Abstract
Regulation of homologous recombination (HR) represents the best-characterized DNA repair function of p53. The role of p53 phosphorylation in DNA repair is largely unknown. Here, we show that wild-type p53 repressed repair of DNA double-strand breaks (DSBs) by HR in a manner partially requiring the ATM/ATR phosphorylation site, serine 15. Cdk-mediated phosphorylation of serine 315 was dispensable for this anti-recombinogenic effect. However, without targeted cleavage of the HR substrate, serine 315 phosphorylation was necessary for the activation of topoisomerase I-dependent HR by p53. Moreover, overexpression of cyclin A1, which mimics the situation in tumors, inappropriately stimulated DSB-induced HR in the presence of oncogenic p53 mutants (not Wtp53). This effect required cyclin A1/cdk-mediated phosphorylation for stable complex formation with topoisomerase I. We conclude that p53 mutants have lost the balance between activation and repression of HR, which results in a net increase of potentially mutagenic DNA rearrangements. Our data provide new insight into the mechanism underlying gain-of-function of mutant p53 in genomic instability.
INTRODUCTION
Because of the central role of p53 as a gatekeeper and a caretaker, the protein must be subject to complex control mechanisms that orchestrate the multiple functions of p53 in transcription, cell-cycle control, apoptosis induction and DNA repair (1,2). Posttranslational modification of p53 by protein phosphorylation has been the most extensively studied potential functional switch mechanism, as it occurs at multiple serine and threonine residues in response to genotoxic stress (3,4). Modification of p53 on serine 15 by ATM and ATR was demonstrated to trigger the cascade of damage-induced phosphorylation and acetylation events that have been implicated in protein stabilization and enhancement of transcriptional transactivation (3,4). However, observations made with knock-in mouse models (5,6) indicated a role for serine 18 in apoptosis, but not in Mdm2-governed protein stability. Moreover, in several studies, no evidence was found for an essential role of the N-terminal casein kinase 1 (CK1) and ATM/ATR phosphorylation sites in damage-induced transcriptional transactivation (7–9). In addition, when DNA replication was blocked, p53 became phosphorylated on serine 15, but this was not accompanied by a rise in key target gene products such as p21 (10–12). This suggested that after replication fork stalling, p53 phosphorylated on serine 15 (p53pSer15) may serve additional functions unrelated to transcriptional transactivation. In support of this hypothesis, colocalization studies indicated that p53pSer15 forms a component of RAD51-specific repair assemblies (11–13).
Over the last few years, a large body of evidence has emerged indicating that p53 is directly involved in DNA repair, particularly in homologous double-strand break (DSB) repair. First, p53 recognizes three-stranded heteroduplex and four-way Holliday junctions and DNA lesions involving mismatches, gaps or DNA ends. The core domain is required for junction DNA-binding and also harbors an exonuclease activity, the extreme C-terminus stimulates these activities upon mismatch recognition (15,2). Second, p53 physically and functionally interacts with critical enzymes and surveillance factors of homologous recombination (HR), namely with RAD51, RAD54, the MRE11 complex, BRCA1, BRCA2 and BLM, and counteracts strand exchange catalyzed by RAD51. Third, using different cell-based test systems, several groups concurrently found that Wtp53 represses inter- and intra-molecular HR, when triggered by DSBs or replication blocking agents. In contrast, hotspot mutants failed to downregulate these HR activities. The identification of separation-of-function mutations, which had lost p53's transcriptional transactivation and cell-cycle regulatory capacity, but retained HR inhibition, and vice versa, provided further evidence for p53's direct role in HR control (15,2). A recent report describes transcriptional repression of RAD51 by direct binding of Wtp53 to a response element within the promoter region (16). This mechanism can only partially explain the role of p53 in HR, because mutations within the p53 interaction site of the RAD51 protein abrogate HR repression by p53 (13). Moreover, p53(138V), which is defective in sequence-specific DNA binding, retains the HR-downregulatory effect (17). The biological meaning of this, at first sight, paradoxical activity directed against a fairly safe DNA repair pathway was unveiled by systematic substrate variation, which indicated a fidelity control mechanism directed against DNA exchange processes between divergent sequences (in 15). Unexpectedly, Wtp53 was more recently found to stimulate recombination in the absence of targeted substrate cleavage in a manner depending on topoisomerase I (topo I) (18,19). Spontaneous recombination events are coupled to the normal DNA metabolism in proliferating cells such as during the bypass of low level, endogenous lesions at replication forks, which are insufficient to activate stress signalling.
Upon exposure to ionizing radiation and generation of highly recombinogenic DNA lesions such as DSBs, the serines 6, 15 and 315 represent the most prominently phosphorylated p53 residues (3). On the other hand, Subramanian and Griffith (20) demonstrated that recognition of Holliday junction DNA by p53 is particularly sensitive to posttranslational phosphorylation at serine 392 as compared to serines 6 or 15. To define the role of phosphorylation in p53-dependent regulation of recombinative repair, we applied an EGFP-based recombination assay in combination with cells expressing the corresponding phosphorylation site mutants. Candidate kinases were manipulated by pharmacological or shRNA-mediated inhibition and by expression of regulatory subunits. We find that serine 15 phosphorylation by ATM/ATR is one contributing mechanism to channel functions of p53 towards DSB repair surveillance. Serine 315 and cdk2 are required for topo I-dependent recombination, which involves dynamic interactions between p53 and topo I in the nucleus. Together, our data indicate that site-specific phosphorylation of p53 plays a critical role in the fine-tuning of repair pathways with important implications for oncogenic p53 mutants.
MATERIAL AND METHODS
Cells and cell-cycle analysis
For HR-measurements KMV clones derived from the human myeloid leukemia cell line K562 and KMV(HR/3′) derivatives with stably integrated HR-EGFP/3′EGFP substrate were raised in RPMI 1640 supplemented with 10% FCS and 2 mM l-glutamine (Biochrom, Berlin, Germany) (21). Stable WTK1(HR/3′) clones for measurements of HR between HR-EGFP and 3′EGFP on chromosomes were established from the lymphoblastoid cell line WTK1 (21) and cultivated in RPMI 1640 with 12% FCS and 3 mM l-glutamine. When using pSVp53her expression plasmids, cells were propagated in estradiol-free medium before transfection and, then, cultivated in the presence of estradiol at a final concentration of 200 nM to fully activate the p53 moiety within the fusion protein of 90 kD (21).
For the analysis of cell-cycle phases and apoptosis induction at the time point of HR measurement, cells were collected by centrifugation, resuspended in 1 ml HBSS (25 mM HEPES, pH 7.4; 173.3 mM NaCl; 5.6 mM KCl; 0.44 mM KH2PO4; 0.34 mM Na2HPO4; 5 mM glucose; 4 mM NaHCO3) and fixed in 10 ml fixing solution (50% acetone; 40% ethanol; 10% ddH2O; at −20°C) on ice for 15 min. For subsequent staining, fixed cells were washed with 2.5 ml ice-cold PBS supplemented with 0.2% EDTA, resuspended in 100 μl propidium iodide staining solution (0.25 mg/ml RNAse A; 50 μg/ml propidium iodide from Sigma in PBS) and incubated for 15 min in the dark. After diluting the suspension by 200 μl PBS with 0.2% EDTA, the stained cells were analyzed in a FACS® Calibur flow cytometer (Becton Dickinson, Heidelberg, Germany). The percentage of cells with sub-G1 DNA content, which is indicative of apoptosis, was determined, and viable cells were subdivided into populations in the cell-cycle phases G1, S and G2.
Plasmids
The following plasmids were previously described: pSUPER-TopoI (19), pSUPER-p53 (22), pcDNA3-cyclin A1 and pCMV-cyclin A2 (23), SMRAD51 chimera expression plasmid pcDNA3.1-Rad51SM (24), pKEX expression constructs for rat p53 (7), pCMV-I-SceI, pCMV-Wtp53, pCMV-p53(248W), pSVp53her and pSVp53(1-333)her (21). pCMV-p53(315A) was constructed from pCMV-p53 (BD Biosciences Clontech, Heidelberg, Germany) by use of Quickchange (Stratagene Europe, Amsterdam, The Netherlands) to introduce the point mutation. pKD-cdk2-v5, pKD-E2F-1-v1 and empty vectors were purchased from Upstate. Plasmid pSUPER-WRN directs the synthesis of siRNA, which had previously been demonstrated to specifically target WRN-mRNA (25). pSUPER-WRN was generated as described for pSUPER-TopoI in Baumann et al. (19) by use of the following oligonucleotides (Thermohybaid, Ulm): anti-WRNsiRNA-fwd, 5′-GATCCCCTTCACTGGATCCATTGTGTTTCAAGAGAACACAATGGATCCAGTGAATTTTTGGAAA-3′; anti-WRNsiRNA-rev, 5′-AGCTTTTCCAAAAATTCACTGGATCCATTGTGTTCTCTTGAAACACAATGGATCCAGTGAAGGG-3′. Additionally, we newly created pSV53(15A) her for the expression of p53her fusion protein mutated at serine 15 using PCR. PCR amplification relied on pSV53her template and the primer 5′-CTGGTACCATGGAGGAGCCGCAGTCAGATCCTAGCGTCGAGCCCCCTCTGGCTCAGGAAACATTTTCAGACC-3′, covering the 5′-end of the p53 cDNA (mutating bases underlined), and 5′-AGATGGCCATGGCGCGGACGCGG-3′, covering an internal NcoI restriction site. The NcoI digested PCR product was inserted into the pUC-subcloned XbaI fragment from pSV53her, which was retransferred into pSV53her after mutagenesis. The complete sequence encoding p53(15A)her was verified by use of the ABi PRISM Big Dye Ready Reaction Cycle Sequencing kit and an ABi 377 sequencer.
