Abstract
The mechanism of mitotic chromosome condensation is poorly understood, but even less is known about the mechanism of formation of the primary constriction, or centromere. A proteomic analysis of mitotic chromosome scaffolds led to the identification of CENP-V, a novel kinetochore protein related to a bacterial enzyme that detoxifies formaldehyde, a by-product of histone demethylation in eukaryotic cells. Overexpression of CENP-V leads to hypercondensation of pericentromeric heterochromatin, a phenotype that is abolished by mutations in the putative catalytic site. CENP-V depletion in HeLa cells leads to abnormal expansion of the primary constriction of mitotic chromosomes, mislocalization and destabilization of the chromosomal passenger complex (CPC) and alterations in the distribution of H3K9me3 in interphase nucleoplasm. CENP-V-depleted cells suffer defects in chromosome alignment in metaphase, lagging chromosomes in anaphase, failure of cytokinesis and rapid cell death. CENP-V provides a novel link between centromeric chromatin, the primary constriction and the CPC.
Keywords: chromosome passenger complex, kinetochore, mitosis, pericentromeric heterochromatin, primary constriction
Introduction
Chromosome behaviour in mitosis is directed by kinetochores, which regulate the attachment and movement of chromosomes on the mitotic spindle, and ensure the fidelity of chromosome segregation (Rieder and Maiato, 2004; Musacchio and Salmon, 2007). The outer kinetochore is a complex proteinaceous structure, with over 80 components (Maiato et al, 2004; Chan et al, 2005; Cheeseman and Desai, 2008). The inner kinetochore is composed of DNA plus proteins that package it into chromatin. In humans, kinetochores are found on the surface of a domain composed of highly repetitive α-satellite sequences (Hyman and Sorger, 1995; Bjerling and Ekwall, 2002) packaged into condensed heterochromatin.
Heterochromatin was first defined (Heitz, 1928) on morphological grounds, but now it is defined in terms of a ‘histone code' of post-translational modifications that influence transitions between chromatin states and the regulation of transcriptional activity (Strahl and Allis, 2000; Jenuwein, 2001; Nightingale et al, 2006; Kouzarides, 2007). For example, histone H4 methylated on lysine 20 (K20) and H3 methylated on lysines 9 or 27 are generally associated with genes the transcription of which is repressed (Bannister et al, 2001; Cao et al, 2002; Peters et al, 2003; Rice et al, 2003; Schotta et al, 2004). Kinetochores also have their own particular chromatin mark, the histone H3 variant CENP-A, which is conserved in eukaryotes from yeast to man (Earnshaw and Rothfield, 1985; Carroll and Straight, 2006; Schueler and Sullivan, 2006).
CENP-A-containing chromatin is usually embedded within a large domain of pericentric heterochromatin, which may also be required for proper sister chromatid cohesion and chromosome segregation during mitosis (Nonaka et al, 2002; Dunleavy et al, 2005). Pericentric heterochromatin is rich in H3K9me2/H3K9me3 and hypoacetylated histones H3 and H4 (Dunleavy et al, 2005). Surprisingly, the chromatin domain immediately adjacent to CENP-A has been reported to contain H3K4me2, a mark of ‘open' chromatin (Sullivan and Karpen, 2004). This has led to the suggestion that the inner kinetochore may possess a specialized form of ‘centromere chromatin' rather than euchromatin or heterochromatin (Schueler and Sullivan, 2006).
In early mitosis, the heterochromatin beneath the kinetochore is associated with the chromosomal passenger complex (CPC), a widely studied regulator of mitosis with functions ranging from the correction of kinetochore attachment errors to the completion of cytokinesis (Vagnarelli and Earnshaw, 2004; Vader et al, 2006; Ruchaud et al, 2007). To accomplish its diverse functions, the CPC occupies characteristic compartments during mitotic progression—the inner centromeres in early mitosis, and the central spindle and equatorial cortex during mitotic exit (Earnshaw and Cooke, 1991). The mechanism of CPC targeting to these compartments remains a major unanswered question—the checkpoint kinase Bub1 is involved (Boyarchuk et al, 2007), but kinetochore assembly itself does not appear to be required (Oegema et al, 2001).
