Abstract
Evasion from apoptotic cell death is a characteristic of cancer; genes that modulate this process may be optimal for therapeutic attack. Identifying key regulators of apoptosis is thus a central goal in cancer therapy. Here, we describe a loss-of-function screen that uses RNA interference libraries to identify genes required for induction of apoptosis. We used a short-hairpin RNA expressing vector with high gene-expression silencing activity that contained fetal brain cDNAs. Survived cells from genotoxic stress were isolated to determine knock-down of molecules that are crucial for induction of apoptosis. We identified TBP-associated factor 1 (TAF1), a gene previously implicated as an essential component of transcription machinery. Depletion of TAF1 was associated with substantial attenuation of apoptosis induced by oxidative as well as genotoxic stress. Microarray analysis further demonstrated that a number of genes were transcriptionally declined in cells silenced for TAF1. Surprisingly, knocking down TAF1 exhibited a marked decrease in p27Kip1 expression, allowing cells resistant from oxidative stress-induced apoptosis. These results suggest that TAF1 regulates apoptosis by controlling p27Kip1 expression. Our system provides a novel approach to identifying candidate genes that modulate apoptosis.
INTRODUCTION
Gene silencing by RNA interference (RNAi) has developed a powerful tool for loss-of-function studies (1). Large-scale RNAi has facilitated the search for genes required for diverse biological processes enabling stepwise dissection of specific signaling pathways. Indeed, in combination with high-throughput assays, genome-wide RNAi studies have uncovered novel gene functions in various biological processes (2). Several of these studies were aimed at the identification of genes essential for cell division, cell cycle progression, endocytosis, tumor transformation and apoptosis (3–10). Through these studies, the power of this approach for the identification of functional modules has been demonstrated.
Regulation of apoptosis is critical in many fundamental cellular processes. Because defective regulation of apoptosis provokes human disease, particularly cancers (11–13), a global survey of genes essential for apoptosis in human cells is thus not only advance the understanding of a fundamental biological process but also delivers novel diagnostic and therapeutic targets for cancer. In particular, the signals that induce apoptosis in response to genotoxic stress are largely unknown.
In this study, we used an improved short-hairpin RNA (shRNA) library with genome-wide coverage (14). We performed transfection of the shRNA library and high-throughput cell-survival analysis to detect cells that escaped from death, which were further analyzed by TUNEL assays. We identified TBP-associated factor 1 (TAF1), an essential component of transcription machinery. Microarray and apoptosis analyses demonstrated that TAF1-mediated p27Kip1 expression is involved in the induction of apoptosis in response to genotoxic stress. Using this approach, we uncovered a new gene implicated in the apoptosis process, including transcriptional regulatory networks that govern cell death in mammalian cells.
MATERIALS AND METHODS
Cell culture
Human 293T embryonal kidney cells, HeLa cervical cancer cells and MCF-7 breast cancer cells were cultured in Dulbecco's modified Eagle medium containing 10% heat-inactivated fetal bovine serum, 100 U/ml penicillin, 100 U/ml streptomycin and 2 mM l-glutamine. U2OS osteosarcoma cells were cultured in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum and antibiotics.
Cell transfection
Plasmid DNA was transfected by using FuGENE 6 transfection reagent (Roche, Basel, Switzerland). TAF1, p27Kip1 and caspase-3 gene-specific siRNAs were purchased by Invitrogen, Carlsbad, CA, USA (Stealth RNAi). Transfection of siRNAs was performed using Lipofectamine RNAi MAX (Invitrogen).
Construction of shRNA expression library
A shRNA library was constructed as previously described (14). The library was generated from human fetal brain cDNAs. DNA fragments in the library theoretically cover a genome-wide transcriptome.
Construction of plasmids
TAF1 cDNA was amplified by PCR using the PfuUltra™ High-Fidelity DNA polymerase (Stratagene, La Jolla, CA, USA) from pBlueBacHis250 and cloned into the pEGFP-C1 vectors. p27Kip1 cDNA was amplified by PCR from 293T cDNA and cloned into pcDNA3-Flag (15,16).
Measurement of cell viability
Cells were cultured in 96-well plates and individually transfected with shRNA expression vector. After treatment with H2O2 or etoposide for 24 h, MTS assays were performed by adding 20 µl CellTiter 96 AQueous One Solution Reagent (Promega, Madison, WI, USA) directly into 80 µl culture media. After incubation for 3 h at 37°C in a humidified 5% CO2 atmosphere, the absorbance was measured at 490 nm with the use of a multilabel counter (PerkinElmer, Waltham, MA, USA).
RT–PCR analysis
Total RNA was isolated from cells using RNeasy kit (Qiagen, Hilden, Germany). First strand cDNA synthesis and following PCR were performed with 500 ng of total RNA using a SuperScript III One-Step RT–PCR System with Platinum Taq DNA Polymerase (Invitrogen) according to the manufacturer's protocol. For TAF1 gene expression, the nucleotide sequence of 5′-GGTATGATATGCTGGGTGTC-3′ was used as the sense primer, and 5′-CAAGAGTGGCTGCAAAACCT-3′ was used as the antisense primer. For GAPDH gene expression, the nucleotide sequence of 5′-AAGGCTGTGGGCAAGGTCATCCCT-3′ was used as the sense primer, and 5′-TTACTCCTTGGAGGCCATGTGGGC-3′ was used as the antisense primer. The reaction products were separated on 2% agarose gels.
Immunoblot analysis
Immunoblot analysis was performed as described elsewhere (17–19). Briefly, cells were suspended with the lysis buffer (50 mM Tris–HCl, pH 7.6, 150 mM NaCl, 1 mM Na3VO4, 1 mM PMSF, 1 mM DTT, 10 µg/ml aprotinin, 1 µg/ml leupeptin, 10 mM NaF, 1 µg/ml Pepstatin A, 0.05% deoxycholic acid and 1% NP-40). Lysates were centrifuged at 15 000g for 5 min at 4°C, and the supernatants were separated by SDS–PAGE and transferred to nitrocellulose membranes. The membranes were incubated with anti-Flag (Sigma-Aldrich, St. Louis, MO, USA), anti-TAF1 (Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-p27Kip1 (Santa Cruz Biotechnology), anti-Notch2 (Developmental Studies Hybridoma Bank, Iowa City, IA, USA), antitubulin (Sigma-Aldrich) or anti-PCNA (Santa Cruz Biotechnology). Immune complexes were incubated with secondary antibodies and visualized by chemiluminescence (PerkinElmer).