Antibody probes
We used the following mAbs: DO1 directed against the N-terminus of p53, C21 against topo I, B88-2 against cyclin A1, BD611168 against WRN and SX118 against p21 from Becton Dickinson, Pab421 against the C-terminus of p53 (Calbiochem, Darmstadt, Germany), 1D6 against topo I (Novocastra Laboratories Ltd, Newcastle upon Tyne, UK), E 67.1 recognizing Cyclin A2 from Cell Signaling, FPS315 against p53pSer315 (MoBiTec/MBL, Göttingen, Germany) and KH20&KH95 against E2F1 (Upstate Biotechnology Incorporated, Lake Placid, NY, USA). For immunodetection of rat p53, we used rabbit serum NCL-p53-CM5p from Novocastra, for RAD51 H-92, for cyclin A1 H-230 from Santa Cruz Biotechnology, Santa Cruz, CA, USA and for cdk2 07-631 from Upstate. For western analyses of actin, we applied the affinity-purified goat polyclonal antibody I-19 from Santa Cruz, for tubulin mAb DM1A, for TBP mAb 1TBP18 and for GAPDH ab9484 from Abcam, Hiddenhausen, Germany. Antibodies used for immunoprecipitation studies were: human Scl-70 serum (Topogen, Port Orange, FL, USA) and mAb 1D6 (Novocastra) against topo I and the rabbit serum CM1 against p53 (Novocastra). Peroxidase-conjugated affinity purified antibodies against goat, rabbit, human and mouse IgG were obtained from Dianova, Hamburg, Germany or Biomol.
Extraction of cells, immunoprecipitation and immunoblot analysis
Total homogenates from 2 × 105 KMV(HR/3′) cells were prepared, subjected to SDS–PAGE (8–15%) and western blot transfer on Immobilon-P membranes (Millipore, Bedford, MA, USA) as described (21). Filters were stained with antibodies in PBS containing 0.2% Tween-20 and 5% nonfat dried milk, followed by incubation with peroxidase-conjugated secondary antibodies and detected by the ECL reagents SuperSignal® West Pico Chemiluminescent Substrate and West Dura Extended Duration Substrate (Pierce, Rockford, IL, USA). Cellular lysates for immunoblot analysis were prepared in lysis buffer (50 mM Tris, pH 7.4; 150 mM NaCl; 2 mM EGTA; 2 mM EDTA; 25 mM NaF; 25 mM β-glycerophosphate; 0.1 mM NaV; proteinase inhibitor, Roche Diagnostics, Mannheim, Germany) including phosphatase inhibitor cocktail (Roche) in the experiment shown in Figure 6B and 100 µg of total protein per sample resolved by SDS–PAGE. Filters were reprobed with antibodies against actin, tubulin or TBP, or Ponceau stained to control for loading differences. Alternatively, signals for a nonspecific band served to verify equal loading.
Figure 6.
Influence of cdk inhibition on HR regulation by cyclin A1 and mutant p53. (A) DSB-induced HR in KMV(HR/3′) cells treated with DMSO (−) or 80 μM olomoucine (+). Recombination frequencies in controls −p53/−cyclin A1/−olomoucine were set to 100% (absolute mean value: 1.1 × 10−3). Columns, mean (n = 12–18); bars, SEM. (B) Immunoblot analysis for p53pSer315 and p53. (C) Cell-cycle analysis. Columns, mean (n = 2); bars, SD.
For immunoprecipitation KMV(HR/3′) cells were lyzed 24 h after electroporation with the indicated plasmids in 50 mM HEPES (pH 7.8), 200 mM NaCl, 0.1% Triton X-100, 20% glycerol and protease inhibitor cocktail (Roche). For isolation of topo I-Wtp53 complexes in vivo cross-linking was performed as described in Zink et al. (26). Centrifuged extracts were precleared with protein A-sepharose (PAS) for 1 h, incubated with anti-topo I or anti-GAPDH antibodies and PAS for 4 h and further processed as in Restle et al. (12).
Densitometric quantification of band intensities was performed using a ChemImager 5500 with software (Alpha Imunotech Corporation, San Leonardo, California, USA). Values for the protein of interest were corrected with the values obtained for the corresponding loading control.
HR-analysis, drug treatment and knockdown
For the determination of HR frequencies with extrachromosomal plasmid substrate, 107 KMV cells were electroporated with a total amount of 40 μg plasmid DNA (10 μg HR-EGFP/3′EGFP, 10 μg pCMV-I-SceI, 10 μg pBS, 10 μg p53 expression plasmid or empty vector controls). For chromosomal HR measurements, 4 × 106 KMV(HR/3′) or 3 × 106 WTK1(HR/3′) cells were electroporated with 30 μg plasmid DNA (10 μg pBS, 10 μg p53 expression plasmid, cyclin A1 or cyclin A2 expression plasmid or empty vector controls) with or without 10 μg pCMV-I-SceI. 48–72 h after electroporation flow cytometric analysis was performed in a FACS® (Becton Dickinson) to determine the fraction of green fluorescent cells within a population of nonfluorescent cells as described previously (21). Each measurement was paralleled by the same coelectroporation including a WtEGFP control plasmid instead of a pBS filler plasmid. The fraction of EGFP-positive cells in these controls was determined each and was used to normalize each single recombination frequency. This calculation procedure also corrects for possible rate deviations related to growth and death regulatory effects. For drug treatment, KMV(HR/3′) cultures were split and treated with kinase inhibitor or solvent starting from 4 h after electroporation until measurements were performed (72 h). Caffeine (Sigma, Deisenhofen, Germany) was dissolved in H2O and included at a final concentration of 400 μM, CGK (Sigma) in DMSO and applied at 2 μM; olomoucine (Calbiochem) was dissolved in DMSO and added at 80 μM. In topo I, p53, WRN, cdk2 and E2F1 knockdown experiments 20 µg of pSUPER-TopoI, pSUPER-p53, pSUPER-WRN, pKD-cdk2-v5, pKD-E2F-1-v1 or empty vector were added to the coelectroporation mixture (transfection efficiency: 40–80%). The statistical significance of differences was calculated by use of the t-test.
RESULTS
Changes in the regulation of homologous DSB repair after mutation of p53 phosphorylation sites
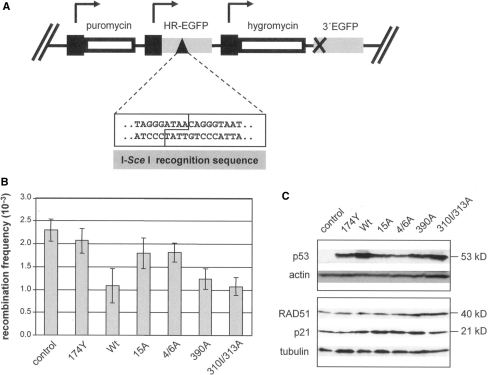
Recombinative repair represents a well-established direct DNA repair function of p53 (15,2) and had been characterized by use of an EGFP-based assay in K562 derived KMV cell clones, which are devoid of endogenous p53 (19,21). In this assay, HR between mutated EGFP sequences, located on the same or the sister chromatid, restores a functional EGFP gene, which can be monitored by flow cytometry via quantitation of green fluorescent cells (Figure 1A). DNA-exchange processes are triggered by DSB formation in vivo, when the rare-cutting meganuclease I-SceI is expressed and introduces a single DSB at the recognition sequence within the repair substrate.
Figure 1.
