Abstract
The mutagenesis of the major DNA adduct N-(deoxyguanosin-8-yl)-1-aminopyrene (C8-AP-dG) formed by 1-nitropyrene was compared with the analogous C8-dG adducts of 2-aminofluorene (AF) and N-acetyl-2-aminofluorene (AAF) in simian kidney (COS-7) cells. The DNA sequence chosen for this comparison contained 5′-CCATCGCTACC-3′ that has been used for solution NMR investigations. The structural and conformational differences among these lesions are well-established (Patel et al., 1998). Accordingly, we found a notable difference in the viability of the progeny, which showed that the AAF adduct was most toxic and the AF adduct was least toxic with the AP adduct exhibiting intermediate toxicity. However, analysis of the progeny showed that translesion synthesis was predominantly error-free. Only low level mutations (<3%) were detected with G→T as the dominant type of mutation by all three DNA adducts. When C8-AP-dG was evaluated in a repetitive 5′-CGCGCG-3′ sequence, higher mutational frequency (~8%) was observed. Again, G→T was the major type of mutations in simian kidney cells, even though in bacteria CpG deletions predominate in this sequence (Hilario et al, 2002). Mutagenesis of C8-AP-dG in a 12-mer containing the local DNA sequence around codon 273 of the p53 tumor suppressor gene, where the adduct was located at the second base of this codon, was also investigated. In this 5′-GTGCGTGTTTGT-3′ site, the mutations were slightly lower but not very different from the progeny derived from the 5′-CGCGCG-3′ sequence. However, the mutational frequency increased by more than 50% when the 5′ C to the adduct was replaced with a 5-methylcytosine (5-MeC). With a 5-MeC, the most notable change in mutation was the enhancement of G→A, which occurred 2.5-times relative to a 5′ C. The C8-AP-dG adduct in codon 273 dodecamer sequence with a 5′ C or 5-MeC was also evaluated in human embryonic kidney (293T) cells. Similar to COS cells, targeted mutations doubled with a 5-MeC 5′ to the adduct. Except for an increase in G→C transversions, the results in 293T were similar to that in COS cells. We conclude that C8-AP-dG mutagenesis depends on the type of cell in which it is replicated, the neighboring DNA sequence, and the methylation status of the 5′ C.
Introduction
Nitropyrenes have been detected in many environmental samples, which include automobile exhaust, urban air particulate, and coal fly ash (1–4). 1-Nitropyrene (1-NP)1, a representative of this class of compounds, is the most abundant nitroaromatic compound in the environment. 1-NP is mutagenic in many bacterial and mammalian assay systems and tumorigenic in experimental animals (5–8). 1-NP is a major constituent in a mixture of more than 200 nitropolycyclic aromatic hydrocarbons in diesel exhaust (9). Furthermore, the content of 1-NP correlates with the mutagenicity of the total dichloromethane extract of diesel exhaust particles (10). 1-NP binds covalently to the C8 position of 2′-deoxyguanosine upon reductive activation to generate N-(deoxyguanosin-8-yl)-1-aminopyrene (C8-AP-dG) (11, 12). This adduct induces many types of mutations including targeted and semi-targeted mutations, although in bacteria small frameshifts predominate (13–15). C8-AP-dG-induced frameshifts include one-base deletions, one-base insertions, and two-base deletions. We have evaluated C8-AP-dG mutagenesis site-specifically in Escherichia coli by introducing the adduct in a dinucleotide repeat sequence, CGCGCG, and a non-repetitive CCATCGCTACC sequence. The CpG repeat sequence was chosen because 1-NP is a very potent mutagen in Salmonella typhimurium frameshift tester strains (such as TA98) and induces CpG deletions in a repetitive CpG sequence near the reversion site (16, 17). The non-repetitive CGC 11-mer sequence was selected because the solution NMR structures and thermodynamic parameters of correct and incorrect base pairing of the adduct in this sequence have been reported (18–20). This sequence has also been used for NMR investigation of the analogous 2-aminofluorene adduct, C8-AF-dG (reviewed in ref. 21). For the C8-AF-dG opposite dC there is a conformational equilibrium between a structure in which AF is positioned externally in the major groove in a B-DNA helix with the modified G in the anti conformation and a modified base-displaced, fluorenyl intercalated conformer in which the modified G is syn. The C8-AP-dG shares many of the structural features of C8-AF-dG. Even so, while 70% of the population mix of C8-AF-dG contained the base-displaced intercalated conformer, the AP adduct contained essentially 100% of this conformer. It appears that the greater aromatic surface of AP compared to AF causes AP to favor stacking more overwhelmingly in this conformer. When these two adducts were studied with an A opposite the adduct, the AP remained in a base-displaced intercalated structure with no hydrogen bond with its partner. By contrast, the smaller AF remains neatly sandwiched into the minor groove and optimal stacking is achieved with the adducted G and the complementary A in an intrahelical position. The AAF adduct has not been studied in the same sequence, and conformational heterogeneity led to uncertainty in the details of the AAF structure in another sequence, but the major conformer in a non-repetitive DNA sequence contained the syn G with displacement of the modified base and insertion of the fluorenyl moiety. How would the replication be influenced by these structures? This is one of the questions we wished to address by incorporating these three related adducts in the same CCATCGCTACC sequence in a plasmid and investigating their replication.
In E. coli the site-specifically situated C8-AP-dG adduct in the CGCGCG sequence induced two-base deletions (22, 23), whereas both base substitutions and one-base deletions occurred in the non-repetitive CCATCGCTACC sequence (24). In each case the adduct is flanked by a 5′ and a 3′ C, yet the mutations in these two sites are very different in E. coli. The two-base deletions in the CGCGCG sequence increases upon induction of SOS functions, and so also the deletions of neighboring 5′ or 3′ C which occurred when the adduct is located in the non-repetitive CGC sequence, but the targeted G→T and G→C transversions at the CCATCGCTACC sequence remains unaltered with SOS. For many carcinogen-DNA adducts, however, mutagenicity in mammalian cells is often different from that in E. coli (25, 26). Therefore, we investigated the C8-AP-dG mutagenesis in these two sequences in simian kidney (COS-7) cells, and indeed we found that the mutational signature of this adduct in COS cells is different from that in E. coli Since. metabolites of many bulky carcinogens such as benzo[a]pyrene diol epoxide, benzo[g]chrysene diol epoxide, etc. bind strongly to codon 273 of the p53 tumor suppressor gene (27), which is a major mutational hotspot in lung cancer (28, 29), we have also determined the mutagenicity of C8-AP-dG located in this site in a dodecamer that contained the DNA sequence of codon 272–275 of the p53 gene. We have determined if a 5-methylcytosine (5-MeC) influences the mutagenicity of this adduct in this site. Finally, we also compared the mutagenicity of the adduct in simian kidney cells with that in human embryonic kidney (293T) cells.