Here, we describe CENP-V, a putative formaldehyde-detoxifying enzyme that is required for normal compaction of heterochromatin and formation of a primary constriction. CENP-V is required for correct localization of the CPC and the centromeric cohesion protector Sgo1. Depletion of CENP-V causes a strong CPC phenotype (difficulties in chromosome bi-orientation and a failure to complete cytokinesis) that is rapidly followed by apoptotic cell death.
Results
CENP-V is conserved among vertebrates and resembles GFA of Paracoccus denitrificans
A proteomic screen for novel human proteins associated with histone-depleted mitotic chromosomes (Gassmann et al, 2005) identified p30, a protein also found in a previous proteomic screen for novel nuclear pore complex components (Cronshaw et al, 2002). p30 was not a nuclear pore component and remained otherwise uncharacterized. As shown below, p30 is a component of the kinetochore of mitotic chromosomes, and for simplicity, we will therefore designate it as CENP-V.
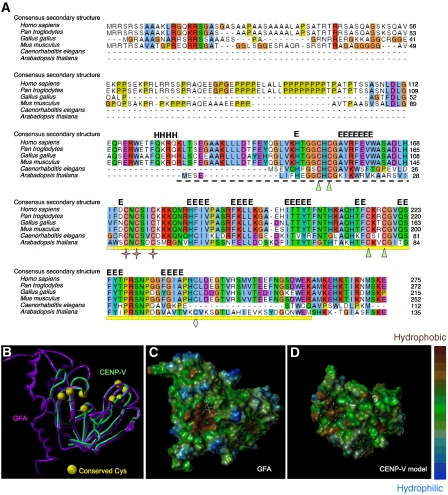
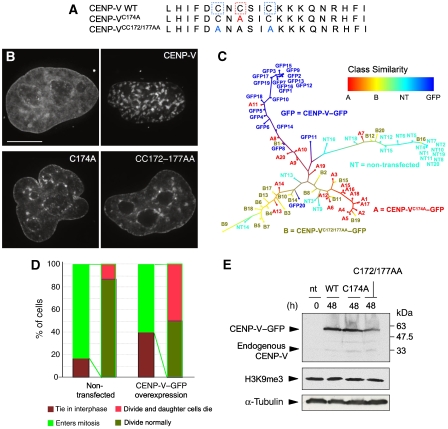
The human CENP-V gene located on chromosome 17 encodes a basic protein (pI 9.78) of 275 amino acids (Uniprot Q7Z7K6∣PRR6_HUMAN). Sequence similarity searches in publicly available databases using BLAST (Altschul et al, 1990) revealed proteins related to CENP-V in all vertebrates as well as in plants and nematodes (Figure 1A). Besides a poly-alanine-rich and a proline-rich region near the N terminus of the protein, a conserved domain annotated as glutathione-dependent formaldehyde-activating enzyme (Gfa) in PFAM (Sonnhammer et al, 1997) (pfam04828, formerly designated duf636) was also identified (yellow bar in Figure 1A). This domain is defined by the presence of five strongly conserved cysteines. No Gfa domain-containing protein is found in Saccharomyces cerevisiae and Drosophila melanogaster, although family members are present in bacteria, Xenopus, Caenorhabditis elegans and in Schizosaccharomyces pombe (Supplementary Figure 1).
Figure 1.
CENP-V is conserved among vertebrates and nematodes and is related to GFA-Pn. (A) Multiple sequence alignment and secondary structure prediction of CENP-V and its closest homologues. The alignment was automatically generated by the program T-Coffee (Notredame et al, 2000) and was visualized using Jalview (Clamp et al, 2004). The consensus secondary structure prediction (H α-helix; E β-strand) shown is based on predictions obtained using the Genesilico metaserver (http://www.genesilico.pl/meta2). NCBI accession codes: Homo sapiens, NP_859067.1 (nuclear protein p30, chromosome17); Pan troglodytes, XP_511783.2 (predicted protein: similar to nuclear protein p30, chromosome 17); Gallus gallus, XP_415846.2 (predicted protein: hypothetical protein, chromosome 19); Mus musculus, XP_921840 (proline-rich protein 6, chromosome 11); Caenorhabditis elegans, NP_741541.1 (F25B4.8b, chromosome 5) and Arabidopsis thaliana, NP_197196.1 (carbon sulphur lyase). The cysteines in the putative catalytic site are represented by stars, the cysteines from the putative structural zinc (II)-binding site by triangles and the extra cysteine conserved only in the most closely related homologues of CENP-V by a diamond. The Gfa domain is indicated by a bright yellow bar—the total region modelled encompasses that plus the striped bar. (B) Human CENP-V Gfa domain structural model (green tube) superimposed onto the homologous GFA-Pn crystal structure (PDB: 1xa8; purple ribbon) that served as a template in the modelling. Cysteine positions conserved in higher eukaryotes are depicted as yellow balls. Model coordinates and figures were generated with SYBYL (Tripos Inc.). (C, D) Hydrophobicity mapped onto the solvent-accessible surfaces of the GFA-Pn structure (C, with co-crystallized glutathione shown in stick representation) and the CENP-V Gfa domain structural model (D) using SYBYL-MOLCAD (Tripos Inc.).