HR analysis after expression of p53 proteins mutated on N- and C-terminal phosphorylation sites. (A) Schematic drawing of the HR assay substrate HR-EGFP/3′EGFP, which allows to quantify homologous DSB repair by FACS after I-SceI-mediated cleavage (21). (B) KMV(HR/3′) cells were electroporated with pCMV-I-SceI, and pKEX (control) or pKEX derivatives for the expression of Wt and mutant (174Y, 15A, 4/6A, 390A, 310/313A) p53 from rat, and cultivated for 72 h. Recombination frequencies were determined as the fraction of green fluorescent cells compared to the total cell population and normalized in each case to the individual transfection efficiency. Columns, mean (n = 6); bars, SEM. (C) Total cell homogenates were prepared from transfected KMV(HR/3′) cells and the amounts of p53, p21 and RAD51 were analyzed by immunoblotting.
To examine the role of p53 phosphorylation in the modulation of HR, we chose to express a series of rat p53 phosphorylation mutants, which in a previous comparative study had been shown to retain transcription activity (7). Thus, we cotransfected KMV(HR/3′) cells, carrying the stably integrated HR-substrate HR-EGFP/3′EGFP (Figure 1A), with expression plasmids for I-SceI (pCMV-I-SceI) and for Wtp53, oncogenic p53(174Y) or phosphorylation mutants p53(15A), p53(4/6A), p53(390) or p53(310/313A), or empty vector. HR was reduced down to 47% by Wtp53, when compared with the p53-negative control (P = 0.034) (Figure 1B). We found similar HR repression by the mutants p53(390A) (P = 0.012) and p53(310/313A) (P = 0.005). p53(174Y), which is devoid of any functions in transcriptional and HR regulation (21), showed loss of function. After expression of p53(15A) or p53(4/6A) minor, statistically not significant, changes in HR frequencies compared to p53-negative controls were noted (P = 0.307 and P = 0.193, respectively). When we analyzed p53 expression, we noticed lower levels for p53(15A) and p53(4/6A) as compared to Wtp53 (Figure 1C). Nevertheless, regarding the products of genes, which are subject to transcriptional transactivation or repression by p53, we observed comparable p21 increases in the presence of Wtp53 as well as for p53(15A), p53(4/6A), p53(390) and p53(310/313A), and no decrease of RAD51.
Repression of homologous DSB repair by p53 is lost after ATM/ATR kinase inhibition
Phosphorylation of p53 at serine 15 by ATM and ATR is thought to initiate the cascade of DNA damage-induced posttranslational modification events on p53 (3,4). To further delineate a potential role of serine 15 phosphorylation in the regulation of DSB repair, we cotransfected KMV(HR/3′) cells with pCMV-I-SceI and constructs for comparable expression of human Wtp53 and the phosphorylation mutant p53(15A) (21) (Figure 2C). Wtp53 caused a 36–51% (P = 0.017–0.046) decrease of chromosomal HR (Figure 2A and B). p53(15A) expressing cells exhibited an intermediate phenotype with a recombination frequency reduction of 10–31% (P = 0.047–0.517). For comparison, the HR regulation defective tetramerization mutant, p53(1-333), caused a statistically not significant 25% decrease (P = 0.126). To study the possible involvement of ATM and/or ATR, we next treated the cells with the ATM and ATR inhibitor caffeine (27). Western blot analysis verified reduced phosphorylation of p53 at serine 15 after caffeine treatment (Figure 2C). Caffeine treatment completely abrogated HR-repression by Wtp53 as well as the residual influence by p53(15A) and p53(1-333) (Figure 2A).
Figure 2.
Influence of ATM/ATR-mediated phosphorylation. (A) Homologous DSB repair of chromosomal substrates was assayed in KMV(HR/3′) cells as in Figure 1 using the following expression plasmids for human p53 fused to the human estrogen receptor hormone-binding domain: pSVp53her (Wt), pSVp53(15A)her, pSVp53(1-333)her or pBS (control) in the absence or presence of 400 μM caffeine (A) or 2 μM CGK (B). Recombination frequencies in cells without p53 were set to 100% [absolute mean value in (A) and (B): 1.0 × 10−3]. Columns, mean (n = 3–9); bars, SEM. (C) Equal expression of p53her and p53(15A)her and decreased phosphorylation of p53her at serine 15 after caffeine and CGK treatment, respectively.
To rule out a major contribution of the cell cycle to the effects shown in Figure 2A, we analyzed by flow cytometry the cellular distribution of G1-, S- and G2-phases. Under the conditions of HR measurements (72 h treatment with 400 μM caffeine, purposefully chosen after titration analysis to exclude cytotoxic effects), all cell types, with or without p53 expression and with or without inhibitor treatment, showed comparable cell-cycle profiles (Supplementary Figure 1A). In addition, the fraction of apoptotic cells identified by sub-G1 DNA content after propidium iodide staining, was neither significantly different in this experiment nor in the experiments described below (Supplementary Figure 1B and data not shown). This is in agreement with the fact that K562 cells are defective in apoptosis induction after p53 expression (28). Since RAD51 was demonstrated to represent the immediate target of p53-dependent HR inhibition (15,2), we wished to compare the influence of Wtp53 with inactivation of RAD51. For that purpose, we expressed SMRAD51, a yeast–mouse chimera, with a dominant-negative effect but which does not modify cell viability in mammalian cells (24). We found repression of DSB-induced HR in KMV(HR/3′) cells down to 63% (P = 0.001), i.e. to a degree similar to that of Wtp53. This is compatible with p53 exerting its anti-recombinogenic effect via inhibition of RAD51-dependent HR pathways such as gene conversion (21,29).
Additionally, when we tested the more potent and selective ATM/ATR inhibitor CGK (30), which also caused reduced phosphorylation of p53 at serine 15 (Figure 2C), we again observed loss of HR repression by Wtp53 and no further change for p53(15A) (Figure 2B). Altogether, our findings suggest that serine 15, which is phosphorylated by ATM and ATR, is required for fully efficient regulation of DSB-induced HR. They also show that cellular ATM and/or ATR kinase activities are needed to uncover differences in HR frequencies between p53-positive and -negative cells.
Stimulation of I-SceI-independent HR by p53 requires the cdk phosphorylation site, serine 315
Given that mutation of phosphorylated amino acids on the C-terminus of rat p53 did not alter DSB repair regulation (Figure 1B), we confirmed these observations for human p53. Therefore, we performed HR measurements on chromosomally integrated substrates after expression of human Wtp53 and cancer-related mutant p53(273H), p53(315A), p53(392A) and p53(392D). p53(315A) and p53(392A) are resistant to phosphorylation on the respective serine, whereas p53(392D) mimics phosphorylation on serine 392. p53(315A), p53(392A) and p53(392D) downregulated homologous DSB repair similar to Wtp53 (P ≤ 0.001) (Figure 3A). For comparison, p53(273H) showed residual chromosomal downregulation activity, however, without statistical significance (P = 0.096). Protein levels of p21 were comparably elevated in Wtp53, p53(315A), p53(392A) and p53(392D), but not p53(273H) producing cells, whereas RAD51 protein levels remained constant (Figure 3B). Thus, phosphorylation of serines 315 and 392 does not play a significant role in p53-mediated repression of DSB-induced HR.
Figure 3.
HR regulation upon mutation of C-terminal p53 phosphorylation sites. (A) Chromosomal homologous DSB repair using human p53 expression plasmid pCMV-p53(273H), pCMV-Wtp53, pCMV-p53(315A), pCMV-p53(392A), pCMV-p53(392D) or empty vector (control). Recombination frequencies in cells without p53 were set to 100% (absolute mean value: 2.0 × 10−3). Columns, mean (n = 6–12); bars, SEM. (B) Western blot analysis of p53, RAD51 and p21.
Since amino acid 315 is located within the topo I-binding site of p53 (31), we next sought to determine the involvement of serine 315 in the stimulation of I-SceI-independent HR, which was shown to be mediated by Wtp53 and topo I (19). HR was monitored using KMV(HR/3′) cells in combination with expression of human Wtp53, p53(315A), or the hotspot mutants, p53(273H) and p53(248W), in the absence of substrate cleavage by I-SceI. Consequently, we scored an average recombination frequency two orders of magnitude below I-SceI-induced HR (see legend to Figure 4A and B). I-SceI-independent HR was increased ∼2-fold by Wtp53 (P = 0.023), p53(273H) (P = 0.113) and p53(248W) (P = 0.001), but not by p53(315A) (Figure 4A). To determine, whether HR enhancement is topo I-dependent, we repressed endogenous topo I expression by transfecting with pSUPER-TopoI (19) (Figure 5D). In these knockdown assays, HR stimulation was abolished for Wtp53, p53(273H) and p53(248W), reaching statistical significance for Wtp53 (P = 0.030) and p53(248W) (P ≤ 0.001). To exclude the possibility that topo I represents an essential component of HR, we performed the same HR measurements with simultaneous I-SceI expression for targeted substrate cleavage. Under these conditions, topo I knockdown had no effect on HR activities, regardless of the p53 status (Figure 4B). FACS analysis of the fraction of cells with G1, S or G2 DNA content or of apoptotic cells failed to detect significant differences between samples without and with expression of Wtp53, p53(273H) or p53(248W), arguing against HR stimulation being indirectly caused by the classical tumor suppressor activities (Supplementary Figure 1B). These results indicated that both Wtp53 and oncogenic mutant p53 stimulate I-SceI-independent HR, topo I-mediated recombination and revealed that serine 315 is essential for the stimulatory effect.