TUNEL assays
Cells cultured in poly-d-lysine-coated 4-well chamber slides were transfected with plasmids or siRNAs and then treated with H2O2 for 24 h. Apoptotic cells were detected by TUNEL assays using a DeadEnd Fluorometric TUNEL System (Promega). To detect apoptotic cells expressing GFP-TAF1, the FluoroLink™ Cy5-dUTP (GE Healthcare, Buckinghamshire, England) was used instead of Fluorescein-12-dUTP.
Microarray analysis
Total RNA was isolated from cells using an RNeasy kit (Qiagen). Total RNA (5 µg) was used to start the protocol of One-Cycle cDNA Synthesis and to label cRNA, following the manufacturer's protocol (Affymetrix, Santa Clara, CA, USA). Before making a cocktail solution, 20 µg of biotin-labeled cRNA was fragmented to 35–200 bases, and 15 µg of cRNA fragment was used to prepare the cocktail solution. The solution was applied into a GeneChip Human Genome U133 plus 2.0 array (Affymetrix) and hybridized for 16 h at 45°C. After hybridization, the arrays were washed and stained using Fluidic station 450 according to protocol EukGE-WS2v5_450 and were then scanned using the Affymetrix GeneChip Scanner 3000. Analysis of the data was performed as previously described (20).
Quantitative real-time RT–PCR analysis
Total RNA (5 µg) was reverse transcribed into cDNA using a Superscript III First-Strand Synthesis System for RT–PCR (Invitrogen), following the manufacturer's protocol. For TAF1 gene expression, the nucleotide sequence of 5′-CAACACAAACCAGTGACCAGAG-3′ was used as the sense primer, and 5′-CCAGAATGCCTTAGCTTCCA-3′ was used as the antisense primer. For Notch2 gene expression, the nucleotide sequence of 5′-TCCTCTTCTGCCTGCCTTTG-3′ was used as the sense primer, and 5′-TACCTTTCCCTTCCCCACCT-3′ was used as the antisense primer. For p27Kip1 gene expression, the nucleotide sequence of 5′-TGGCATGTTTTGTGCATTTG-3′ was used as the sense primer, and 5′-TTGGCTCAGTATGCAACCTTTT-3′ was used as the antisense primer. For Actin gene expression, the nucleotide sequence of 5′-GTGGCCGAGGACTTTGATTG-3′ was used as the sense primer, and 5′-TGGACTTGGGAGAGGACTGG-3′ was used as the antisense primer. The PCR reaction was performed by Power SYBR Green Master Mix (15 µl Power SYBR Green Master Mix, 0.3 µl each of 5 µM primer, 5 µl cDNA, 9.4 µl water) using Power SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA). The PCR program was as follows; after incubation for 10 min at 95°C, denaturation for 15 s at 95°C, annealing for 60 s at 60°C and extension for 30 s at 72°C. Cumulative fluorescence was measured at the end of the extension phase of each cycle. Quantification was based on standard curves from serial dilution of 293T total RNA. The results were normalized for the level of actin.
RESULTS
shRNA screen for regulators of apoptosis in response to genotoxic stress
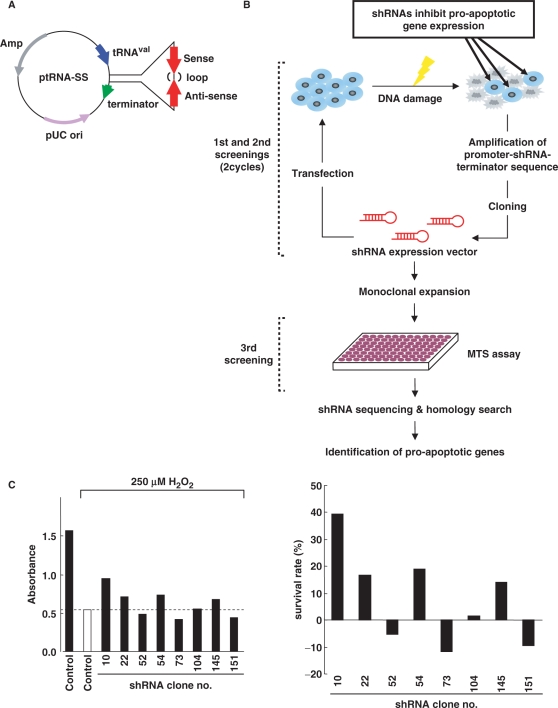
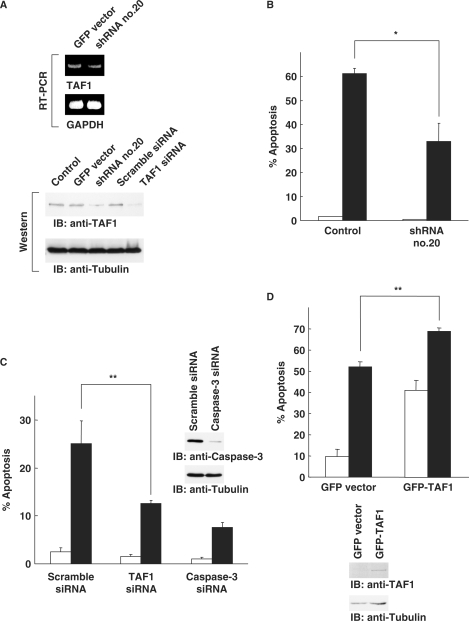
In order to identify regulators of apoptosis by a genetic screen, we have developed shRNAs that can theoretically target a genome-wide trasncriptome (Figure 1A). The shRNA expression library was constructed as described elsewhere (14). Cells were transfected with the shRNA library, then treated with H2O2 or etoposide for 24 h. Adherent cells, judged as survived, were harvested to extract total cellular DNA (Figure 1B). The cassettes containing promoter-hairpin-terminator were amplified by PCR then ligated into the pUC19. After repeating this cycle, selected shRNAs were individually transfected into cells in 96-well plates, followed by treatment with H2O2 or etoposide for 24 h. Viability of cells was monitored by MTS assays. We have evaluated >1000 shRNA clones with respect to escape from cell death in response to genotoxic stress. When the absorbance obtained from cells transfected with GFP vector was determined as a control, 75 clones, showing higher absorbance than control, were selected in this screening step (Figure 1C). After sequencing the short-hairpin region, putative target genes for each shRNA were determined by homology search. In this step, 30 clones were annotated as putative targets for at least one gene (Table 1). To determine if each shRNAs are capable for knock-down of target genes, cells were transfected with each of the shRNAs in 96-well plates. Analysis of mRNA expression by RT–PCR revealed that 8 of the 30 clones were associated with substantial attenuation of putative target genes. To further assess the effects on apoptosis by knock-down with shRNAs, cells were transfected with each shRNA, followed by treatment with H2O2 or etoposide for 24 h. Apoptotic cells were monitored by TUNEL assays. The results demonstrated that transfection of cells with clone No. 20, which targets TAF1 (Figure 2A), substantially attenuated H2O2-induced apoptosis (Figure 2B). To confirm these results, we prepared siRNAs that target TAF1 (Figure 2A and Supplementary Figure 1A and B). 293T cells were transfected with TAF1 siRNAs, followed by treatment with H2O2. As a control, the effect of transfection with scramble siRNA or caspase-3 siRNA was also evaluated. Knocking down TAF1 significantly attenuated H2O2-induced apoptosis to a level similar to that obtained with caspase-3 siRNA (Figure 2C and Supplementary Figure1C). Similar results were obtained in HeLa, MCF-7 and U2OS cells (Supplementary Figure 2). Comparable results were also obtained with other stimuli, such as treatment with adriamycin (Supplementary Figure 3). To establish whether TAF1 induces apoptosis in response to genotoxic stress, U2OS cells were transfected with GFP vector or GFP-TAF1. Upon exposure to H2O2, the ratio of apoptosis increased in control cells (Figure 2D). More importantly, increased apoptosis was significantly augmented by ectopic expression of TAF1 (Figure 2D). These results collectively demonstrated that TAF1 is a key regulator of apoptosis in response to genotoxic stress.