Materials and Methods
Materials
[γ-32P] ATP was from Du Pont New England Nuclear (Boston, MA). EcoRV restriction endonuclease, T4 DNA ligase, and T4 polynucleotide kinase were obtained from New England Bioloabs (Beverly, MA). Escherichia coli DH10B was purchased from Life Technologies, Inc. (Grand Island, NY). The simian kidney (COS-7) cell line available in our laboratory was originally obtained from Peter Glazer, Yale University, and the pMS2 phagemid was a gift of Maasaki Moriya, SUNY, Stony Brook. The human embryonic kidney cell line 293T/17 was purchased from American Type Culture Collection (ATCC).
Methods
Synthesis and characterization of oligonucleotides
The adduct containing oligonucleotides have been synthesized and characterized as reported (18, 30, 31). Unmodified oligonucleotides were analyzed by MALDI-TOF MS analysis, which gave a molecular ion with a mass within 0.005% of theoretical, whereas adducted oligonucleotides were analyzed by ESI-MS in addition to digestion followed by HPLC analysis.
Construction and characterization of pMS2 vectors containing a single C8-AP-dG, C8-AF-dG, or C8-AAF-dG
The single stranded pMS2 shuttle vector, which contains its only EcoRV site in a hairpin region, was prepared as described (26). The pMS2 DNA (58 pmols, 100 μg) was digested with a large excess of EcoRV (300 pmol, 4.84 μg) for 1 h at 37°C followed by room temperature overnight. A 58-mer scaffold oligonucleotide was annealed overnight at 9°C to form the gapped DNA. The control and lesion containing oligonucleotides were phosphorylated with T4 polynucleotide kinase, hybridized to the gapped pMS2 DNA, and ligated overnight at 16°C. Unligated oligonucleotides were removed by passing through Centricon-100 and the DNA was precipitated with ethanol. The scaffold oligonucleotide was digested by treatment with T4 DNA polymerase and exonuclease III, the proteins were extracted with phenol/chloroform, and the DNA was precipitated with ethanol. The final construct was dissolved in 1 mM Tris-HCl-0.1 mM EDTA, pH 8, and a portion was subjected to electrophoresis on 1% agarose gel in order to assess the amount of circular DNA.
Replication and analysis in simian kidney cells
COS-7 cells were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum. The cells were seeded at 5 × 105 cells per 60-mm plate. Following overnight incubation, the cells were transfected with 50 ng of ss DNA by electroporation. The culture was incubated for 2 days, and the progeny plasmid was recovered by the method of Hirt (32). It was then used to transform E. coli DH10B, and transformants were analyzed by oligonucleotide hybridization (33, 34). Oligonucleotide probes containing the complementary 15-mer sequence were used to analyze progeny phagemids. Two 14-mer left and right probes were used to select phagemids containing the correct insert, and transformants that did not hybridize with both the left and right probes were omitted. Any transformant that hybridized with the left and right probes but failed to hybridize with the 15-mer wild-type probe were subjected to DNA sequence analysis.
Replication and analysis in human embryonic kidney (293T/17) cells
The 293T/17 cell line is a derivative of the 293T (293tsA1609neo). It is a highly transfectable derivative of the 293 cell line into which the temperature sensitive gene for simian virus 40 (SV40) T-antigen was inserted. As described by Pear et al. (35), the 293T cell line was cloned by limiting dilution to ensure that the starting population was uniform, and several independent clones were isolated and screened. To determine which clone was capable of producing the highest titer, seven highly transfectable clones were cotransfected with equimolar amounts of a replication-defective retroviral construct containing the lacZ gene (pBND) and a replication-competent Moloney murine leukemia virus (pZAP). One of these clones, referred to as 293T/17, was chosen because of its ability to produce high titers, as determined by infection into NIH 3T3 cells and staining for β-gal activity (35). These cells constitutively express the SV40 large T antigen.
The 293T/17 cells were maintained in Dulbecco’s modified Eagle’s medium supplemented with 4 mM L-glutamine, and adjusted to contain 1.5 g/L sodium bicarbonate, 4.5 g/L glucose, and 10% fetal bovine serum. The cells were grown to ~90% confluency and transfected with 50 ng of each construct using 6 μL of Lipofectamine cationic lipid reagent (Invitrogen, Carlsbad, CA). Following transfection with modified or unmodified pMS2, the cells were allowed to grow at 37°C in 5% CO2 for 2 days and then the plasmid DNA was collected and purified by the method of Hirt (32). Subsequent transformation in E. coli DH10B and analysis were performed similarly as with the plasmid from COS cells.