Only prokaryotic enzymes containing the Gfa domain have been characterized functionally. GFA enzymes (EC no. 4.4.1.22) catalyse the first step of the glutathione-linked oxidation pathway for the conversion of formaldehyde—the condensation of formaldehyde and glutathione to S-hydroxymethylglutathione. In methylotrophic bacteria such as Paracoccus denitrificans and Rhodobacter sphaeroides, GFA is involved in the complete oxidation of methanol to carbon dioxide. In non-methylotrophic bacteria, such as Escherichia coli and in higher organisms, glutathione-linked oxidation serves to detoxify formaldehyde.
The crystallographic structure of GFA from P. denitrificans (GFA-Pn) has been solved (Neculai et al, 2005). We used this structure together with multiple alignment information to predict a structure of the conserved region of CENP-V containing the Gfa domain (amino acids 126–260). To produce a three-dimensional (3D) model of this structure, we first generated a target-to-template sequence alignment of CENP-V plus its closest homologues with the GFA subfamily containing the P. denitrificans enzyme sequence. The automated alignment from PFAM required only minor adjustments, and local ambiguities were resolved by considering tertiary structural features in the crystal structure, subfamily-specific conservation in both subfamilies and the predicted secondary structure for the CENP-V subfamily. Using this alignment as input, atomic coordinates were produced with the SYBYL software package (Tripos Inc.), yielding a robust model for the region of CENP-V containing the Gfa domain (Figure 1D). The N- and C-terminal portions of CENP-V (aa1–126 and 261–275) were not modelled, as they appear to contain non-conserved secondary structure and are unlikely to be intrinsic to the function of the Gfa domain.
The three cysteines present in the catalytic site of GFA-Pn (Figure 1A, stars) are highly conserved, as are four cysteines forming a structural zinc (II)-binding site (two within the Gfa domain and two upstream of it—triangles in Figure 1A). Another cysteine found only in the most closely related homologues of CENP-V is buried inside the structure according to our model (Figure 1A, diamond). The conserved cysteine residues in GFA-Pn form part of a redox switch that regulates the activity of the protein (Neculai et al, 2005).
We predict no notable differences between the surface charge characteristics of CENP-V and the GFA-Pn domains (data not shown). However, a hydrophobic pocket present in the catalytic site of the bacterial protein (Figure 1C) is not reproduced in our CENP-V model (Figure 1D). This pocket is important for glutathione binding in GFA-Pn. Its absence in the model suggests that if CENP-V is an enzyme, it is unlikely to utilize glutathione in a mechanism exactly similar to that of GFA-Pn.
CENP-V localizes to kinetochores in early mitosis
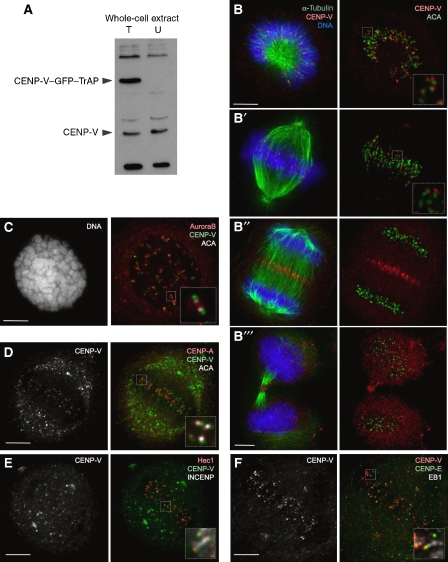
Affinity-purified rabbit polyclonal antibody Ra4552 raised against full-length CENP-V recognizes a protein of the expected size (30 kDa) on immunoblots of HeLa cell lysates (Figure 2A). Specificity of the antibody is supported by the fact that this band disappears when CENP-V is depleted by RNA interference (RNAi) (Figure 3A, Supplementary Figure 2B) as does the specific immunostaining pattern observed in mitotic cells, as described below (Supplementary Figure 3B–B′′). The purified antibody also recognizes a novel 63-kDa band in immunoblots of extracts from HeLa cells stably expressing CENP-V–GFP–TrAP (Figure 2A).