Figure 1.
shRNA library screen for pro-apoptotic genes. (A) Construction of shRNA expression library. The ptRNA-SS vector contains a tRNAval promoter and terminator in the pUC19 backbone, including ampicillin-resistance markers. The shRNA consists of a target transcript-specific 25–35 base double-stranded stem, connected by a 6-base loop sequence. (B) A schematic diagram of screening for identifying pro-apoptotic genes. 293T, HeLa or MCF-7 cells transfected with shRNA expression libraries (8 µg) were treated with H2O2 (200 µM for 293T and HeLa cells, and 500 µM for MCF-7 cells) or etoposide (100 µM for 293T cells, and 50 µM for HeLa cells) for 24 h. Adherent cells, judged as survived, were harvested, and total genomic DNA were isolated. The promoter-shRNA-terminator region was amplified from total genomic DNA. PCR products were digested by SphI and EcoRI, then ligated into pUC19. This process was repeated, and then the selected shRNAs were monoclonally expanded. Cells were plated on 96-well dishes and transfected with each of the shRNA clones, followed by treatment with H2O2 or etoposide for 24 h. MTS assays were performed to detect surviving cells. Sequencing of shRNA regions was carried out to identify putative target genes by homology search with BLAST. (C) MCF-7 cells transfected with individual shRNA clones were treated with 250 µM H2O2 for 24 h. MTS assays were performed to detect cells escaped from apoptosis. Absorbance obtained from cells transfected with pEGFP-C1 vector was determined as a control (left panel; open bar). Cells showing higher absorbance than the control value were selected as positive clones. A closed bar represents the value for untreated cells transfected with the empty vector (left panel). Survival rates were defined by calculating the absorbance obtained from the control cells with or without treatment, as 0 or 100%, respectively (right panel).
Table 1.
Putative apoptosis-related genes identified by the RNAi screen
| Cell line | Reagent | Clone no. | Gene | mRNA knockdown |
|---|---|---|---|---|
| HEK-293T | H2O2 | 20 | TAF1 RNA polymerase II, TATA box-binding protein (TBP)-associated factor, 250kDa | +++ |
| 101 | Ribosomal protein S27a | +++ | ||
| Etoposide | 13 | Procollagen C-endopeptidase enhancer 2 | ± | |
| 32 | McKusick-Kaufman syndrome | – | ||
| 82 | Signal transducing adaptor molecule (SH3 domain and ITAM motif) 1 | – | ||
| 115 | Protein-l-isoaspartate (d-aspartate) O-methyltransferase | – | ||
| 149 | TAR DNA-binding protein | +++ | ||
| HeLa S3 | H2O2 | 8 | N-acetylglucosamine-1-phosphodiester alpha-N-acetylglucosaminidase | – |
| 101 | Reticulon 4 | – | ||
| 174 | Metallothionein 1F (functional) | – | ||
| Etoposide | 21 | Splicing factor, arginine/serine-rich 3 | – | |
| 22 | Vacuolar protein sorting 33 homolog A (Saccharomyces cerevisiae) | – | ||
| Leukocyte-associated immunoglobulin-like receptor 2 | ± | |||
| 25 | Pituitary tumor-transforming 1 interacting protein | – | ||
| Surface glycoprotein | ND | |||
| 67 | Calmodulin 2 (phosphorylase kinase, delta) | – | ||
| 72 | Protein tyrosine phosphatase, nonreceptor type 11 (Noonan syndrome 1) | ++ | ||
| 77 | Solute carrier family 4, sodium bicarbonate transporter-like, member 10 | ND | ||
| Eukaryotic translation elongation factor 1 alpha 1 | – | |||
| 101 | Heat shock protein 90 | – | ||
| 102 | Peroxiredoxin 1 | – | ||
| 108 | Tumor necrosis factor (ligand) superfamily, member 14 | – | ||
| 171 | DC31 | – | ||
| MCF-7 | H2O2 | 3 | Aldehyde dehydrogenase 3 family, member B1 | – |
| 10 | DnaJ (Hsp40) homolog, subfamily B, member 11 | ++ | ||
| 22 | Craniofacial development protein 1 | +++ | ||
| 51 | Solute carrier family 11 (proton-coupled divalent metal ion transporters), member 1 | ++ | ||
| 54 | Galactose-3-O-sulfotransferase 4 variant protein | ± | ||
| 104 | Autosomal highly conserved protein | – | ||
| 145 | Histone deacetylase 2 | – | ||
| Fibroblast growth factor receptor 1 (fms-related tyrosine kinase 2, Pfeiffer syndrome) | +++ | |||
| PRP4 pre-mRNA processing factor 4 homolog (yeast) | ND | |||
| Polyribonucleotide nucleotidyltransferase 1 | ND | |||
| Caspase 6, apoptosis-related cysteine peptidase | + | |||
| 151 | Mesoderm development candidate 2 | ND | ||
| Cathepsin B | + | |||
| Ring finger protein 125 | + | |||
| Nicotinamide nucleotide adenylyltransferase 1 | ND | |||
| POU domain transcription factor Oct-3 alternative transcript 6.3 (POU5F1) pseudogene mRNA | ND | |||
| 185 | Activated RNA polymerase II transcription cofactor 4 [SUB1 homolog (S. cerevisiae)] | ± | ||
| 199 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, 4, 9kDa (NDUFA4), nuclear gene-encoding mitochondrial protein | – |
Figure 2.