Results
Construction and characterization of ss pMS2 vector containing the C8-dG adducts and their replication in mammalian cells
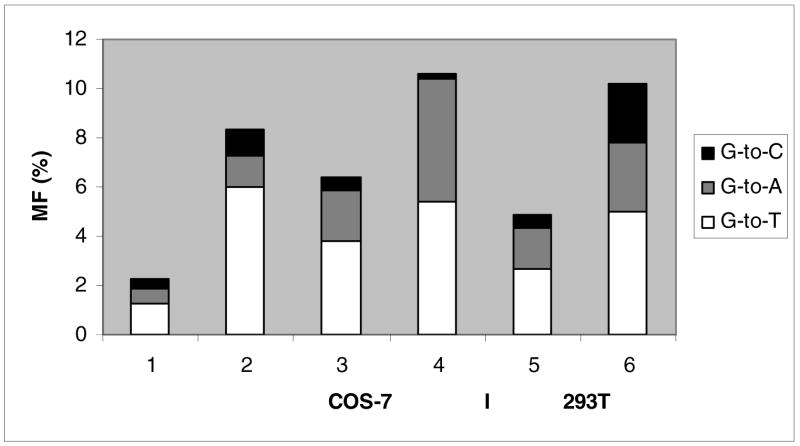
In order to investigate translesion synthesis in SV-40-transformed simian kidney cell line COS-7 and human embryonic kidney cell line 293T, we employed a site-specifically-modified single stranded vector, pMS2, which confers neomycin and ampicillin resistance (26). Biological effects of many DNA damages have been studied by using this vector (25, 26, 36, 37 and references therein) and the strategy for employing this plasmid is shown in Scheme 1. Briefly, the double stranded hairpin region of pMS2 ss DNA was digested with EcoRV and the linear DNA was hybridized with a scaffold (57-mer, 52-mer, and 58-mer, respectively, for the 11-mer, 6-mer, and 12-mer lesion containing oligonucleotides) to yield a gapped DNA. The oligonucleotides containing unmodified (dG) or modified (C8-AP-dG, C8-AF-dG, or C8-AAF-dG) nucleosides were ligated to this gap. The control and lesion containing constructs were treated with exonuclease III and T4 DNA polymerase to remove the scaffold. A portion of each of these vectors was run on a 1% agarose gel. As shown in Figure 1, lanes 1 & 2 show migration characteristics of pMS2 DNA before and after digestion with EcoRV. Lanes 3, 4, 5, and 6 show ligation of AAF-, AF-, AP-containing and control 11-mer, respectively, to pMS2 followed by enzymatic removal of the scaffold. Lanes 7 & 8 represent, before and after removal of the scaffold, respectively, of a “mock” ligation mixture, in which no oligonucleotide was added. It is evident from lanes 7 & 8 that end to end ligation of the scaffolded linear DNA in the absence of appropriate insert was negligible. Estimation of relative intensity of the circular and linear DNA indicated 30–40% ligation of the 11-mers occurred on both sides and that ligation efficiencies of the control and lesion containing oligonucleotides were approximately the same. The results with the AP-containing oligonucleotides of other sequences were similar (data not shown).
Scheme 1.
General protocol for making the pMS2 construct.
Figure 1.
Agarose gel electrophoresis of the CCATCGCTACC constructs. Lanes 1 & 2: pMS2 DNA before and after digestion with EcoRV. Lanes 3, 4, 5, and 6 show pMS2 constructs containing C8-AAF-dG, C8-AF-dG, C8-AP-dG, and dG, respectively, after enzymatic removal of the scaffold. Lanes 7 & 8 represent, before and after the removal of the scaffold, respectively, of a “mock” ligation mixture, which did not contain any 11-mer.
The vectors containing C8-AF-dG, C8-AAF-dG, or C8-AP-dG were used to transfect COS-7 cells. Progeny phagemids were recovered and used to transform E. coli DH10B. Transformants were analyzed by oligonucleotide hybridization followed by DNA sequencing in order to confirm the number of progeny derived that contained the oligonucleotide insert and the mutational outcome of the lesions.
Inhibition of replication by the C8 guanine adducts
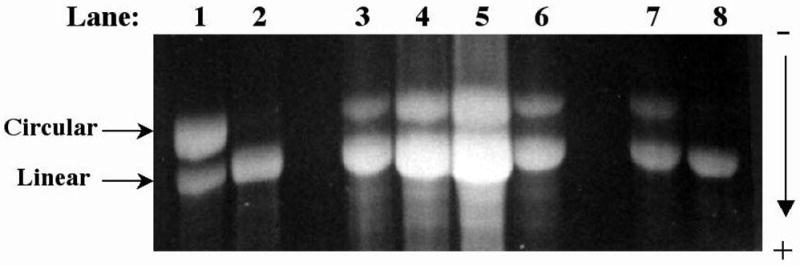
Most bulky carcinogen-DNA adducts have been found to be replication blocking lesions. It is therefore of interest to determine the yield of progeny plasmids in order to assess the relative toxicity of the adducts. Using the same amount of circular DNA for transfection, and based on the number of ampicillin-resistant progeny that contained the insert, we were able to assess the extent of translesion synthesis of the modified vectors. As shown in Figure 2, the average survival of the three adducts were 13±4% for AAF, 51±4% for AP, and 97±12% for AF. The survival of the AAF adduct was the lowest, even though AP contained a larger aromatic ring system.
Figure 2.
Percent viability of C8-2′-deoxyguanosine adducts of AP, AF, and AAF. The control (dG) and adducted (C8-AP-dG, C8-AF-dG, or C8-AAF-dG) 11-mers were ligated to a gapped pMS2 plasmid and the scaffold was removed by enzymatic digestion. Single-stranded control or adducted pMS2 DNA construct (50 ng) was transfected into COS-7 cells. Progeny plasmid was isolated after 2 days and used to transform E. coli DH10B for ampicillin resistance. Percent viability was determined as the % transformants, which contained the oligonucleotide insert as determined by hybridization with the left and right probes, relative to the number of transformants arising from the control construct. The result shown here was derived from three separate experiments that employed independently constructed plasmids.
Mutational specificity
Most of the progeny derived from the adducted vectors in the CCATCGCTACC sequence contained a guanine at the adduct site, indicating that the three C8 adducts have been replicated without significant errors. Because of the low mutational frequency (MF) the data from three separate transformations have been combined in Table 1. The targeted mutations induced by the three C8 guanine adducts in this site in COS-7 cells did not show a significant difference. For the three adducts the most common base substitution was G→T, which occurred in approximately 1% frequency. Semi-targeted mutations occurred in low frequency in the control and the adducted vector. Of the semi-targeted mutations, it seems noteworthy that only C8-AAF-dG showed prevalence of a one-base deletions 3 bases 3′ to the adduct (Table 1), whereas several types of different base substitutions near the adduct site occurred with C8-AF-dG, C8-AP-dG, and control.
Table 1.
Mutational specificity of AAF, AF, and AP adducts in CCATCGCTACC sequence in simian kidney (COS-7) cellsa
| Construct | Total colonies screened | G*→ G | C | A | T | Total targeted mutations | Semi-targeted mutations |
|---|---|---|---|---|---|---|---|
| CGC | 698 | 695 (99.6)b | 0 | 0 | 0 | 0 (0) | 3c (0.4) |
| CGAPC | 660 | 639 (96.8) | 3 (0.4) | 4 (0.6) | 9 (1.3) | 16 (2.4) | 5d (0.7) |
| CGAFC | 740 | 730 (98.6) | 2 (0.3) | 0 | 6 (0.8) | 8 (1.1) | 2e (0.3) |
| CGAAFC | 590 | 572 (96.9) | 2 (0.3) | 4f (0.3) | 6 (1.0) | 12 (2.0) | 6g (1.0) |
Data from several transformations have been combined.
The number in parenthesis shows the frequency in %.
A T→G two bases 5′, an A→C three bases 3′, and a C→T five bases 3′ to G; i.e., CCATCGCTACC was changed to CCAGCGCTACC, CCATCGCTCCC, and CCATCGCTACT.