Figure 4.
p53-mediated regulation of HR after knockdown of topo I. (A) and (B) KMV(HR/3′) cells were transfected with pCMV-Wtp53, pCMV-p53(315A), pCMV-p53(273H), pCMV-p53(248W) or empty vector (control) and pSUPER (−) or pSUPER-TopoI (+) for topo I-specific RNA interference, and subjected to HR-measurements. For I-SceI-mediated substrate cleavage in (B), pCMV-I-SceI was included in the transfection mixture. Recombination frequencies of p53-negative cells with pSUPER were set to 100% (absolute mean values: 2.7 × 10−5 without I-SceI and 2.0 × 10−3 with I-SceI). Columns, mean values of 12 to 41 (A) and six (B) measurements, respectively; bars, SEM.
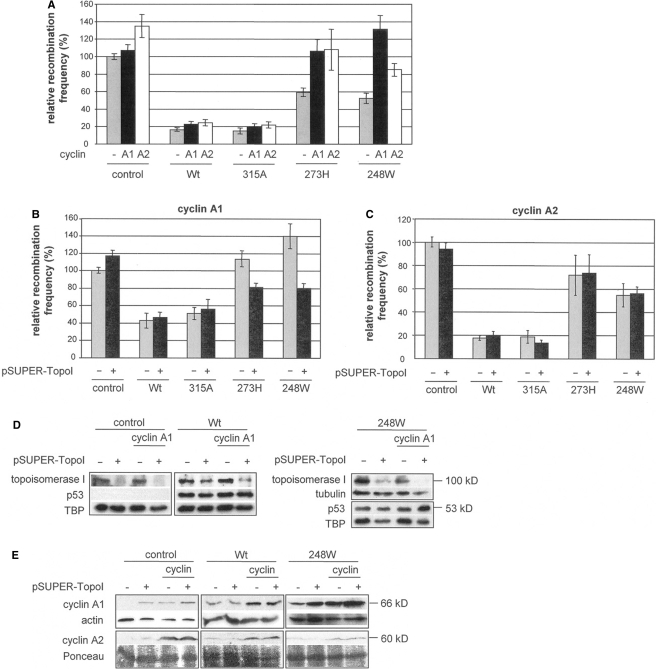
Figure 5.
Effect of cyclin A1 and A2 on homologous DSB repair in cells differing in their p53 and topo I status. (A) DSB-induced HR was determined in KMV(HR/3′) cells cotransfected with pCMV-I-SceI, and pCMV-Wtp53, pCMV-p53(315A), pCMV-p53(273H), pCMV-p53(248W) or empty vector (control), and pcDNA3-cyclin A1 (A1), pCMV-cyclin A2 (A2) or empty vector (−). Columns, mean (n = 12–27); bars, SEM. (B) DSB-induced HR in cells overexpressing cyclin A1, cotransfected with pSUPER (−) or pSUPER-TopoI (+). Columns, mean (n = 12–18); bars, SEM. (C) DSB-induced HR in cells overexpressing cyclin A2, cotransfected with pSUPER (−) or pSUPER-TopoI (+). Columns, mean (n = 6); bars, SEM. Recombination frequencies in controls were set to 100% (absolute mean value in (A) −p53/−cyclins: 2.1 × 10−3; in (B) −p53/+cyclin A1/−pSUPER-TopoI: 2.2 × 10−3; in (C) −p53/+cyclin A2/−pSUPER-TopoI: 2.8 × 10−3). (D) pSUPER-TopoI-mediated downregulation of topo I protein compared to p53 expression. (E) Western blotting of cyclin A1 and cyclin A2.
Oncogenic p53 mutants stimulate cyclin A1-induced, topo I-dependent homologous DSB repair
Cyclin A1, an alternative, cdk2 associated A-type cyclin, has been linked to DSB repair, and its promoter represents a transcriptional transactivation target of p53 (23). To test a potential contribution of cyclin A1 in the regulation of homologous DSB repair by p53, we simultaneously expressed cyclin A1, p53 proteins and I-SceI in KMV(HR/3′) cells. In parallel, we examined the influence of cyclin A2. We found a 69–83% enhancement of homologous DSB repair after cyclin A1 expression in cells with p53(273H) (P ≤ 0.004) and of 65–148% for p53(248W) (P ≤ 0.006), whereas in p53-negative controls frequencies were only 7–18% higher (P > 0.05) (Figures 5A, 6A and 7A). In cells expressing mutant p53(175H), we measured 78% higher HR frequencies (P = 0.013). After cyclin A2 expression, relative increases were 47–83% for p53(273H) (P = 0.009–0.025), 42–60% for p53(248W) (P = 0.003–0.014), and 35–49% for controls (P ≥ 0.019). As expected, in cells with Wtp53 and p53(315A), DSB-induced HR was strongly suppressed, namely on average to 20% of control levels (Figure 3A), with increments of 35–41% and 17–33% upon cyclin A1 and of 13–48% and 46–47% upon cyclin A2 production (P > 0.05), respectively (Figures 5A, 6A and 7A). Flow cytometric analysis revealed that cells expressing cyclin A1 or cyclin A2 showed similar cell-cycle distributions compared with controls (Supplementary Figure 1C). Moreover, we measured recombination in the presence of the pancaspase inhibitor zVAD-fmk (50 µM) for Wtp53, p53(315A) and p53(273H) versus control (± cyclin A1 each). Recombination results did not differ from the initial results, thus, arguing against an involvement of apoptosis in the HR regulation (data not shown).
Figure 7.
Regulation of HR by cyclin A1 and mutant p53 after knockdown of cdk2. (A) DSB-induced HR in KMV(HR/3′) cells cotransfected with empty vector or pKD-cdk2-v5 (pKD-cdk2) for cdk2-specific RNA interference. Recombination frequencies in controls −p53/−cyclin A1/−pKD-cdk2 were set to 100% (absolute mean value: 1.2 × 10−3). Columns, mean (n = 6); bars, SEM. (B) Western blot analysis of cdk2.
We noticed that exogenous p53(273H) exerted residual HR-repression activities to a varying extent (Figures 5A, 6A and 7A) (21,32). Therefore, we wished to compare cyclin A1-mediated HR stimulation in cells stably expressing endogenous mutant p53 and cell derivatives without p53. Consequently, we analyzed HR activities in the human B-lymphoblastoid cell line WTK1(HR/3′) carrying p53(237I) (21) and chromosomally integrated HR substrate. HR assays were performed with and without cyclin A1 overexpression and concomitantly with or without knockdown of endogenous p53 by pSUPER-p53 (Supplementary Figure 2B). This approach revealed a statistically significant (25%, P = 0.001) rise in HR frequencies from mutant p53-deficient (+ pSUPER-p53) to -proficient cells (+pSUPER) only after cyclin A1 overexpression (Supplementary Figure 2A). Cell-cycle changes were not observed (Supplementary Figure 2C).
We next reapplied pSUPER-TopoI to analyze the involvement of topo I in the cyclin A1 and cyclin A2-dependent stimulation of homologous DSB repair. Interestingly, after topo I knockdown in KMV(HR/3′) cells we did not find significant differences in HR frequencies except for the cells expressing cyclin A1 in combination with the oncogenic p53 mutants (Figure 5B and C). Thus, with p53(273H) HR was reduced by 28% (P = 0.004), with p53(248W) by 43% (P = 0.001). We verified topoisomerase downregulation as depicted in Figure 5D for cultures with and without cyclin A1 expression. In the same samples, p53 levels did not follow the topo I expression pattern when compared to the TBP control. Taken together, both cyclin A1 and cyclin A2 overexpression (Figure 5E) caused an enhancement of DSB-induced HR, particularly in cells expressing a p53 hotspot mutant. When compared to p53-negative controls, mutant p53-mediated HR stimulation was much more pronounced in cells with cyclin A1 rather than cyclin A2. This indicated a cyclin A1- and mutant p53-specific component. Most importantly, HR enhancement was abolished by topo I knockdown solely in cells with cyclin A1 and mutant p53.