Identification of a target gene responsible for induction of apoptosis by genotoxic stress. (A) The identified shRNA (No. 20) suppresses TAF1 gene. 293T cells were transfected with pEGFP-C1 vector or the shRNA clone No. 20. Total RNA was subjected to RT–PCR analysis using primer sets for TAF1 or GAPDH. 293T cells were also transfected with scramble siRNA or siRNA targeting TAF1. Cell lysates were subjected to immunoblot analysis with anti-TAF1 or antitubulin. (B) TAF1 gene-specific shRNA or siRNA inhibits induction of apoptosis by genotoxic stress. 293T cells transfected with pUC19 vector or the shRNA clone No. 20 were left untreated (open bar) or treated (closed bar) with H2O2 for 24 h. The percentage of apoptotic cells was determined by TUNEL assays. The data represent the mean ± SD from three independent experiments, each performed in triplicate. An asterisk indicates P < 0.05. (C) 293T cells transfected with scramble siRNA, TAF1 siRNA or caspase-3 siRNA were left untreated (open bar) or treated (closed bar) with H2O2 for 24 h. The percentage of apoptotic cells was analyzed as described earlier. The data represent the means ± SD from three independent experiments, each performed in triplicate. Two asterisks indicate P < 0.01. Cell lysates were subjected to immunoblot analysis with anti-caspase-3 or antitubulin. (D) Ectopic expression of TAF1 induces apoptosis. U2OS cells were transfected with pEGFP-C1 vector or GFP-TAF1 and untreated (open bar) or treated (closed bar) with H2O2 for 24 h. The percentage of apoptotic cells was determined by TUNEL assays. The data indicate the means ± SD from three independent experiments, each performed in triplicate. Two asterisks indicate P < 0.01. Cell lysates were subjected to immunoblot analysis with anti-GFP or antitubulin.
TAF1 controls numerous gene expressions including apoptosis- and cell cycle-associated genes
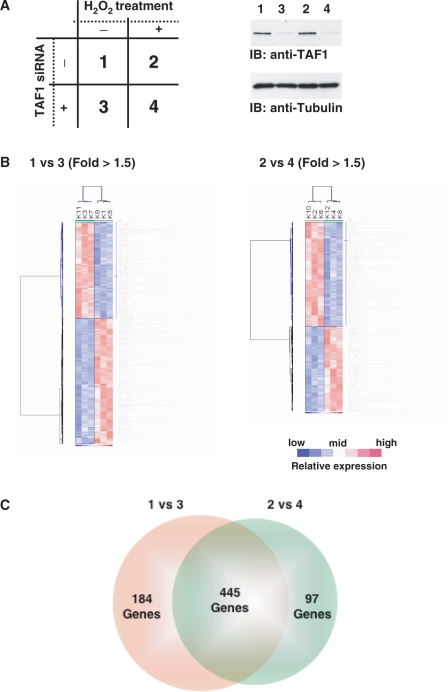
To explore potential molecular mechanisms by which TAF1 regulates induction of apoptosis, we examined TAF1-mediated transcriptional control by microarray analysis. 293T cells were transfected with scramble siRNA or TAF1 siRNA, then left untreated or treated with H2O2 (Figure 3A). Total RNA was extracted and mRNA expression was analyzed with a GeneChip system. Knocking down TAF1 affected a number of gene expressions in both cells left untreated (1 versus 3; 629 genes) and those treated with H2O2 (2 versus 4; 542 genes) (Figure 3B). The number of merged genes from both categories was 445 (Figure 3C). In contrast, treatment with H2O2 had little if any effect on gene expression, at least at the transcriptional level (1 versus 2; 35 genes, 3 versus 4; 32 genes). These results indicate that TAF1 modulates a number of genes at the transcriptional level, regardless of oxidative stress.
Figure 3.
GeneChip analysis for responsive gene modulated by transcription factor TAF1. (A) Experimental conditions of the GeneChip analysis. 1: Cells transfected with scramble siRNA; 2: cells transfected with scramble siRNA and treated with H2O2; 3: cells transfected with TAF1 siRNA; 4: cells transfected with TAF1 siRNA and treated with H2O2. Cell lysates were subjected to immunoblot analysis with indicated antibodies. (B) Hierarchical clustering depicting expression profiles of differentially regulated genes by depletion of TAF1 gene. The data indicate the means from three independent experiments, each performed in triplicate. 1 versus 3, the value for the expression level of condition 1 divided by that of condition 3; 2 versus 4, the value for the expression level of condition 2 divided by that of condition 4. Up- and down-regulated genes are depicted in red and blue, respectively. (C) The number of GeneChip probes more than 1.5-fold up- or down-regulated by knocking down TAF1 is indicated.