A C→A four bases 5′, a T→G two bases 5′, two C→T one base 5′, and an A→C three bases 3′ to GAP; i.e., CCATCGAPCTACC was changed to CAATCGCTACC, CCAGCGCTACC, CCATTGCTACC, and CCATCGCTCCC.
A C→T and a C→A one base 5′ to GAF; i.e., CCATCGAFCTACC was changed to CCATTGCTACC and CCATAGCTACC.
Two of these included CGAAF→TA double mutations.
Six A deletions three bases 3′ to GAAF; i.e., CCATCGAAFCTACC was changed to CCATCGCT_CC.
Table 2 shows the mutagenicity of C8-AP-dG in the repetitive CpG sequence CGCGCG in which the MF increased 3–4-fold relative to CCATCGCTACC. However, like the non-repetitive CGC sequence, G→T transversion (6%) was the major type of mutation here as well. It is interesting to note that, unlike in E. coli where this sequence largely gave CpG deletions, the only frameshifts we detected involved a 5′C deletion adjacent to the adducted G, a +A 5′ to the adducted G, and two double mutations in which targeted G→T transversions also accompanied an adjacent 3′C deletion.
Table 2.
Mutational specificity of C8-AP-dG in CGCGCG sequence in simian kidney (COS-7) cells
| Expt. # | Construct | Total colonies screened | G*→ G | C | A | T | Total targeted mutations | Semi-targeted mutations |
|---|---|---|---|---|---|---|---|---|
| 1 | CGC | 168 | 168 (100)a | 0 | 0 | 0 | 0 (0) | 0 |
| 2 | CGC | 119 | 118 (99.2) | 0 | 0 | 0 | 0 (0) | 1b (0.8) |
| Total | CGC | 287 | 286 | 0 | 0 | 0 | 0 (0) | 1 (0.4) |
| 1 | CGAPC | 139 | 128 (92.1) | 1 (0.7) | 2 (1.4) | 8 (5.8) | 11 (7.9) | 0 |
| 2 | CGAPC | 161 | 145 (90.1) | 2 (1.2) | 2 (1.2) | 10c (6.2) | 14 (8.7) | 2d (1.2) |
| Total | CGAPC | 300 | 273 (91.0) | 3 (1.0) | 4 (1.3) | 18 (6.0) | 25 (8.3) | 2 (0.7) |
The number in parenthesis shows the frequency in %.
G deletion at the 3′ ligation site; i.e., CGCGCG was changed to CGCGC_.
Two of these GAP→T transversions also accompanied an adjacent 3′C deletion; i.e., CGCGAPCG was changed to CGCT_G.
One 5′ C→A adjacent to the adducted G and one +A 5′ to the adducted G; i.e., CGCGAPCG was changed to CGAGCG and CGCaGCG (where the inserted base was shown as an underlined lower case base).
Mutagenicity of C8-AP-dG in a dodecamer GTGCGTGTTTGT carrying the local sequence around codon 273 of the p53 gene is shown in Table 3. In this case we have compared the mutagenesis of the adduct located in a CGT sequence with that of C5-MeGT sequence. In COS-7 cells the frequency of targeted mutations in the p53 codon 273 CGT (~7%) was not remarkably different from what we determined in the repetitive CGCGCG site (~8%). However, both the MF and the types of mutations changed significantly when the 5′C was replaced with 5-MeC. The MF increased by more than 50% (to ~11%) and specifically G→A transitions increased 2.5-fold.
Table 3.
Mutational specificity of C8-AP-dG in p53 codon 273 sequence in simian kidney (COS-7) and human embryonic kidney (293T) cells
| Expt. # (cell line) | Construct | Total colonies screened | G*→G | C | A | T | Δa | Total targeted mutations | Semi-targeted or complex mutations |
|---|---|---|---|---|---|---|---|---|---|
| 1 (COS-7) | CGT | 167 | 168 (100)b | 0 | 0 | 0 | 0 | 0 (0) | 0 |
| 2 (COS-7) | CGT | 132 | 132 (100) | 0 | 0 | 0 | 0 | 0 (0) | 0 |
| Total (COS-7) | CGT | 299 | 299 (100) | 0 | 0 | 0 | 0 | 0 (0) | 0 |
| 1 (COS-7) | CGAPT | 206 | 189 (91.7) | 1 (0.5) | 4c (1.9) | 6d (2.9) | 1e (0.5) | 12 (5.8) | 5f (2.4) |
| 2 (COS-7) | CGAPT | 219 | 202 (92.2) | 1 (0.5) | 5g (2.3) | 10h (4.6) | 1 (0.5) | 17 (7.8) | 0 |
| Total (COS-7) | CGAPT | 425 | 391 (92.0) | 2 (0.5) | 9 (2.1) | 16 (3.8) | 2 (0.5) | 29 (6.8) | 5 (1.2) |
| 1 (COS-7) | C5-MeGAPT | 252 | 227 (90.1) | 0 (0) | 13 (5.2) | 12 (4.8) | 0 (0) | 25 (9.9) | 0 |
| 2 (COS-7) | C5-MeGAPT | 208 | 184 (88.5) | 1 (0.5) | 10 (4.8) | 13i (6.3) | 0 (0) | 24 (11.5) | 0 |
| Total (COS-7) | C5-MeGAPT | 460 | 411 (89.3) | 1 (0.2) | 23 (5.0) | 25 (5.4) | 0 (0) | 49 (10.6) | 0 |
| 1 (293T) | CGT | 294 | 294 (100) | 0 | 0 | 0 | 0 | 0 (0) | 0 |
| 1 (293T) | CGAPT | 387 | 360 (93.0) | 4j (1.0) | 4 (1.0) | 14k (3.6) | 0 | 22 (5.7) | 5l (1.3) |
| 2 (293T) | CGAPT | 250 | 235 (94.0) | 0 (0) | 6m (2.4) | 3n (1.2) | 1 (0.4) | 10 (4.0) | 5o (2.0) |
| Total (293T) | CGAPT | 637 | 595 (93.4) | 4 (0.6) | 10 (1.6) | 17 (2.7) | 1 (0.2) | 32 (5.0) | 10 (1.6) |
| 1 (293T) | C5-MeGAPT | 328 | 283 (86.3) | 15 (4.6) | 7p (2.1) | 16 (4.9) | 0 | 38 (11.6) | 7q (2.1) |
| 2 (293T) | C5-MeGAPT | 287 | 258 (89.9) | 0 (0) | 10r (3.5) | 15s (5.2) | 2t (0.7) | 27 (9.4) | 2u (0.7) |
| Total (293T) | C5-MeGAPT | 615 | 541 (88.0) | 15 (2.4) | 17 (2.8) | 31 (5.0) | 2 (0.3) | 65 (10.6) | 9 (1.5) |
Indicates targeted one-base deletion.