Mutant p53-mediated stimulation of homologous DSB repair requires cdk2
We next used two independent approaches to analyze the role of cdk2 in regulating HR after cyclin A1 and mutant p53 expression. First, we included the cdk inhibitor, olomoucine, into KMV(HR/3′) cultures after transfection with I-SceI, cyclin A1, and p53 expression plasmids. Olomoucine treatment abolished the recombination stimulatory effect of cyclin A1 in mutant p53 expressing cells (Figure 6A; P ≤ 0.016). Unexpectedly, in p53-negative controls, HR enhancement by cyclin A1 was more pronounced after drug treatment. Without cyclin A1 expression, no significant differences were observed between any of the mock- and olomoucine-treated samples (Figure 6A). Immunoblotting verified the effectiveness of the cdk inhibitor, because signals specific for p53 phosphorylated on serine 315 were reduced down to 37–51% after olomoucine treatment in p53(248W) samples, although total levels of p53 remained unaltered (Figure 6B). Phosphorylation of serine 315 in Wtp53 was affected to a lesser extent (64–100% residual levels) and additionally accompanied by a decrease in total p53 levels. Flow cytometric analysis did not reveal major changes in cell cycle distribution for p53-positive cells (Figure 6C). However, for p53-negative cells expressing cyclin A1, we noticed that the percentage of cells residing in S-phase rose from 20% to 38% in response to olomoucine treatment, which was consistent with observations previously made in breast cancer cells (33). Considering that cells are highly active in HR during S-phase (34), cell cycle regulatory effects may underly the unexpected HR increase that we observed in treated, cyclin A1 expressing, p53-negative cells.
In the second experimental setup, we examined DSB-induced HR after cdk2 knockdown. As depicted in Figure 7A, downregulation of cdk2 (Figure 7B) in assays with cyclin A1 and p53(273H) or p53(248W) significantly reduced HR frequencies to levels without cyclin A1 (P = 0.000, P = 0.014, respectively). Although corresponding mean values for p53-negative controls also indicated a HR decrease, statistical significance was not reached (P = 0.066). Cell-cycle profiles did not show major differences after cdk2 knockdown (Supplementary Figure 1D). We further tested a possible involvement of E2F1 in the cyclin A1- and mutant p53-dependent HR stimulation, since nuclear retention of p53 by the transcription factor E2F1 was reported to require phosphorylation of p53 at serine 315 (35) and since cyclin A1 was shown to bind to E2F1 (36). In addition, we checked the influence of Werner's syndrome protein (WRN), which was described to bind both p53 (37) and topo I (38,39). Recently, Song et al. (40) reported that mutants p53(248W) and p53(273H) displayed tumor-promoting characteristics in a humanized p53 knockin mouse model and caused genetic instability by inactivating ATM signaling of DSBs. Consequently, we also examined links to ATM. However, unlike the situation observed after olomoucine treatment or cdk2 knockdown, neither downregulation of E2F1, WRN or ATM nor caffeine treatment reversed recombination enhancement mediated by cyclin A1 and mutant p53 (data not shown). In summary, cdk2 inactivation by pharmacological inhibition or RNA interference suppressed the HR-stimulatory effect of mutant p53, which suggests that phosphorylation by cyclin A1-cdk2 is a critical step in this process.
Cyclin A1 stimulates association of topo I with p53
Given that cyclin A1-mediated stimulation of homologous DSB repair depends on topo I and mutant p53, we examined whether formation of protein complexes containing oncogenic mutant p53 and topo I is involved in cyclin A1-mediated recombination stimulation. For this purpose, we performed coimmunoprecipitation analysis using Scl70-antiserum, which had previously been established for specific precipitation of topo I complexes (41). Topo I-bound proteins were recovered from extracts of cells transfected and treated for DSB-repair measurements as in Figure 6. p53(248W) bound to topo I, particularly in cells overexpressing cyclin A1 (Figure 8). Under these conditions, we were unable to detect Wtp53 signals in topo I precipitates, unless chromatin complexes were cross-linked in vivo (Figure 8), indicating weak or transient interactions with topo I. Additional treatment of these cells with olomoucine reduced the association of Wtp53 and particularly of p53(248W), with topo I, supporting the hypothesis that cyclin A1/cdk-mediated phosphorylation, plays an important role in the stable association of oncogenic p53 with topo I.
Figure 8.
Coimmunoprecipitation of topo I and p53 is enhanced by cyclin A1. KMV(HR/3′) cells were electroporated with pCMV-I-SceI plus empty expression vector (control), pCMV-p53(248W) or pCMV-Wtp53 plus empty vector or pcDNA3-cyclin A1, and treated with DMSO (−) or 80 μM olomoucine, exactly as described in Figure 6. Twenty-four hours later, whole-cell extracts were prepared without [p53(248W)] or with (Wtp53) cross-linking of chromatin complexes in vivo and topo I immunoprecipitated with Scl-70 serum followed by western blotting for p53 or topo I. As a negative control for nonspecific topo I interactions, the extract was immunoprecipitated with anti-GAPDH antibodies (IgG). Nonspecific signals in the western blot from the heavy chain with a molecular mass similar to p53 were excluded by the empty vector controls.
DISCUSSION
The p53 is phosphorylated on several residues in response to genotoxic stress, in particular to insults such as DSBs (15,2). Because of the well-established regulatory functions of p53 in DSB repair pathways, we and others previously proposed that damage-induced phosphorylation may modulate repair-related p53 activities. In this study, we systematically examined this hypothesis, demonstrating that phosphorylation of p53 at different positions modulates distinct DNA repair-related functions of p53.
Functional proof of a partial involvement of serine 15 in DSB repair
Phosphorylation of p53 at serine 15 by ATM represents the initial p53 modification induced by DSBs (4). In chromatin immunoprecipitation experiments ATM and p53 were found to localize to DSBs preceding repair (42). Moreover, we and others previously demonstrated that p53pSer15 represents the p53 subpopulation colocalizing with the MRE11 complex, RAD51 and RAD54 after replication blockage (11–13). However, so far, only indirect evidence suggested HR-related functions for this subpopulation of posttranslationally modified p53. In this work, we demonstrate that serine 15 of p53 is necessary for fully efficient repression of homologous DSB repair. On the other hand, with C-terminal phosphorylation site mutants, changes in HR downregulation were not detectable. We noticed that the phosphorylation-resistant mutant p53(15A) displayed residual HR-repression activities, whereas chemical inhibition of ATM and ATR completely abrogated p53-dependent HR downregulation. This can be explained by the involvement of additional N-terminal sites which, like serines 20 and 37, are directly or indirectly modified after ATM/ATR activation (3,4). However, we cannot exclude that ATM and/or ATR are required for p53-mediated HR repression independently of p53 phosphorylation.
Under the conditions of the DSB repair test, we observed no alterations in transcriptional transactivation nor in repression by p53(15A), when monitored via p21 and RAD51 expression. Additionally, we largely excluded that the influence of serine 15 phosphorylation on HR was caused by major p53 activities in cell cycle control or apoptosis induction. Our observations strengthen earlier conclusions of a direct involvement of p53 in RAD51-dependent gene conversion (15,2). Physical interactions with heteroduplex DNA recombination intermediates and RAD51 do not involve the N-terminus of p53 (15). RPA binding, which like RAD51 binding is required for HR repression by p53, was mapped around aa 48–54, and, therefore, is more likely to be modulated by N-terminal phosphorylation (43). Importantly, colocalization and immunoprecipitation experiments showed that BLM facilitates physical interactions between p53 and the sensor proteins 53BP1 and H2AX, and thereby leads to efficient localization of p53 to RAD51 foci and to a net increase in p53pSer15 (11,44). Therefore, phosphorylation of N-terminal p53 amino acids by ATM, ATR and downstream kinases may stabilize one or multiple contacts with recombination factors, enabling the HR repression function of p53.
Links between p53 and cyclin A1 in recombination
Consistent with our previous observations (18,19), we showed here that Wtp53 activates HR in the absence of targeted substrate cleavage in a topo I-dependent manner. We have now discovered the same to be true for the hotspot mutants p53(273H) and p53(248W). Serine 315 turned out to be necessary for topo I-dependent HR stimulation, whereas it was dispensable for the repression of DSB-induced HR.
Cyclin A-cdk2 was previously shown to phosphorylate serine 315 of human p53 (45,46). Interestingly, we found that even DSB-induced HR was stimulated by mutant p53 in a manner depending on topo I, when cyclin A1 was overexpressed (up to 20-fold further increase as compared to the p53-negative control). Under the same conditions, Wtp53 caused repression of homologous DSB repair. In contrast to cyclin A1, overexpression of cyclin A2 was accompanied by a mostly p53- and topo I-independent increase in homologous DSB repair. This general increase in HR is in line with a report describing that efficient DSB-induced ATM/ATR-signaling to Chk1 requires cdk kinase activity (47). Since Chk1 is required for RAD51 activation, this implies that cdks promote HR (48).