TAF1-mediated regulation of p27Kip1 is associated with induction of apoptosis
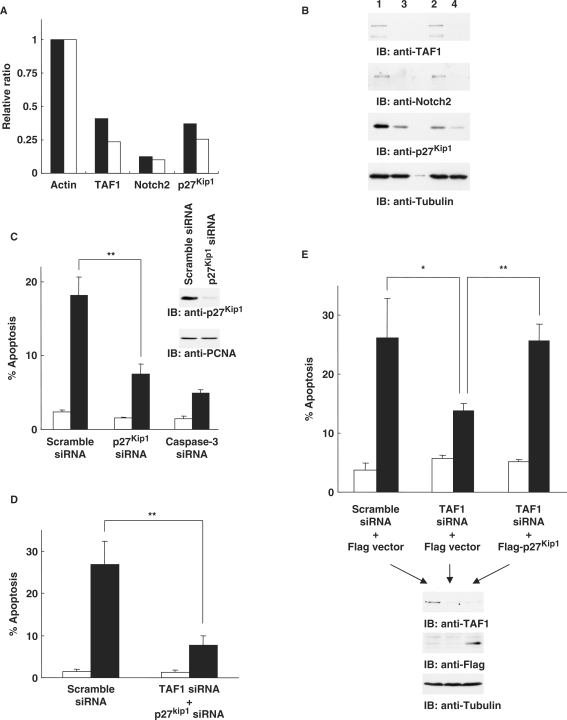
On the basis of the gene clusters obtained from expression profiles (Figure 3B), we assessed genes categorized as ‘apoptosis’ and ‘cell cycle’ in terms of gene ontology. In particular, we focused on p27Kip1 and Notch2 since the expressions of these genes were markedly reduced in cells silenced for TAF1 (data not shown). To confirm the results of microarray analysis, gene expression was explored by real-time RT–PCR. As shown for expression profiles by microarray, knock-down of TAF1 in 293T cells was associated with substantial attenuation of Notch2 and p27Kip1 (Figure 4A). With regard to p27Kip1 expression, similar results were obtained in various cells, such as MCF-7 and HeLa cells (Supplementary Figure 4A). To further examine the expressions of these genes at the protein level, TAF1-depleted 293T cells were analyzed by immunoblotting. As expected, expression levels of Notch2 and p27Kip1 decreased in cells transfected with TAF1 siRNA (Figure 4B), indicating that knock-down of TAF1 attenuates both Notch2 and p27Kip1 at the transcriptional and subsequent protein levels. With regard to p27Kip1 expression, similar findings were obtained in MCF-7 and HeLa cells (Supplementary Figure 4B). To determine if attenuation of Notch2 or p27Kip1 expression affects the induction of apoptosis, cells were transfected with scramble siRNA, Notch2 siRNA, p27Kip1 siRNA or caspase-3 siRNA, followed by treatment with H2O2. Assessment of apoptosis by TUNEL assays revealed that p27Kip1-depleted cells were substantially resistant to H2O2-induced cell death, similar to caspase-3-depleted cells (Figure 4C). In contrast, there was no significant effect on cells silenced for Notch2 (data not shown), suggesting that p27Kip1 is involved in apoptosis induced by oxidative stress. To exclude the possibility that the apoptotic effect with p27Kip1 is due to its mRNA stability, 293T cells were transfected with scramble siRNA or TAF1 siRNA then treated with transcription inhibitor actinomycin D. Analysis of real-time RT–PCR for p27Kip1 expression revealed that knocking down TAF1 is no effect on decrease of p27Kip1 mRNA stability after the inhibition of transcription (Supplementary Figure 5). These findings support the involvement of p27Kip1 expression on apoptosis induction at the level of transcription rather than mRNA stability. Interestingly, p27Kip1 expression at the protein levels slightly decreased after H2O2 stimulation (Figure 4B). In this context, previous studies have shown that p27Kip1 expression is down-regulated after H2O2 exposure whereas the mechanism is uncertain (21). To define whether this is controlled at the transcriptional level or the posttranslational level, we have explored real-time RT–PCR analysis. The results demonstrated that there is no significant difference on p27Kip1 mRNA between the presence and the absence of H2O2 (Supplementary Figure 6A; 1 versus 2 and 3 versus 4). This strongly suggests that the expression level of p27Kip1 mRNA remains unchanged even after exposure to H2O2. Thus, the expression of p27Kip1 should be controlled at the posttranslational level. Indeed, it is well known that p27Kip1 is degraded by the ubiquitin–proteasome pathway (22,23). To determine this is the case, 293T cells were pretreated with the proteasome inhibitor MG-132 followed by treatment with H2O2. As shown previously, p27Kip1 expression slightly decreased upon exposure to H2O2. In contrast, pretreatment with MG-132 abrogated oxidative stress-induced down-regulation of p27Kip1 (Supplementary Figure 6B). Taken together, these results demonstrate that p27Kip1 is partially degraded after H2O2 exposure by the ubiquitin–proteasome machinery. Importantly, knocking down TAF1 significantly attenuated p27Kip1 expression at the transcriptional level (Figure 4A and Supplementary Figure 6A). In this regard, it is conceivable that substantial transcriptional suppression, but not partial posttranslational suppression, of p27Kip1 is necessary for attenuation of apoptosis. To examine the synergic effect of TAF1 and p27Kip1 on apoptosis, 293T cells were transfected with TAF1 and p27Kip1 siRNAs together followed by H2O2 treatment. Analysis of TUNEL assays indicated that silencing of both TAF1 and p27Kip1 confers more pronounced effect on attenuation of apoptosis as compared to that of each knocking down alone (Figure 4D). To further determine if induction of apoptosis by TAF1 is mediated via p27Kip1 expression, cells were transfected with TAF1 siRNA, followed by forced expression of Flag vector or Flag-p27Kip1. After stimulation of cells with H2O2, we performed TUNEL assays to assess apoptotic cell death. As shown previously, depletion of TAF1 was associated with a marked attenuation of apoptosis (Figure 4E). In contrast, forced expression of p27Kip1 in TAF1-depleted cells increased H2O2-induced apoptosis (Figure 4E). These results demonstrated that p27Kip1 is, at least in part, responsible for TAF1-induced cell death in response to genotoxic stress.
Figure 4.