The number in parenthesis shows the frequency in %.
In addition to GAP→A, one contained one-base deletion 2 bases 5′ and one contained A & C additions as shown using the underlined lower case bases 5′-GaTGCATGcTTTGT-3′.
One of these GAP→T transversions also accompanied an one-base G deletion 2 bases 5′.
The GAP deletion accompanied deletion of one of the three consecutive 3′-T bases.
The dodecamer sequence 5′-GTGCGAPTGTTTGT-3′ was changed as follows: 5′-GTGCGTGTTAGT-3′, 5′-GTGCG_GTTAGT-3′, 5′-GTGCTCGACGAT-3′, 5′-CGGCATTTTGT-3′, 5′-GGCATTTGT-3′.
One GAP→A accompanied two 5′G→T transversions to 5′-TTTCATGTTTGT-3′.
Two GAP→T accompanied 5′G→T transversion to 5′-GTTCTTGTTTGT-3′.
One also contained C5-Me→T transition, and one contained C5-Me→T and 5′ G→C substitutions to 5′-GTCTTTGTTTGT-3′.
One GAP→C transversion accompanied a 5′C → T transition.
Three GAP → T transversions also accompanied a G → T transversion 2 bases 5′.
The dodecamer sequence 5′-GTGCGAPTGTTTGT-3′ was changed as follows: 5′-GTTCGTGTTTGT-3′, 5′-CTG__TGTTTG_-3′, 5′-GTG__ATTTGT-3′ (2), 5′-TAATTAATGT-3′
In addition to GAP→A, one contained an addition of A as shown in the underlined lower case base, a 5′-G → A transition, and deletion of one of the consecutive 3′-T bases as follows: 5′-GaTACATGTT_GT-3′. The other one contained an addition of T as shown in the underlined lower case base, a 5′-G → T transversion, and deletion of one of the consecutive 3′-T bases 5′-GTTtCATGTT_GT-3′.
One GAP→ T transversion contained a deletion of one of the consecutive 3′-T bases.
A 5′-C → T transition 5′-GTGTGTGTTTGT- 3′, a 5′-G deletion and 5′-C → T transition to 5′-GT_ TGTGTTTGT-3′, a 5′-C → A transversion to 5′-GTGAGTGTTTGT- 3′, one was changed from 5′-GTGCGAPTGTTTGT-3′ to 5′-G_GCGctTGTTGGT-3′, and the other contained an addition of T as shown using the underlined lower case base: 5′-GTGtCGTGTTTGT- 3′.
In addition to GAP→A, one contained an addition of C as shown using the underlined lower case base 5′-GTGCAcTGTTTGT-3′.
The dodecamer sequence 5′-GTGC5-MeGAPTGTTTGT-3′ was changed as follows: 5′-GGGCGTGTTTGT-3′, 5′-GTG__TGTTTGT-3′ (2), 5′-TTGTTTGTTTGT-3′ (4).
One GAP→ A transition accompanied deletion of one of the three consecutive 3′-T bases.
One GAP→T contained a 5′-G → T, C5-Me → T, and deletion of one of the three consecutive 3′-T bases: 5′-GTT TTT GT_TGT- 3′.
Both GAP deletions accompanied the deletion of one of the three consecutive 3′-T bases.
C5-Me→T transition, i.e., the sequence 5- GTGC5-MeGAPTGTTTGT-3′ for both was changed to: 5′-GTGAGTGTTTGT- 3′.
The effect of 5-MeC on mutational frequency was more pronounced in 293T cells, in which the targeted mutations doubled (5.0% vs. 10.6%) when the 5′-C was replaced with a 5-MeC. All types of targeted base substitutions increased with 5-MeC, whereas the overall frequency of various types of semi-targeted or complex mutations remained approximately the same.
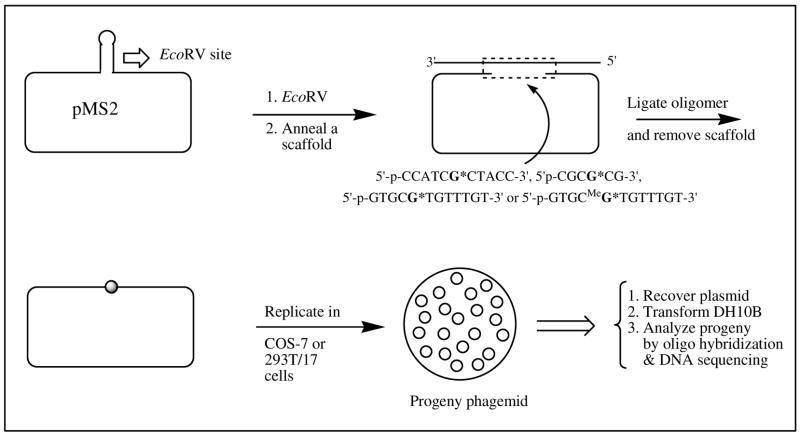
A comparison of the various targeted mutations in the different sequences by C8-AP-dG has been provided in Figure 3, which indicates that the mutational frequency and the type of mutations of this adduct in mammalian cells depend significantly on the sequence context and the methylation status of the 5′ cytosine. However, in contrast to our earlier results in E. coli, which contained a large number of frameshifts, in each case in simian and human embryonic kidney cells G→T was the major type of mutations observed.
Figure 3.
Sequence context effects of C8-AP-dG mutagenesis in mammalian cells. 1–4 represent targeted mutations in COS-7 cells, whereas 5 & 6 are results from 293T cells. 1 = CCATCGAPCTACC, 2 = CGCGAPCG, 3 & 5 = GTGCGAPTGTTTGT, and 4 & 6 = GTGC5-MeGAPTGTTTGT; in the bar graph, targeted G→T, G→A, and G→C mutations are represented by white, gray, and black, respectively.
