Chromosome Mutation and Neoplasia. Alan R. Liss: Inc, New York; 1983. pp. 135–168. [Google Scholar]
- 12.Karow JK, Chakraverty RK, Hickson ID. The Bloom's syndrome gene product is a 3′-5′ DNA helicase. J. Biol. Chem. 1997;272:30611–30614. doi: 10.1074/jbc.272.49.30611. [DOI] [PubMed] [Google Scholar]
- 13.Mohaghegh P, Karow JK, Brosh R.M., Jr, Bohr VA, Hickson ID. The Bloom's and Werner's syndrome proteins are DNA structure-specific helicases. Nucleic Acids Res. 2001;29:2843–2849. doi: 10.1093/nar/29.13.2843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bachrati CZ, Borts RH, Hickson ID. Mobile D-loops are a preferred substrate for the Bloom's syndrome helicase. Nucleic Acids Res. 2006;34:2269–2279. doi: 10.1093/nar/gkl258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bischof O, Kim SH, Irving J, Beresten S, Ellis NA, Campisi J. Regulation and localization of the Bloom syndrome protein in response to DNA damage. J. Cell Biol. 2001;153:367–380. doi: 10.1083/jcb.153.2.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yankiwski V, Marciniak RA, Guarente L, Neff NF. Nuclear structure in normal and Bloom syndrome cells. Proc. Natl Acad. Sci. USA. 2000;97:5214–5219. doi: 10.1073/pnas.090525897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lillard-Wetherell K, Machwe A, Langland GT, Combs KA, Behbehani GK, Schonberg SA, German J, Turchi JJ, Orren DK, Groden J. Association and regulation of the BLM helicase by the telomere proteins TRF1 and TRF2. Hum. Mol. Genet. 2004;13:1919–1932. doi: 10.1093/hmg/ddh193. [DOI] [PubMed] [Google Scholar]
- 18.Wu L, Hickson ID. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- 19.Hand R, German J. A retarded rate of DNA chain growth in Bloom's syndrome. Proc. Natl Acad. Sci. USA. 1975;72:758–762. doi: 10.1073/pnas.72.2.758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lönn U, Lönn S, Nylen U, Winblad G, German J. An abnormal profile of DNA replication intermediates in Bloom's syndrome. Cancer Res. 1990;50:3141–3145. [PubMed] [Google Scholar]
- 21.Brosh RM, Jr, Li JL, Kenny MK, Karow JK, Cooper MP, Kureekattil RP, Hickson ID, Bohr VA. Replication protein A physically interacts with the Bloom's syndrome protein and stimulates its helicase activity. J. Biol. Chem. 2000;275:23500–23508. doi: 10.1074/jbc.M001557200. [DOI] [PubMed] [Google Scholar]
- 22.Sharma S, Sommers JA, Wu L, Bohr VA, Hickson ID, Brosh R.M., Jr Stimulation of flap endonuclease-1 by the Bloom's syndrome protein. J. Biol. Chem. 2004;279:9847–9856. doi: 10.1074/jbc.M309898200. [DOI] [PubMed] [Google Scholar]
- 23.Jiao R, Bachrati CZ, Pedrazzi G, Kuster P, Petkovic M, Li JL, Egli D, Hickson ID, Stagljar I. Physical and functional interaction between the Bloom's syndrome gene product and the largest subunit of chromatin assembly factor 1. Mol. Cell Biol. 2004;24:4710–4719. doi: 10.1128/MCB.24.11.4710-4719.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanz MM, Proytcheva M, Ellis NA, Holloman WK, German J. BLM, the Bloom's syndrome protein, varies during the cell cycle in its amount, distribution, and co-localization with other nuclear proteins. Cytogenet. Cell Genet. 2000;91:217–223. doi: 10.1159/000056848. [DOI] [PubMed] [Google Scholar]
- 25.Dutertre S, Ababou M, Onclercq R, Delic J, Chatton B, Jaulin C, Amor-Gueret M. Cell cycle regulation of the endogenous wild type Bloom's syndrome DNA helicase. Oncogene. 2000;19:2731–2738. doi: 10.1038/sj.onc.1203595. [DOI] [PubMed] [Google Scholar]
- 26.Davies SL, North PS, Dart A, Lakin ND, Hickson ID. Phosphorylation of the Bloom's syndrome helicase and its role in recovery from S-phase arrest. Mol. Cell Biol. 2004;24:1279–1291. doi: 10.1128/MCB.24.3.1279-1291.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 28.Jiao R, Harrigan JA, Shevelev I, Dietschy T, Selak N, Indig FE, Piotrowski J, Janscak P, Bohr VA, Stagljar I. The Werner syndrome protein is required for recruitment of chromatin assembly factor 1 following DNA damage. Oncogene. 2007;26:3811–3822. doi: 10.1038/sj.onc.1210150. [DOI] [PubMed] [Google Scholar]
- 29.Wu L, Davies SL, North PS, Goulaouic H, Riou JF, Turley H, Gatter KC, Hickson ID. The Bloom's syndrome gene product interacts with topoisomerase III. J. Biol. Chem. 2000;275:9636–9644. doi: 10.1074/jbc.275.13.9636. [DOI] [PubMed] [Google Scholar]