From our data and given that cyclin A1 is a target gene product of Wtp53 (23), we would like to put forward the following hypothesis (Figure 9): WTp53 may support rare HR processes via cyclin A1 and topo I in normal, proliferating cells, particularly during S-phase, when active DNA synthesis is associated with replication fork stalling and recombinative bypass events. However, in the presence of severe damage such as DSBs, ATM-signaling may dominantly promote HR-repression functions of Wtp53, i.e. overcome HR stimulation. Oncogenic mutant p53 has lost the HR repression but retained the HR-stimulation function resulting in a net increase of recombination.
Figure 9.
Proposed model of a cyclin A1-induced mechanism leading to genomic instability by oncogenic mutant p53.
Dynamic p53-topo I interactions as the mechanistic basis of cyclin A1-induced recombination
Topo I is known to bind p53 between aa 302 and 321 (31), i.e. in a region containing serine 315, which is phosphorylated in response to cyclin A1 expression. Immunoprecipitation experiments revealed that overexpression of cyclin A1 promoted physical interactions between topo I and p53(248W). For Wtp53, we noticed comparatively weak or transient topo I binding, which is in agreement with observations made by Gobert et al. (31). Cyclin A1-enhanced p53–topo I interactions were reduced after pharmacological inhibition of cdks, particularly for p53(248W). Thus, we propose that cyclin A1/cdk2-mediated phosphorylation of p53 enables stable complex formation with topo I, thereby causing hyper-recombination in p53 mutant cells (Figure 9).
Topo I has been recognized as a DNA metabolizing enzyme that—once localized to DNA lesions by other proteins—is necessary for the recruitment of DNA repair enzymes. On the other hand, topo I is potentially dangerous due to its nicking–closing activity (49). Topo I facilitates initiation of genetic recombination through strand breakage after strand pairing and/or resolution of DNA junctions (50–52). p53, and oncogenic mutant p53 in particular, was previously demonstrated to stimulate topo I relaxation activity (54,31) and the formation of topo I double cleavage complexes causing oligonucleotide excision coupled to recombinative gap filling (55). From this, it is conceivable that stable complex formation between topo I and mutant p53 after cdk-mediated p53 phosphorylation is sufficient to inappropriately stimulate recombinase activities through recruitment to genomic DNA breaks and enzymatic activation of topo I.
Gain-of-function in p53 mutants and genome destabilization
It has been well-established that cancer-related p53 mutants, including p53(273H) and p53(248W), not only lost tumor suppressor functions, but also acquired additional oncogenic activities (56). In this study, we utilized cellular systems derived from p53-negative or mutant p53 cells to exclude dominant negative effects on endogenous Wtp53. p53 gain-of-function effects on genomic instability, in particular, are manifested by aberrant ploidy, increased mutation and gene amplification rates (56–58). We and others proposed that topo I mediates gain-of-function of mutant p53 (18,31,41,54). To our knowledge this is the first report that demonstrates that cyclin A1 stimulates DSB-triggered HR in cells that express mutant p53 but not in cells lacking p53, which indicates a gain-of-function. This effect is mediated by cdk2-dependent phosphorylation and by topo I. Cyclin A1 overexpression revealed a gain-of-function of mutant p53 even when directly comparing HR frequencies in cells with versus without mutant p53. This was evident after knockdown of endogenous mutant p53 (Supplementary Figure 2).
Increased topo I activities were detected in several cancer types and found to correlate with abnormal p53 function in melanomas (49). Since mutant p53 is transcriptionally defective, it is important to note that p53 is not the only regulator of cyclin A1 expression. Rather, upon mutation of TP53, alternative transcription factors, such as the oncoproteins c-myb, Six1 and PML-RARα, can induce cyclin A1 expression (59–61). In fact, elevated cyclin A1 levels have been detected in several cancers including male germ cell tumors (62), breast cancer (61) and in acute myeloid leukemia (63), whereas it is normally suppressed in most somatic cells (Figure 9). Evidence for the important role of cyclin A1 in the initiation of acute myeloid leukemia was provided by a transgenic mouse model with targeted cyclin A1 in the bone marrow (64). Most intriguingly, with respect to the observed synergistic effect of mutant p53 and cyclin A1 on recombination, Tokumaru and colleagues (65) found a negative correlation between cyclin A1 hypermethylation, with robust cyclin A1 protein expression and p53 mutation in head and neck cancer. From our results, mutant p53 is predicted to promote genome instability in tumors with cyclin A1 overexpression.
In summary, we have demonstrated that phosphorylation of p53 at distinct amino acids does differentially regulate direct DNA repair-related activities. We provide functional data, which unveil a role of the ATM/ATR-target serine 15 in fully enabling p53 to downregulate HR and of cdk2-dependent phosphorylation in activating HR. Phosphorylation of p53 at serine 15 forms part of the ATM/ATR-controlled DNA damage response network, which is activated early in tumorigenesis before genomic instability occurs (66,67). p53Ser15 may, therefore, contribute at least in part through repair regulatory functions to the barrier against cancer, and indeed, recent in vivo data demonstrate that serine 15 is required for p53-mediated tumor suppression (68). On the other hand, we provide evidence that oncogenic p53 mutant proteins have lost the balance between activation and repression of recombinative repair, which under conditions such as cyclin A1 overexpression may result in an increase of mutagenic DNA rearrangements such as gene amplification or loss of heterozygosity. Our data provide new answers to the longstanding question of the mechanism underlying the gain-of-function in oncogenic p53 mutants in genome instability.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
We are grateful to Bernard S. Lopez (Fontenay aux Roses, France) for providing plasmid SMRAD51 chimera expression plasmid and Klaus Römer (Homburg/Saar, Germany) for pCMV-p53(392D) and pCMV-p53(392A). We thank Nuray Akyüz and Christine Janz for initial help with HR-measurements and Gisa S. Boehden for cloning pSV53(15A)her. This work was supported by Deutsche Forschungsgemeinschaft (Wi 1376/3-2, Wi 3099/7-1); Landesstiftung Baden-Württemberg (10-1907-Wi 2); Dr Mildred Scheel Stiftung für Krebsforschung (107744). Funding to pay the Open Access publication charges for this article was provided by the University of Ulm, Germany.
Conflict of interest statement. None declared.