TAF1 controls oxidative stress-induced apoptosis by regulating p27Kip1 expression. (A) Quantitation of TAF1, Notch2 and p27Kip1 mRNAs by real-time RT–PCR analysis. Closed bars represent the value for the expression level of condition 3 (Figure 3A) divided by that of condition 1; open bars represent the value for the expression level of condition 4 divided by that of condition 2. The value is normalized by the expression level of actin. (B) 293T cells transfected with scramble siRNA or the TAF1 siRNA were left untreated or treated with H2O2 for 24 h. Cell lysates were subjected to immunoblot analysis with anti-TAF1, anti-Notch2, anti-p27Kip1 or antitubulin. (C) p27Kip1 gene-specific siRNA attenuates induction of apoptosis in response to DNA damage. 293T cells transfected with scramble, p27Kip1, or caspase-3 siRNA were left untreated (open bar) or treated (closed bar) with H2O2 for 24 h. The percentage of apoptotic cells was determined by TUNEL assays. The data indicate the means ± SD from three independent experiments, each performed in triplicate. Two asterisks indicate P < 0.01. Cell lysates were subjected to immunoblot analysis with anti-p27Kip1 or anti-PCNA. (D) 293T cells were transfected with scramble siRNA or the TAF1 and p27Kip1 siRNAs together, then left untreated or treated with H2O2 for 24 h. Apoptotic cells were analyzed as described above. (E) 293T cells pretransfected with TAF1 siRNA were transfected with Flag vector or Flag-p27Kip1, then left untreated or treated with H2O2 for 24 h. The percentage of apoptotic cells was determined by TUNEL assays. The data indicate the means ± SD from three independent experiments, each performed in triplicate. One asterisk and two asterisks indicate P < 0.05 and P < 0.01, respectively. Cell lysates were subjected to immunoblot analysis with anti-TAF1, anti-Flag or antitubulin.
DISCUSSION
The cellular response to genotoxic stress that damages DNA includes cell cycle arrest, activation of DNA repair and in the event of irreparable damage, induction of apoptosis. However, the signals that determine cell fate, that is, survival or apoptosis, are largely unknown. In order to identify genes required for apoptosis, we have chosen a genome-scale RNAi screening strategy. The emergence of RNAi as a mechanism to silence gene expression has enabled loss-of-function analysis in mammalian cells in a potentially genome-wide manner (24–26). We have utilized such an RNAi-based forward genetic approach to identify genes that are involved in induction of apoptosis in human cell lines. We identified TAF1 as a candidate inducer of apoptosis. These findings support the utility of this novel approach for the identification of genes relevant to apoptosis.
TAF1 is the largest subunit of TFIID, which is composed of TBP and 13 TAFs (27,28). Binding of TFIID to the core promoter elements is required for assembly of a functional transcription initiation complex. TFIID also serves as a coactivator by directly transmitting signals from sequence-specific activators to other components of the basal transcription machinery (29–31). Moreover, TAFs function to directly activate selected genes in vivo (32–34). TAF1 appears to function as a major scaffold by which TBP and other TAFs interact in the assembly of TFIID. TAF1 plays a critical role in the regulation of cell growth (32). TAF1 possesses intrinsic protein kinase activity (35), histone acetyltransferase activity (36) and ubiquitin-activating and conjugating activity (37). The TAF1 kinase is bipartite, consisting of N- and C-terminal kinase domains. Previous work has demonstrated that kinase activity of TAF1 is important for the progression through the G1 phase (38). Another study has shown that the retinoblastoma protein Rb interacts directly with TAF1 and inhibits the kinase activity of TAF1 (39), suggesting that the kinase activity of TAF1 may have a pivotal role in tumor suppression. Whereas TAF1 affects cell cycle regulation, little is known about its effect on apoptosis control. To our knowledge, our study is the first to provide evidence suggesting that TAF1 functions as an inducer of apoptosis in response to genotoxic stress. The molecular mechanisms by which TAF1 exerts apoptotic cell death are largely unclear. Further studies are needed to clarify the precise roles for TAF1 exposed to genotoxic stress. Nevertheless, our findings demonstrate that transcriptional regulation of p27Kip1 by TAF1 is, at least in part, involved in induction of apoptosis. In this regard, function as a transcription factor is apparently required for TAF1-mediated induction of apoptosis.
The Cdk-inhibitor p27Kip1 is ubiquitously expressed and binds cyclin-Cdk2 to inhibit kinase activity (40). Human cancers often express very low levels of p27Kip1, which is associated with poor prognosis. Recent studies implied that p27Kip1 is a tumor suppressor. Indeed, reduced p27Kip1 levels in primary cancers strongly correlate with decreased patient survival (41). Germline mutations of p27Kip1 have also recently described in a subset of patients with multiple endocrine neoplasia syndrome (42). Finally, p27Kip1-null mice develop organomegaly and pituitary adenomas (43–45). The p27Kip1 level is frequently controlled by regulated translation and proteolysis (22,23). However, little is known about the transcriptional control of p27Kip1. In this regard, the present study revealed that TAF1 controls p27Kip1 expression. Importantly, such control affects induction of apoptosis in response to genotoxic stress. Given that the down-regulation of p27Kip1 markedly impaired apoptosis, it is conceivable that low levels of p27Kip1 expression in cancer are associated with efficient escape from apoptosis induced by anticancer agents.
In conclusion, our findings provide evidence that phenotypic RNAi profiling can be useful for assigning potential functions to novel genes, in combination with microarray analysis. This was highlighted by the identification of a putatively conserved transcriptional network governing apoptosis in response to genotoxic stress.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
ACKNOWLEDGEMENTS
The authors thank Dr Masami Horikoshi for providing TAF1 cDNA. This work was supported by grants from the Ministry of Education, Science and Culture of Japan (to K.Y. and Y.M.), K.Y. was also supported by the Sagawa Foundation for Promotion of Cancer Research, the Sumitomo Foundation, the Yasuda Memorial Foundation, Astellas Foundation for Research on Metabolic Disorders, Life Science Foundation of Japan, Osaka Cancer Research Foundation, Kato Memorial Bioscience Foundation and Uehara Memorial Foundation. Funding to pay the Open Access publication charges for this article was provided by the Ministry of Education, Science and Culture of Japan.
Conflict of interest statement. None declared.