Discussion
A single stranded vector was replicated in simian kidney cells to compare three C8-guanine adducts. NMR data in the same sequence have been reported, which established that only AF has the ability to form Watson-Crick hydrogen bonds with a cytosine resulting in protrusion of the fluorine moiety into the major groove (21). Whereas 100% of the AP adduct remains in the base displaced intercalated conformer, 70% of the population mix of the AF adduct remains in this conformer in equilibrium with 30% of the external major groove conformer. It appears that this flexibility allows the DNA polymerase to accommodate the external major groove conformer of AF better during replication, which was manifested in high viability of the progeny from the AF construct. Even though the NMR data did not show any hydrogen bond between the syn-guanine of C8-AP-dG and a cytosine (or adenine) opposite it (18, 19), bypass occurred at approximately 50% efficiency. The AAF adduct, which not only remains in syn-guanine but also exhibits conformational heterogeneity (21), was most toxic with 10–15% viability. This is consistent with other studies indicating that it requires specialized bypass polymerases for translesion synthesis (38). It has been suggested that AF can undergo transition from syn to anti at the insertion site where it can pair with an incoming dCTP, whereas AAF cannot maneuver this transition as it is restricted from adopting the anti orientation owing to the steric hindrance imposed by the N-acetyl group which essentially interfered with the polymerase’s ability to bypass this adduct (39). Our data indicating ~50% survival of the C8-AP-dG vector suggest that AP is significantly more flexible than AAF in this regard. Despite the stark difference in the viability of these adducted vectors, each showed predominantly (>97%) error-free bypass, and the type of mutations induced by the adducts was not significantly different. For each C8 adduct, G→T was the major type of mutation. We believe that the NMR solution structure may suggest the polymerases’ ability to bypass the adduct, but it is far more difficult to relate it to polymerase errors. From the cellular replication data, it is yet to be established if the same (or same group of) DNA polymerase(s) bypass these three structurally related adducts in COS cells. In E. coli, C8-AP-dG induces both base substitutions and one-base deletions in the CCATCGCTACC sequence, which include SOS-dependent one-base deletions of an adjacent C and G→T and G→C transversions (24). In addition to E. coli, mutagenicity of C8-AP-dG in the CCATCGCTACC sequence in duplex form has been determined in vitro in normal human fibroblasts (20). Mutational frequency (~2 × 10−3) of the adduct is low in this system, but all the mutants show a targeted G:C base pair deletion. Even in the same sequence context, the C8-AP-dG, therefore, induces different types of mutation in different organisms or cells.
The G→T substitutions remained the main type of mutations for C8-AP-dG when this CGC sequence was altered to a repetitive CpG sequence CGCGCG, although frequency of each type of base substitution increased and the total targeted mutations climbed to ~8%. An interesting aspect of this result is that in E. coli this sequence gave high frequency of CpG deletions, which did not occur at all in COS cells. We suspect that this difference stems from a difference in the DNA polymerase(s) that take part in translesion synthesis in the two systems. In a random mutation study, high frequency of base substitutions rather than frameshifts in the HPRT gene of human T-lymphocytes by treatment with 1-nitrosopyrene has been reported, which also suggests that mutagenesis by the adducts in mammalian cells could be significantly different from that in bacteria (7). With the AAF adduct also it was shown that bypass of this adduct located within the NarI mutation hotspot requires Pol II for -2 frameshifts but Pol V for error-free translesion synthesis (38). In another study, when the AAF adduct was located in the GGCGCC site of NarI, >90% two-base deletions occurred in SOS-induced E. coli, whereas only base substitutions (total MF ~19%) could be detected in the same site in COS-7 cells (40).
C8-AP-dG in GTGCGTGTTTGT carrying the DNA sequence of codon 272–275 of the p53 gene in which the adduct was situated in codon 273 exhibited similar mutations as in the CGCGCG site. This DNA sequence is of interest because in the p53 gene it is a hotspot for mutations in many human cancers. We also determined if the mutagenesis of the C8-AP-dG is influenced when the adduct is situated 3′ to a 5-MeC. It was earlier shown that C5 cytosine methylation may significantly increase DNA adduction at CpG sites by N-hydroxy-4-aminobiphenyl and benzo[a]pyrene diol epoxide (41, 42). However, to our knowledge, there is no study that showed if either the type or the frequency of mutagenicity of a lesion might change when the 5′ cytosine is replaced with a 5-MeC. This is the first demonstration that a 5-MeC 5′ to C8-AP-dG in the p53 codon 273 increased the MF by 50% and a major change was the increased frequency of G→A transitions. It is noteworthy that in tobacco-smoking related lung cancer patients, G→T transversions predominate in this site of the p53 gene, whereas G→A transitions occur at a high frequency in lung cancers of non-smokers (28, 43). The progeny from 293T cells showed a similar pattern in that ~5% targeted base substitutions in the CGT sequence increased to 11% in the C5-MeGT sequence. A structural rationale for this increase due to 5-MeC is yet to be developed. Nevertheless, it is not inconceivable that enhanced mutagenesis of DNA adducts with a 5-MeC 5′ could be a contributing factor in p53 mutational hotspots in human cancers. In conclusion, C8-AP-dG mutagenicity in mammalian cells is significantly different from that in E. coli, but the type and frequency are dependent on DNA sequence context and methylation status of 5′ cytosine in addition to the type of cells in which it is replicated.
Acknowledgments
This study was supported by NIEHS grants ES09127 and ES013324.
Footnotes
Abbreviations: 1-NP, 1-nitropyrene; C8-AP-dG, N-(deoxyguanosin-8-yl)-1-aminopyrene; AF, 2-aminofluorene; AAF, N-acetyl-2-aminofluorene; 5-MeC, 5-methylcytosine; MF, mutational frequency.