REFERENCES
- 1.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 2.Sengupta S, Harris CC. p53: traffic cop at the crossroads of DNA repair and recombination. Nat. Rev. Mol. Cell Biol. 2005;6:44–55. doi: 10.1038/nrm1546. [DOI] [PubMed] [Google Scholar]
- 3.Saito S, Yamaguchi H, Higashimoto Y, Chao C, Xu Y., Jr, Fornace AJ, Appella E, Anderson CW. Phosphorylation site interdependence of human p53 post-translational modifications in response to stress. J. Biol. Chem. 2003;278:37536–37544. doi: 10.1074/jbc.M305135200. [DOI] [PubMed] [Google Scholar]
- 4.Meek DW. The p53 response to DNA damage. DNA Repair. 2004;3:1049–1056. doi: 10.1016/j.dnarep.2004.03.027. [DOI] [PubMed] [Google Scholar]
- 5.Chao C, Hergenhahn M, Kaeser MD, Wu Z, Saito S, Iggo R, Hollstein M, Appella E, Xu Y. Cell type- and promoter-specific roles of Ser18 phosphorylation in regulating p53 responses. J. Biol. Chem. 2003;278:41028–41033. doi: 10.1074/jbc.M306938200. [DOI] [PubMed] [Google Scholar]
- 6.Sluss HK, Armatam H, Gallant J, Jones SN. Phosphorylation of serine 18 regulates distinct p53 functions in mice. Mol. Cell Biol. 2004;24:976–984. doi: 10.1128/MCB.24.3.976-984.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fuchs B, O’Connor D, Fallis L, Scheidtmann KH, Lu X. p53 phosphorylation mutants retain transcription activity. Oncogene. 1995;10:789–793. [PubMed] [Google Scholar]
- 8.Thompson T, Tovar C, Yang H, Carvajal D, Vu BT, Xu Q, Wahl GM, Heimbrook DC, Vassilev LT. Phosphorylation of p53 on key serines is dispensable for transcriptional activation and apoptosis. J. Biol. Chem. 2004;279:53015–53022. doi: 10.1074/jbc.M410233200. [DOI] [PubMed] [Google Scholar]
- 9.Jackson MW, Agarwal MK, Agarwal ML, Agarwal A, Stanhope-Baker P, Williams BRG, Stark GR. Limited role of N-terminal residues in the activation of transcription by p53. Oncogene. 2004;23:4477–4487. doi: 10.1038/sj.onc.1207575. [DOI] [PubMed] [Google Scholar]
- 10.Gottifredi V, Shieh S, Taya Y, Prives C. p53 accumulates but is functionally impaired when DNA synthesis is blocked. Proc. Natl Acad. Sci. USA. 2001;98:1036–1041. doi: 10.1073/pnas.021282898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sengupta S, Linke SP, Pedeux R, Yang Q, Farnsworth J, Garfield SH, Valerie K, Shay JW, Ellis NA, Wasylyk B, et al. BLM helicase-dependent transport of p53 to sites of stalled DNA replication forks modulates homologous recombination. EMBO J. 2003;22:1210–1222. doi: 10.1093/emboj/cdg114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Restle A, Janz C, Wiesmüller L. Differences in the association of p53 phosphorylated on serine 15 and key enzymes of homologous recombination. Oncogene. 2005;24:4380–4387. doi: 10.1038/sj.onc.1208639. [DOI] [PubMed] [Google Scholar]
- 13.Linke SP, Sengupta S, Khabie N, Jeffries BA, Buchhop S, Miska S, Henning W, Pedeux R, Wang XW, Hofseth LJ, et al. p53 interacts with hRAD51 and hRAD54, and directly modulates homologous recombination. Cancer Res. 2003;63:2596–2605. [PubMed] [Google Scholar]
- 14.Bertrand P, Saintigny Y, Lopez BS. P53's double life: transactivation-independent repression of homologous recombination. Trends Genet. 2004;20:235–243. doi: 10.1016/j.tig.2004.04.003. [DOI] [PubMed] [Google Scholar]
- 15.Gatz SA, Wiesmüller L. p53 in recombination and repair. Cell Death Differ. 2006;13:1003–1016. doi: 10.1038/sj.cdd.4401903. [DOI] [PubMed] [Google Scholar]
- 16.Arias-Lopez C, Lazaro-Trueba I, Kerr P, Lord CJ, Dexter T, Iravani M, Ashworth A, Silva A. p53 modulates homologous recombination by transcriptional regulation of the RAD51 gene. EMBO Rep. 2006;7:219–224. doi: 10.1038/sj.embor.7400587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Boehden GS, Baumann C, Siehler S, Wiesmüller L. Wild-type p53 stimulates homologous recombination upon sequence-specific binding to the ribosomal gene cluster repeat. Oncogene. 2005;24:4183–4192. doi: 10.1038/sj.onc.1208592. [DOI] [PubMed] [Google Scholar]
- 18.Boehden GS, Restle A, Marschalek R, Stocking C, Wiesmüller L. Recombination at chromosomal sequences involved in leukaemogenic rearrangements is differentially regulated by p53. Carcinogenesis. 2004;25:1305–1313. doi: 10.1093/carcin/bgh092. [DOI] [PubMed] [Google Scholar]
- 19.Baumann C, Boehden GS, Bürkle A, Wiesmüller L. Poly(ADP-RIBOSE) polymerase-1 (Parp-1) antagonizes topoisomerase I-dependent recombination stimulation by p53. Nucleic Acids Res. 2006;34:1036–1049. doi: 10.1093/nar/gkj509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Subramanian D, Griffith JD. Modulation of p53 binding to Holliday junctions and 3-cytosine bulges by phosphorylation events. Biochemistry. 2005;44:2536–2544. doi: 10.1021/bi048700u. [DOI] [PubMed] [Google Scholar]
- 21.Akyüz N, Boehden GS, Süsse S, Rimek A, Preuss U, Scheidtmann KH, Wiesmüller L. DNA substrate dependence of the p53-mediated regulation of double-strand break repair. Mol. Cell Biol. 2002;22:6306–6317. doi: 10.1128/MCB.22.17.6306-6317.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brummelkamp TR, Bernards R, Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. doi: 10.1126/science.1068999. [DOI] [PubMed] [Google Scholar]
- 23.Müller-Tidow C, Ji P, Diederichs S, Potratz J, Baumer N, Kohler G, Cauvet T, Choudary C, van der Meer T, Chan WY. The cyclin A1-CDK2 complex regulates DNA double-strand break repair. Mol. Cell Biol. 2004;24:8917–8928. doi: 10.1128/MCB.24.20.8917-8928.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lambert S, Lopez BS. Characterization of mammalian RAD51 double strand break repair using non-lethal dominant-negative forms. EMBO J. 2000;19:3090–3099. doi: 10.1093/emboj/19.12.3090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Grandori C, Wu KJ, Fernandez P, Ngouenet C, Grim J, Clurman BE, Moser MJ, Oshima J, Russell DW, Swisshelm K, et al. Werner syndrome protein limits MYC-induced cellular senescence. Genes Dev. 2003;17:1569–1574. doi: 10.1101/gad.1100303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zink D, Mayr C, Janz C, Wiesmüller L. Association of p53 and MSH2 with recombinative repair complexes during S-phase. Oncogene. 2002;21:4788–4800. doi: 10.1038/sj.onc.1205614. [DOI] [PubMed] [Google Scholar]
- 27.Sarkaria JN, Busby EC, Tibbetts RS, Roos P, Taya Y, Karnitz LM, Abraham RT. Inhibition of ATM and ATR kinase activities by the radiosensitizing agent, caffeine. Cancer Res. 1999;59:4375–4382. [PubMed] [Google Scholar]
- 28.Mahdi T, Alcalay D, Cognard C, Tanzer J, Kitzis A. Rescue of K562 cells from MDM2-modulated p53-dependent apoptosis by growth factor-induced differentiation. Biol. Chem. 1998;90:15–27. [PubMed] [Google Scholar]
- 29.Saintigny Y, Rouillard D, Chaput B, Soussi T, Lopez BS. Mutant p53 proteins stimulate spontaneous and radiation-induced intrachromosomal homologous recombination independently of the alteration of the transactivation activity and of the G1 checkpoint. Oncogene. 1999;18:3553–3565. doi: 10.1038/sj.onc.1202941. [DOI] [PubMed] [Google Scholar]
- 30.Won J, Kim M, Kim N, Ahn JH, Lee WG, Kim SS, Chang KY, Yi YW, Kim TK. Small molecule-based reversible reprogramming of cellular lifespan. Nat. Chem. Biol. 2006;2:369–374. doi: 10.1038/nchembio800. [DOI] [PubMed] [Google Scholar]
- 31.Gobert C, Skladanowski A, Larsen AK. The interaction between p53 and DNA topoisomerase I is regulated differently in cells with wild-type and mutant p53. Proc. Natl Acad. Sci. USA. 1999;96:10355–10360. doi: 10.1073/pnas.96.18.10355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boehden GS, Akyüz N, Roemer K, Wiesmüller L. p53 mutated in the transactivation domain retains regulatory functions in homology-directed double-strand break repair. Oncogene. 2003;22:4111–4117. doi: 10.1038/sj.onc.1206632. [DOI] [PubMed] [Google Scholar]
- 33.Wesierska-Gadek J, Gueorguieva M, Wojciechowski J, Horky M. Cell cycle arrest induced in human breast cabcer celly by cyclin-dependent kinase inhibitors: a comparison of the effects exerted by roscovitine and olomoucine. Pol. J. Pharmacol. 2004;56:635–641. [PubMed] [Google Scholar]