REFERENCES
- 1.Hannon GJ. RNA interference. Nature. 2002;418:244–251. doi: 10.1038/418244a. [DOI] [PubMed] [Google Scholar]
- 2.Echeverri CJ, Perrimon N. High-throughput RNAi screening in cultured cells: a user's guide. Nat. Rev. Genet. 2006;7:373–384. doi: 10.1038/nrg1836. [DOI] [PubMed] [Google Scholar]
- 3.Bjorklund M, Taipale M, Varjosalo M, Saharinen J, Lahdenpera J, Taipale J. Identification of pathways regulating cell size and cell-cycle progression by RNAi. Nature. 2006;439:1009–1013. doi: 10.1038/nature04469. [DOI] [PubMed] [Google Scholar]
- 4.Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, Field CM. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bettencourt-Dias M, Giet R, Sinka R, Mazumdar A, Lock WG, Balloux F, Zafiropoulos PJ, Yamaguchi S, Winter S, Carthew RW, et al. Genome-wide survey of protein kinases required for cell cycle progression. Nature. 2004;432:980–987. doi: 10.1038/nature03160. [DOI] [PubMed] [Google Scholar]
- 6.Westbrook TF, Martin ES, Schlabach MR, Leng Y, Liang AC, Feng B, Zhao JJ, Roberts TM, Mandel G, Hannon GJ, et al. A genetic screen for candidate tumor suppressors identifies REST. Cell. 2005;121:837–848. doi: 10.1016/j.cell.2005.03.033. [DOI] [PubMed] [Google Scholar]
- 7.Kolfschoten IG, van Leeuwen B, Berns K, Mullenders J, Beijersbergen RL, Bernards R, Voorhoeve PM, Agami R. A genetic screen identifies PITX1 as a suppressor of RAS activity and tumorigenicity. Cell. 2005;121:849–858. doi: 10.1016/j.cell.2005.04.017. [DOI] [PubMed] [Google Scholar]
- 8.Nicke B, Bastien J, Khanna SJ, Warne PH, Cowling V, Cook SJ, Peters G, Delpuech O, Schulze A, Berns K, et al. Involvement of MINK, a Ste20 family kinase, in Ras oncogene-induced growth arrest in human ovarian surface epithelial cells. Mol. Cell. 2005;20:673–685. doi: 10.1016/j.molcel.2005.10.038. [DOI] [PubMed] [Google Scholar]
- 9.Ngo VN, Davis RE, Lamy L, Yu X, Zhao H, Lenz G, Lam LT, Dave S, Yang L, Powell J, et al. A loss-of-function RNA interference screen for molecular targets in cancer. Nature. 2006;441:106–110. doi: 10.1038/nature04687. [DOI] [PubMed] [Google Scholar]
- 10.Wu W, Hodges E, Hoog C. Thorough validation of siRNA-induced cell death phenotypes defines new anti-apoptotic protein. Nucleic Acids Res. 2006;34:e13. doi: 10.1093/nar/gnj015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kastan MB, Bartek J. Cell-cycle checkpoints and cancer. Nature. 2004;432:316–323. doi: 10.1038/nature03097. [DOI] [PubMed] [Google Scholar]
- 12.Massague J. G1 cell-cycle control and cancer. Nature. 2004;432:298–306. doi: 10.1038/nature03094. [DOI] [PubMed] [Google Scholar]
- 13.Rajagopalan H, Lengauer C. Aneuploidy and cancer. Nature. 2004;432:338–341. doi: 10.1038/nature03099. [DOI] [PubMed] [Google Scholar]
- 14.Fukano H, Hayatsu N, Goto R, Suzuki Y. A technique to enzymatically construct libraries which express short hairpin RNA of arbitrary stem length. Biochem. Biophys. Res. Commun. 2006;347:543–550. doi: 10.1016/j.bbrc.2006.05.124. [DOI] [PubMed] [Google Scholar]
- 15.Taira N, Nihira K, Yamaguchi T, Miki Y, Yoshida K. DYRK2 is targeted to the nucleus and controls p53 via Ser46 phosphorylation in the apoptotic response to DNA damage. Mol. Cell. 2007;25:725–738. doi: 10.1016/j.molcel.2007.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Yoshida K, Yamaguchi T, Natsume T, Kufe D, Miki Y. JNK phosphorylation of 14-3-3 proteins regulates nuclear targeting of c-Abl in the apoptotic response to DNA damage. Nat. Cell Biol. 2005;7:278–285. doi: 10.1038/ncb1228. [DOI] [PubMed] [Google Scholar]
- 17.Yoshida K, Kharbanda S, Kufe D. Functional interaction between SHPTP1 and the Lyn tyrosine kinase in the apoptotic response to DNA damage. J. Biol. Chem. 1999;274:34663–34668. doi: 10.1074/jbc.274.49.34663. [DOI] [PubMed] [Google Scholar]
- 18.Yoshida K, Kufe D. Negative regulation of the SHPTP1 protein tyrosine phosphatase by protein kinase C delta in response to DNA damage. Mol. Pharmacol. 2001;60:1431–1438. doi: 10.1124/mol.60.6.1431. [DOI] [PubMed] [Google Scholar]
- 19.Yoshida K, Wang HG, Miki Y, Kufe D. Protein kinase Cdelta is responsible for constitutive and DNA damage-induced phosphorylation of Rad9. EMBO J. 2003;22:1431–1441. doi: 10.1093/emboj/cdg134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nguyen ST, Hasegawa S, Tsuda H, Tomioka H, Ushijima M, Noda M, Omura K, Miki Y. Identification of a predictive gene expression signature of cervical lymph node metastasis in oral squamous cell carcinoma. Cancer Sci. 2007;98:740–746. doi: 10.1111/j.1349-7006.2007.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deng X, Gao F, May W.SJr. Bcl2 retards G1/S cell cycle transition by regulating intracellular ROS. Blood. 2003;102:3179–3185. doi: 10.1182/blood-2003-04-1027. [DOI] [PubMed] [Google Scholar]
- 22.Hengst L, Reed SI. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- 23.Pagano M, Tam SW, Theodoras AM, Beer-Romero P, Del Sal G, Chau V, Yew PR, Draetta GF, Rolfe M. Role of the ubiquitin-proteasome pathway in regulating abundance of the cyclin-dependent kinase inhibitor p27. Science. 1995;269:682–685. doi: 10.1126/science.7624798. [DOI] [PubMed] [Google Scholar]
- 24.Berns K, Hijmans EM, Mullenders J, Brummelkamp TR, Velds A, Heimerikx M, Kerkhoven RM, Madiredjo M, Nijkamp W, Weigelt B, et al. A large-scale RNAi screen in human cells identifies new components of the p53 pathway. Nature. 2004;428:431–437. doi: 10.1038/nature02371. [DOI] [PubMed] [Google Scholar]