References
- 1.Rosenkranz HS, McCoy EC, Sanders DR, Butler M, Kiriazides DK, Mermelstein R. Nitropyrenes: isolation, identification, and reduction of mutagenic impurities in carbon black and toners. Science. 1980;209:1039–1043. doi: 10.1126/science.6996095. [DOI] [PubMed] [Google Scholar]
- 2.International Agency for Research on Cancer. IARC Monographs on the Evaluation of Carcinogenic Risk to Humans. Vol. 39. IARC; Lyon, France: 1989. Diesel and gasoline engine exhausts and some nitroarenes; pp. 1–458. [PMC free article] [PubMed] [Google Scholar]
- 3.Beland FA, Kadlubar FF. Metabolic activation and DNA adducts of aromatic amines and nitroaromatic hydrocarbons. In: Cooper CS, Grover PL, editors. Handbook of Experimental Pharmacology. Springer-Verlag; Heidelberg: 1990. pp. 267–325. [Google Scholar]
- 4.Purohit V, Basu AK. Mutagenicity of nitroaromatic compounds. Chem Res Toxicol. 2000;13:673–692. doi: 10.1021/tx000002x. [DOI] [PubMed] [Google Scholar]
- 5.Rosenkranz HS, Mermelstein R. Mutagenicity and genotoxicity of nitroarenes. All nitro-containing chemicals were not created equal. Mutat Res. 1983;114:217–267. doi: 10.1016/0165-1110(83)90034-9. [DOI] [PubMed] [Google Scholar]
- 6.Rosenkranz HS, Mermelstein R. The genotoxicity, metabolism, and carcinogenicity of nitrated polycyclic aromatic hydrocarbons. J Environ Sci Health C. 1985;3:221–272. [Google Scholar]
- 7.McGregor WG, Maher VM, McCormick JJ. Kinds and locations of mutations induced in the hypoxanthine-guanine phosphoribosyltransferase gene of human T-lymphocytes by 1-nitrosopyrene, including those caused by V(D)J recombinase. Cancer Res. 1994;54:4207–4213. [PubMed] [Google Scholar]
- 8.Hirose M, Lee MS, Wang CY, King CM. Induction of rat mammary gland tumor by 1-nitropyrene, a recently recognized environmental mutagen. Cancer Res. 1984;44:1158–1162. [PubMed] [Google Scholar]
- 9.Scheepers PTJ, Velders DD, Martens MHJ, Noordhoek J, Bos RP. Gas chromatographic-mass spectrometric determination of nitro polycyclic aromatic hydrocarbons in airborne particulate matter from workplace atmosphere contaminated with diesel exhaust. J Chromatogr A. 1994;677:107–121. [Google Scholar]
- 10.Scheepers PTJ, Martens MHJ, Velders DD, Fijneman P, Van Karkhoven M, Noordhoek J, Bos RP. 1-Nitropyrene as the marker for mutagenicity of diesel exhaust-derived airborne particulate matter in workplace atmosphere. Environ Mol Mutagen. 1995;25:134–147. doi: 10.1002/em.2850250207. [DOI] [PubMed] [Google Scholar]
- 11.Howard PC, Heflich RH, Evans FE, Beland FA. Formation of DNA adducts in vitro and in Salmonella typhimurium upon metabolic reduction of the environmental mutagen 1-nitropyrene. Cancer Res. 1983;43:2052–2058. [PubMed] [Google Scholar]
- 12.Djuric Z, Fifer EK, Ymazoe y, Beland FA. DNA binding by 1-nitropyrene and 1,6-dinitropyrene in vitro and in vivo: effects of nitroreductase induction. Carcinogenesis. 1988;9:357–364. doi: 10.1093/carcin/9.3.357. [DOI] [PubMed] [Google Scholar]
- 13.Stanton CA, Garner RC, Martin CN. The mutagenicity and DNA base sequence changes induced by 1-nitroso- and 1-nitropyrene in the cI gene of lambda prophage. Carcinogenesis. 1988;9:1153–1157. doi: 10.1093/carcin/9.7.1153. [DOI] [PubMed] [Google Scholar]
- 14.Melchoir WB, Jr, Marques MM, Beland FA. Mutations induced by aromatic amine DNA adducts in pBR322. Carcinogenesis. 1994;15:889–899. doi: 10.1093/carcin/15.5.889. [DOI] [PubMed] [Google Scholar]
- 15.Malia SA, Basu AK. Mutagenic specificty of reductively activated 1-nitropyrene in Escherichia coli. Biochemistry. 1995;34:96–104. doi: 10.1021/bi00001a012. [DOI] [PubMed] [Google Scholar]
- 16.Bell DA, Levin JG, DeMarini DM. DNA sequence analysis of revertants of the hisD3052 allele of Salmonella typhimurium TA98 using the polymerase chain reaction and direct sequencing: application to 1-nitropyrene induced revertants. Mutat Res. 1991;252:35–44. doi: 10.1016/0165-1161(91)90249-8. [DOI] [PubMed] [Google Scholar]
- 17.Nohmi T, Yamada M, Matsui M, Matsui K, Watanabe M, Sofuni T. Involvement of umuDCst genes in nitropyrene-induced -CG frameshift mutagenesis of the repetitive CG sequence in the hisD3052 allele of Salmonella typhimurium. Mol Gen Genet. 1995;247:7–16. doi: 10.1007/BF00425816. [DOI] [PubMed] [Google Scholar]
- 18.Mao B, Vyas RR, Hingerty BE, Broyde S, Basu AK, Patel DJ. Solution conformation of N-(deoxyguanosin-8-yl)-1-aminopyrene opposite dC in a DNA duplex. Biochemistry. 1996;35:12659–12670. doi: 10.1021/bi961078o. [DOI] [PubMed] [Google Scholar]
- 19.Gu Z, Gorin A, Krishnasamy R, Hingerty BE, Basu AK, Broyde S, Patel DJ. Solution structure of the N-(deoxyguanosin-8-yl)-1-aminopyrene ([AP]dG) adduct opposite dA in a DNA duplex. Biochemistry. 1999;38:10843–10854. doi: 10.1021/bi9912138. [DOI] [PubMed] [Google Scholar]
- 20.Nolan SJ, McNulty JM, Krishnasamy R, McGregor WG, Basu AK. C8-Guanine adduct-induced stabilization of a -1 frameshift intermediate in a nonrepetitive DNA sequence. Biochemistry. 1999;38:14056–14062. doi: 10.1021/bi991342o. [DOI] [PubMed] [Google Scholar]
- 21.Patel DJ, Mao B, Gu Z, Hingerty BE, Gorin A, Basu AK, Broyde S. NMR Solution structures of covalent aromatic amine-DNA adducts and their mutagenic relevance. Chem Res Toxicol. 1998;11:391–407. doi: 10.1021/tx9702143. [DOI] [PubMed] [Google Scholar]