- 34.Rothkamm K, Krüger I, Thompson LH, Löbrich M. Pathways of DNA double-strand break repair during the mammalian cell cycle. Mol. Cell Biol. 2003;23:5706–5715. doi: 10.1128/MCB.23.16.5706-5715.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fogal V, Hsieh JK, Royer C, Zhong S, Lu X. Cell cycle-dependent nuclear retention of p53 by E2F1 requires phosphorylation of p53 at Ser315. EMBO J. 2005;24:2768–2782. doi: 10.1038/sj.emboj.7600735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang R, Muller C, Huynh V, Fung YK, Yee AS, Koeffler HP. Functions of cyclin A1 in the cell cycle and its interactions with transcription factor E2F-1 and the Rb family of proteins. Mol. Cell Biol. 1999;19:2400–2407. doi: 10.1128/mcb.19.3.2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blander G, Kipnis J, Leal JF, Yu CE, Schellenberg GD, Oren M. Physical and functional interaction between p53 and the Werner's syndrome protein. J. Biol. Chem. 1999;274:29463–29469. doi: 10.1074/jbc.274.41.29463. [DOI] [PubMed] [Google Scholar]
- 38.Lebel M, Spillare EA, Harris CC, Leder P. The Werner syndrome gene product co-purifies with the DNA replication complex and interacts with PCNA and topoisomerase I. J. Biol. Chem. 1999;274:37795–37799. doi: 10.1074/jbc.274.53.37795. [DOI] [PubMed] [Google Scholar]
- 39.Laine JP, Opresko PL, Indig FE, Harrigan JA, von Kobbe C, Bohr VA. Werner protein stimulates topoisomerase I DNA relaxation activity. Cancer Res. 2003;63:7136–7146. [PubMed] [Google Scholar]
- 40.Song H, Hollstein M, Xu Y. p53 gain-of-function cancer mutants induce genetic instability by inactivating ATM. Nat. Cell Biol. 2007;9:573–580. doi: 10.1038/ncb1571. [DOI] [PubMed] [Google Scholar]
- 41.El-Hizawi S, Lgowski JP, Kulesz-Martin M, Albor A. Induction of gene amplification as a gain-of-function phenotype of mutant p53 proteins. Cancer Res. 2002;62:3264–3270. [PubMed] [Google Scholar]
- 42.Perkins EJ, Nair A, Cowley DO, Van Dyke T, Chang Y, Ramsden DA. Sensing of intermediates in V(D)J recombination by ATM. Genes Dev. 2002;16:159–164. doi: 10.1101/gad.956902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Romanova LY, Willers H, Blagosklonny MV, Powell SN. The interaction of p53 with replication protein A mediates suppression of homologous recombination. Oncogene. 2004;23:9025–9033. doi: 10.1038/sj.onc.1207982. [DOI] [PubMed] [Google Scholar]
- 44.Sengupta S, Robles AI, Linke SP, Sinogeeva NI, Zhang R, Pedeux R, Ward IM, Celeste A, Nussenzweig A, Chen J, et al. Functional interaction between BLM helicase and 53BP1 in a Chk1-mediated pathway during S-phase arrest. J. Cell Biol. 2004;166:801–813. doi: 10.1083/jcb.200405128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- 46.Luciani MG, Hutchins JRA, Zheleva D, Hupp TR. The C-terminal regulatory domain of p53 contains a functional docking site for cyclin A. J. Mol. Biol. 2000;300:503–518. doi: 10.1006/jmbi.2000.3830. [DOI] [PubMed] [Google Scholar]
- 47.Jazayeri A, Falck J, Lukas C, Bartek J, Smith GC, Lukas J, Jackson SP. ATM- and cell cycle-dependent regulation of ATR in response to DNA double-strand breaks. Nat. Cell Biol. 2006;8:37–45. doi: 10.1038/ncb1337. [DOI] [PubMed] [Google Scholar]
- 48.Sorensen CS, Hansen LT, Dziegielewski J, Syljuasen RG, Lundin C, Bartek J, Helleday T. The cell-cycle checkpoint kinase Chk1 is required for mammalian homologous recombination repair. Nat. Cell Biol. 2005;7:195–201. doi: 10.1038/ncb1212. [DOI] [PubMed] [Google Scholar]
- 49.Larsen AK, Gobert C. DNA topoisomerase I in oncology: Dr Jekyll or Mr Hyde? Pathol. Oncol. Res. 1999;5:171–178. doi: 10.1053/paor.1999.0209. [DOI] [PubMed] [Google Scholar]
- 50.Champoux JJ. Strand breakage by the DNA untwisting enzyme results in covalent attachment of the enzyme to DNA. Proc. Natl Acad. Sci. USA. 1977;74:3800–3804. doi: 10.1073/pnas.74.9.3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sekiguchi J, Seeman NC, Shuman S. Resolution of Holliday junctions by eukaryotic DNA topoisomerase I. Proc. Natl Acad. Sci. USA. 1996;93:785–789. doi: 10.1073/pnas.93.2.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liao S, Mao C, Birktoft JJ, Shuman S, Seeman NC. Resolution of undistorted symmetric immobile DNA junctions by vaccinia topoisomerase I. Biochemistry. 2004;43:1520–1531. doi: 10.1021/bi0358061. [DOI] [PubMed] [Google Scholar]
- 53.Gobert C, Bracco LB, Rossi F, Olivier M, Tazi J, Lavelle F, Larsen AKI, Riou JF. Modulation of DNA topoisomerase I activity by p53. Biochemistry. 1996;35:5778–5786. doi: 10.1021/bi952327w. [DOI] [PubMed] [Google Scholar]
- 54.Albor A, Kaku S, Kulesz-Martin M. Wildtype and mutant forms of p53 activate human topoisomerase I: a possible mechanism for gain of function in mutants. Cancer Res. 1998;5:2091–2094. [PubMed] [Google Scholar]
- 55.Stephan H, Grosse F, Soe K. Human topoisomerase I cleavage complexes are repaired by a p53-stimulated recombination-like reaction in vitro. Nucleic Acids Res. 2002;30:5087–5093. doi: 10.1093/nar/gkf659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Roemer K. Mutant p53: gain-of-function oncoproteins and wild-type p53 inactivators. Biol. Chem. 1999;380:879–887. doi: 10.1515/BC.1999.108. [DOI] [PubMed] [Google Scholar]
- 57.Liu PK, Kraus E, Wu TA, Strong LC, Tainsky MA. Analysis of genomic instability in Li-Fraumeni fibroblasts with germline p53 mutations. Oncogene. 1996;12:2267–2278. [PMC free article] [PubMed] [Google Scholar]
- 58.Murphy KL, Rosen JM. Mutant p53 and genomic instability in a transgenic mouse model of breast cancer. Oncogene. 2000;19:1045–1051. doi: 10.1038/sj.onc.1203274. [DOI] [PubMed] [Google Scholar]
- 59.Müller C, Yang R, Idos G, Tidow N, Diederichs S, Koch OM, Verbeek W, Bender TP, Koeffler HP. c-myb transactivates the human cyclin A1 promoter and induces cyclin A1 gene expression. Blood. 1999;94:4255–4262. [PubMed] [Google Scholar]
- 60.Müller C, Yang R, Park DJ, Serve H, Berdel WE, Koeffler HP. The aberrant fusion proteins PML-RAR alpha and PLZF-RAR alpha contribute to the overexpression of cyclin A1 in acute promyelocytic leukemia. Blood. 2000;96:3894–3899. [PubMed] [Google Scholar]
- 61.Coletta RD, Christensen K, Reichenberger KJ, Lamb J, Micomonaco D, Huang L, Wolf DM, Müller-Tidow C, Golub TR, Kawakami K, et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc. Natl Acad. Sci. USA. 2004;101:6478–6483. doi: 10.1073/pnas.0401139101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Müller-Tidow C, Diederichs S, Schrader MG, Vogt U, Miller K, Berdel WE, Serve H. Cyclin A1 is highly expressed in aggressive testicular germ cell tumors. Cancer Lett. 2003;190:89–95. doi: 10.1016/s0304-3835(02)00582-7. [DOI] [PubMed] [Google Scholar]
- 63.Yang R, Nakamaki T, Lubbert M, Said J, Sakashita A, Freyaldenhoven BS, Spira S, Huynh V, Muller C, Koeffler HP. Cyclin A1 expression in leukemia and normal hematopoietic cells. Blood. 1999;93:2067–2074. [PubMed] [Google Scholar]
- 64.Liao C, Wang XY, Wei HQ, Li SQ, Merghoub T, Pandolfi PP, Wolgemuth DJ. Altered myelopoiesis and the development of acute myeloid leukemia in transgenic mice overexpressing cyclin A1. Proc. Natl Acad. Sci. USA. 2001;98:6853–6858. doi: 10.1073/pnas.121540098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tokumaru Y, Yamashita K, Osada M, Nomoto S, Sun DI, Xiao Y, Hoque MO, Westra WH, Califano JA, Sidransky D. Inverse correlation between cyclin A1 hypermethylation and p53 mutation in head and neck cancer identified by reversal of epigenetic silencing. Cancer Res. 2004;64:5982–5987. doi: 10.1158/0008-5472.CAN-04-0993. [DOI] [PubMed] [Google Scholar]
- 66.Bartkova J, Horejsi Z, Koed K, Kramer A, Tort F, Zieger K, Guldberg P, Sehested M, Nesland JM, Lukas C, et al. DNA damage response as a candidate anti-cancer barrier in early human tumorigenesis. Nature. 2005;434:864–870. doi: 10.1038/nature03482. [DOI] [PubMed] [Google Scholar]
- 67.Gorgoulis VG, Vassiliou LV, Karakaidos P, Zacharatos P, Kotsinas A, Liloglou T, Venere M, Ditullio RA, Kastrinakis NG, Levy B, et al. Activation of the DNA damage checkpoint and genomic instability in human precancerous lesions. Nature. 2005;434:907–913. doi: 10.1038/nature03485. [DOI] [PubMed] [Google Scholar]
- 68.Armata HL, Garlick DS, Sluss HK. The ataxia telangiectasia-mutated target site ser18 is required for p53-mediated tumor suppression. Cancer Res. 2007;67:11696–11703. doi: 10.1158/0008-5472.CAN-07-1610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.