- 25.Brummelkamp TR, Berns K, Hijmans EM, Mullenders J, Fabius A, Heimerikx M, Velds A, Kerkhoven RM, Madiredjo M, Bernards R, et al. Functional identification of cancer-relevant genes through large-scale RNA interference screens in mammalian cells. Cold Spring Harb. Symp. Quant. Biol. 2004;69:439–445. doi: 10.1101/sqb.2004.69.439. [DOI] [PubMed] [Google Scholar]
- 26.Paddison PJ, Silva JM, Conklin DS, Schlabach M, Li M, Aruleba S, Balija V, O'Shaughnessy A, Gnoj L, Scobie K, et al. A resource for large-scale RNA-interference-based screens in mammals. Nature. 2004;428:427–431. doi: 10.1038/nature02370. [DOI] [PubMed] [Google Scholar]
- 27.Burley SK, Roeder RG. Biochemistry and structural biology of transcription factor IID (TFIID) Annu. Rev. Biochem. 1996;65:769–799. doi: 10.1146/annurev.bi.65.070196.004005. [DOI] [PubMed] [Google Scholar]
- 28.Tora L. A unified nomenclature for TATA box binding protein (TBP)-associated factors (TAFs) involved in RNA polymerase II transcription. Genes Dev. 2002;16:673–675. doi: 10.1101/gad.976402. [DOI] [PubMed] [Google Scholar]
- 29.Albright SR, Tjian R. TAFs revisited: more data reveal new twists and confirm old ideas. Gene. 2000;242:1–13. doi: 10.1016/s0378-1119(99)00495-3. [DOI] [PubMed] [Google Scholar]
- 30.Green MR. TBP-associated factors (TAFIIs): multiple, selective transcriptional mediators in common complexes. Trends Biochem. Sci. 2000;25:59–63. doi: 10.1016/s0968-0004(99)01527-3. [DOI] [PubMed] [Google Scholar]
- 31.Thomas MC, Chiang CM. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- 32.Sekiguchi T, Nohiro Y, Nakamura Y, Hisamoto N, Nishimoto T. The human CCG1 gene, essential for progression of the G1 phase, encodes a 210-kilodalton nuclear DNA-binding protein. Mol. Cell Biol. 1991;11:3317–3325. doi: 10.1128/mcb.11.6.3317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruppert S, Wang EH, Tjian R. Cloning and expression of human TAFII250: a TBP-associated factor implicated in cell-cycle regulation. Nature. 1993;362:175–179. doi: 10.1038/362175a0. [DOI] [PubMed] [Google Scholar]
- 34.Wang EH, Tjian R. Promoter-selective transcriptional defect in cell cycle mutant ts13 rescued by hTAFII250. Science. 1994;263:811–814. doi: 10.1126/science.8303298. [DOI] [PubMed] [Google Scholar]
- 35.Dikstein R, Ruppert S, Tjian R. TAFII250 is a bipartite protein kinase that phosphorylates the base transcription factor RAP74. Cell. 1996;84:781–790. doi: 10.1016/s0092-8674(00)81055-7. [DOI] [PubMed] [Google Scholar]
- 36.Mizzen CA, Yang XJ, Kokubo T, Brownell JE, Bannister AJ, Owen-Hughes T, Workman J, Wang L, Berger SL, Kouzarides T, et al. The TAF(II)250 subunit of TFIID has histone acetyltransferase activity. Cell. 1996;87:1261–1270. doi: 10.1016/s0092-8674(00)81821-8. [DOI] [PubMed] [Google Scholar]
- 37.Pham AD, Sauer F. Ubiquitin-activating/conjugating activity of TAFII250, a mediator of activation of gene expression in Drosophila. Science. 2000;289:2357–2360. doi: 10.1126/science.289.5488.2357. [DOI] [PubMed] [Google Scholar]
- 38.O’Brien T, Tjian R. Functional analysis of the human TAFII250 N-terminal kinase domain. Mol. Cell. 1998;1:905–911. doi: 10.1016/s1097-2765(00)80089-1. [DOI] [PubMed] [Google Scholar]
- 39.Siegert JL, Robbins PD. Rb inhibits the intrinsic kinase activity of TATA-binding protein-associated factor TAFII250. Mol. Cell. Biol. 1999;19:846–854. doi: 10.1128/mcb.19.1.846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sherr CJ, Roberts JM. CDK inhibitors: positive and negative regulators of G1-phase progression. Genes Dev. 1999;13:1501–1512. doi: 10.1101/gad.13.12.1501. [DOI] [PubMed] [Google Scholar]
- 41.Slingerland J, Pagano M. Regulation of the cdk inhibitor p27 and its deregulation in cancer. J. Cell Physiol. 2000;183:10–17. doi: 10.1002/(SICI)1097-4652(200004)183:1<10::AID-JCP2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 42.Pellegata NS, Quintanilla-Martinez L, Siggelkow H, Samson E, Bink K, Hofler H, Fend F, Graw J, Atkinson MJ. Germ-line mutations in p27Kip1 cause a multiple endocrine neoplasia syndrome in rats and humans. Proc. Natl Acad. Sci. USA. 2006;103:15558–15563. doi: 10.1073/pnas.0603877103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fero ML, Rivkin M, Tasch M, Porter P, Carow CE, Firpo E, Polyak K, Tsai LH, Broudy V, Perlmutter RM, et al. A syndrome of multiorgan hyperplasia with features of gigantism, tumorigenesis, and female sterility in p27(Kip1)-deficient mice. Cell. 1996;85:733–744. doi: 10.1016/s0092-8674(00)81239-8. [DOI] [PubMed] [Google Scholar]
- 44.Kiyokawa H, Kineman RD, Manova-Todorova KO, Soares VC, Hoffman ES, Ono M, Khanam D, Hayday AC, Frohman LA, Koff A. Enhanced growth of mice lacking the cyclin-dependent kinase inhibitor function of p27(Kip1) Cell. 1996;85:721–732. doi: 10.1016/s0092-8674(00)81238-6. [DOI] [PubMed] [Google Scholar]
- 45.Nakayama K, Ishida N, Shirane M, Inomata A, Inoue T, Shishido N, Horii I, Loh DY, Nakayama K. Mice lacking p27(Kip1) display increased body size, multiple organ hyperplasia, retinal dysplasia, and pituitary tumors. Cell. 1996;85:707–720. doi: 10.1016/s0092-8674(00)81237-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.