- 22.Malia SA, Vyas RR, Basu AK. Site-specific frame-shift mutagenesis by the 1-nitropyrene adduct N-(deoxyguanosin-8-yl)-1-aminopyrene located in the (CG)3 sequence: effects of SOS, proofreading, and mismatch repair. Biochemistry. 1996;35:4568–4577. doi: 10.1021/bi9525132. [DOI] [PubMed] [Google Scholar]
- 23.Hilario P, Yan S, Hingerty BE, Broyde S, Basu AK. Comparative mutagenesis of the C8-guanine adducts of 1-nitropyrene, and 1, 6- and 1, 8-dinitropyrene in a CpG repeat sequence: A slipped frameshift intermediate model for dinucleotide deletion. J Biol Chem. 2002;277:45068–45074. doi: 10.1074/jbc.M208103200. [DOI] [PubMed] [Google Scholar]
- 24.Bacolod MD, Krishnasamy R, Basu AK. Mutagenesis of N-(deoxyguanosin-8-yl)-1-aminopyrene in a nonrepetitive CGC sequence in Escherichia coli. Chem Res Toxicol. 2000;13:523–528. doi: 10.1021/tx000023r. [DOI] [PubMed] [Google Scholar]
- 25.Moriya M, Zhang W, Johnson F, Grollman AP. Mutagenic potency of exocyclic DNA adducts: marked differences between Escherichia coli and simian kidney cells. Proc Natl Acad Sci USA. 1994;91:11899–11903. doi: 10.1073/pnas.91.25.11899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pandya G, Moriya M. 1, N6-Ethenodeoxyadenosine, a DNA adduct highly mutagenic in mammalian cells. Biochemistry. 1996;35:11487–11492. doi: 10.1021/bi960170h. [DOI] [PubMed] [Google Scholar]
- 27.Smith LE, Denissenko MF, Bennett WP, Li H, Amin S, Tang M-s, Pfeifer GP. Targeting of lung cancer mutational hotspots by polycyclic aromatic hydrocarbons. J Natl Cancer Inst. 2000;92:803–811. doi: 10.1093/jnci/92.10.803. [DOI] [PubMed] [Google Scholar]
- 28.Greenblatt MS, Bennett WP, Hollstein M, Harris CC. Mutations in the p53 tumor suppressor gene: clues to cancer etiology and molecular pathogenesis. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 29.Hollstein M, Shomer B, Greenblatt MS, Soussi T, Hovig E, Montesano R, Harris CC. Somatic point mutations in the p53 gene of human tumors and cell lines: updated compilation. Nucleic Acids Res. 1996;24:141–146. doi: 10.1093/nar/24.1.141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vyas RR, Nolan SJ, Basu AK. Synthesis and characterization of oligodeoxynucleotides containing N-(deoxyguanosin-8-yl)-1-aminopyrene. Tetrahedron Lett. 1993;34:2247–2250. [Google Scholar]
- 31.Luo C, Krishnasamy R, Basu AK, Zou Y. Recognition and incision of site-specifically modified C8 guanine adducts formed by 2-aminofluorene, N-acetyl-2-aminofluorene, and 1-nitropyrene by the UvrABC nuclease. Nucleic Acids Res. 2000;28:3719–3724. doi: 10.1093/nar/28.19.3719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 33.Ramos LA, Lipman R, Tomasz M, Basu AK. The major mitomycin C-DNA monoadduct is cytotoxic but not mutagenic in Escherichia coli. Chem Res Toxicol. 1998;11:64–69. doi: 10.1021/tx970163+. [DOI] [PubMed] [Google Scholar]
- 34.Kalam MA, Basu AK. Mutagenesis of 8-oxoguanine adjacent to an abasic site in simian kidney cells: Tandem mutations and enhancement of G→T transversions. Chem Res Toxicol. 2005;18:1187–1192. doi: 10.1021/tx050119r. [DOI] [PubMed] [Google Scholar]
- 35.Pear WS, Nolan GP, Scott ML, Baltimore D. Production of high-titer helper-free retroviruses by transient transfection. Proc Natl Acad Sci USA. 1993;90:8392–8386. doi: 10.1073/pnas.90.18.8392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fernandes A, Liu T, Amin S, Geacintov NE, Grollman AP, Moriya M. Mutagenic potential of stereoisomeric bay region (+)- and (−)-cis-anti-benzo[a]pyrene diol epoxide-N2-2′-deoxyguanosine adducts in Escherichia coli and simian kidney cells. Biochemistry. 1998;37:10164–10172. doi: 10.1021/bi980401f. [DOI] [PubMed] [Google Scholar]
- 37.Sanchez AM, Minko IG, Kurtz AJ, Kanuri M, Moriya M, Lloyd RS. Comparative evaluation of the bioreactivity and mutagenic spectra of acrolein-derived α-HOPdG and γ-HOPdG regioisomeric deoxyguanosine adducts. Chem Res Toxicol. 2003;16:1019–1028. doi: 10.1021/tx034066u. [DOI] [PubMed] [Google Scholar]
- 38.Napolitano R, Janel-Bintz R, Wagner J, Fuchs RPP. All three SOS-inducible DNA polymerases (Pol II, Pol IV and Pol V) are involved in induced mutagenesis. EMBO J. 2000;19:6259–6265. doi: 10.1093/emboj/19.22.6259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hsu GW, Kiefer JR, Burnouf D, Becherel OJ, Fuchs RPP, Beese LS. Observing translesion synthesis of an aromatic amine DNA adduct by a high-fidelity DNA polymerase. J Biol Chem. 2004;279:50280–50285. doi: 10.1074/jbc.M409224200. [DOI] [PubMed] [Google Scholar]
- 40.Tan X, Suzuki N, Grollman AP, Shibutani S. Mutagenic events in Escherichia coli and mammalian cells generated in response to acetylaminofluorene-derived DNA adducts positioned in the Nar I restriction enzyme site. Biochemistry. 2002;41:14255–14262. doi: 10.1021/bi0202878. [DOI] [PubMed] [Google Scholar]
- 41.Tang MS, Zheng JB, Denissenko MF, Pfeifer GP, Zheng Y. Use of UvrABC nuclease to quantify benzo[a]pyrene diol epoxide-DNA adduct formation at methylated CpG sites in the p53 gene. Carcinogenesis. 1999;20:1085–1089. doi: 10.1093/carcin/20.6.1085. [DOI] [PubMed] [Google Scholar]
- 42.Feng Z, Hu W, Rom WN, Beland FA, Tang MS. N-Hydroxy-4-aminobiphenyl-DNA binding in human p53 gene: sequence preference and the effect of C5 cytosine methylation. Biochemistry. 2002;41:6414–6421. doi: 10.1021/bi020093s. [DOI] [PubMed] [Google Scholar]
- 43.Pfeifer GP, Denissenko MF, Tang MS. p53 mutations, benzo[a]pyrene and lung cancer: a reply. Mutagenesis. 1998;13:537–538. doi: 10.1093/mutage/13.6.537. [DOI] [PubMed] [Google Scholar]