- 30.Pedrazzi G, Perrera C, Blaser H, Kuster P, Marra G, Davies SL, Ryu GH, Freire R, Hickson ID, Jiricny J, et al. Direct association of Bloom's syndrome gene product with the human mismatch repair protein MLH1. Nucleic Acids Res. 2001;29:4378–4386. doi: 10.1093/nar/29.21.4378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petkovic M, Dietschy T, Freire R, Jiao R, Stagljar I. The human Rothmund-Thomson syndrome gene product, RECQL4, localizes to distinct nuclear foci that coincide with proteins involved in the maintenance of genome stability. J. Cell Sci. 2005;118:4261–4269. doi: 10.1242/jcs.02556. [DOI] [PubMed] [Google Scholar]
- 32.Bachrati CZ, Hickson ID. Analysis of the DNA unwinding activity of RecQ family helicases. Methods Enzymol. 2006;409:86–100. doi: 10.1016/S0076-6879(05)09005-1. [DOI] [PubMed] [Google Scholar]
- 33.Pedrazzi G, Bachrati CZ, Selak N, Studer I, Petkovic M, Hickson ID, Stagljar I. The Bloom's syndrome helicase directly interacts with the human mismatch repair protein MSH6. Biol. Chem. 2003;384:1155–1164. doi: 10.1515/BC.2003.128. [DOI] [PubMed] [Google Scholar]
- 34.Ababou M, Dutertre S, Lecluse Y, Onclercq R, Chatton B, Amor-Gueret M. ATM-dependent phosphorylation and accumulation of endogenous BLM protein in response to ionizing radiation. Oncogene. 2000;19:5955–5963. doi: 10.1038/sj.onc.1204003. [DOI] [PubMed] [Google Scholar]
- 35.von Kobbe C, Karmakar P, Dawut L, Opresko P, Zeng XM, Brosh R.M., Jr, Hickson ID, Bohr VA. Colocalization, physical, and functional interaction between Werner and Bloom syndrome proteins. J. Biol. Chem. 2002;277:22035–22044. doi: 10.1074/jbc.M200914200. [DOI] [PubMed] [Google Scholar]
- 36.Ralf C, Hickson ID, Wu L. The Bloom's syndrome helicase can promote the regression of a model replication fork. J. Biol. Chem. 2006;281:22839–22846. doi: 10.1074/jbc.M604268200. [DOI] [PubMed] [Google Scholar]
- 37.Liberi G, Maffioletti G, Lucca C, Chiolo I, Baryshnikova A, Cotta-Ramusino C, Lopes M, Pellicioli A, Haber JE, Foiani M. Rad51-dependent DNA structures accumulate at damaged replication forks in sgs1 mutants defective in the yeast ortholog of BLM RecQ helicase. Genes Dev. 2005;19:339–350. doi: 10.1101/gad.322605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang S, Zhou Y, Trusa S, Meng X, Lee EY, Lee MY. A novel DNA damage response: rapid degradation of the p12 subunit of DNA polymerase δ. J. Biol. Chem. 2007;282:15330–15340. doi: 10.1074/jbc.M610356200. [DOI] [PubMed] [Google Scholar]
- 39.Podust VN, Hubscher U. Lagging strand DNA synthesis by calf thymus DNA polymerases alpha, beta, delta and epsilon in the presence of auxiliary proteins. Nucleic Acids Res. 1993;21:841–846. doi: 10.1093/nar/21.4.841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Maga G, Villani G, Tillement V, Stucki M, Locatelli GA, Frouin I, Spadari S, Hubscher U. Okazaki fragment processing: modulation of the strand displacement activity of DNA polymerase delta by the concerted action of replication protein A, proliferating cell nuclear antigen, and flap endonuclease-1. Proc. Natl Acad. Sci. USA. 2001;98:14298–14303. doi: 10.1073/pnas.251193198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kamath-Loeb AS, Johansson E, Burgers PMJ, Loeb LA. Functional interaction between the Werner syndrome protein and DNA polymerase δ. Proc. Natl Acad. Sci. USA. 2000;97:4603–4608. doi: 10.1073/pnas.97.9.4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kamath-Loeb AS, Loeb LA, Johansson E, Burgers PMJ, Fry M. Interactions between the Werner syndrome helicase and DNA polymerase δ specifically facilitate copying of tetraplex and hairpin structures of the D(CGG)n trinucleotide repeat sequence. J. Biol. Chem. 2001;276:16439–16446. doi: 10.1074/jbc.M100253200. [DOI] [PubMed] [Google Scholar]
- 43.Fry M, Loeb LA. Human Werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 44.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom's syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 46.Szekely AM, Chen YH, Zhang C, Oshima J, Weissman SM. Werner protein recruits DNA polymerase δ to the nucleolus. Proc. Natl Acad. Sci. USA. 2000;97:11365–11370. doi: 10.1073/pnas.97.21.11365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schawalder J, Paric E, Neff NF. Telomere and ribosomal DNA repeats are chromosomal targets of the bloom syndrome DNA helicase. BMC Cell Biol. 2003;4:15. doi: 10.1186/1471-2121-4-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.