Figure 2.
CENP-V localizes to kinetochores throughout mitosis. (A) Immunoblot of HeLa cell lysate probed with serum Ra4552. T, lysate from HeLa cells expressing CENP-V–GFP–TrAP; U, lysate from untransfected HeLa cells. (B–B′′′) Localization of endogenous CENP-V (red) to kinetochores in (B, B′) prometaphase and metaphase, (B′′) to the mid-zone in anaphase and (B′′′) the mid-body in cytokinesis in HeLa cells (microtubules, green; DNA, blue). Right panels show CENP-V (red) co-stained with anticentromere antibody (ACA, green). (C) Localization of endogenous CENP-V (green) to kinetochores co-stained with Aurora B (red) and ACA (white), in cells treated with colcemid (2 h); (D–F) Localization of endogenous CENP-V (green (D, E), red (F)) co-stained with various centromere/kinetochore markers as follows: (D) CENP-A (red), (E) Hec1 (red), INCENP (white), (F) CENP-E (green) and the plus end MAP EB1 (white). Insets show higher magnification views of a single optical section of the same region. Bars, 5 μm.
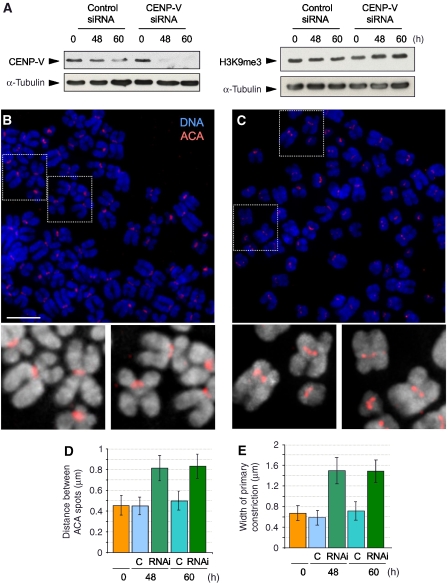
Figure 3.
Depletion of CENP-V by RNA interference causes broadening of the primary constriction without affecting heterochromatin levels. (A) Levels of endogenous CENP-V decrease 48 h after transfection with a specific siRNA oligo. Levels of H3K9me3 are not affected when CENP-V is depleted. (B) At 48 h after transfection with control siRNA, chromosomes from cells show a prominent primary constriction. (C) At the same time point, cells transfected with a specific CENP-V siRNA show abnormal centromeric structure. Insets show higher magnification views of chromosomes with ACA, red. (D, E) Quantification of the distance between ACA spots (D) and the width of primary constriction of chromosomes (E). Both values are increased when CENP-V is depleted. Bar, 5 μm.
Indirect immunofluorescence using this antibody revealed that CENP-V is localized to kinetochores from prometaphase to metaphase (Figure 2B and B′). CENP-V staining is external to CENP-A and CENP-B (Figure 2D, data not shown), internal to CENP-E and EB1 (Figure 2F) and shows most overlap with Hec1 (Figure 2E). Thus, CENP-V is localized to the outer kinetochore. This specific localization is further supported by the pattern of kinetochore staining in cells treated with colcemid (Figure 2C) and the absence of centromere staining in interphase cells (Supplementary Figure 1B).
At anaphase onset, CENP-V transfers to the spindle mid-zone and then the mid-body in telophase and cytokinesis (Figure 2B′′ and B′′′). Of known chromosomal proteins, the distribution of CENP-V most closely resembles that of CENP-E (Yen et al, 1991).
CENP-V is required for a normal primary constriction of mitotic chromosomes
To analyse the role of CENP-V in chromosome structure and function, we depleted the protein by RNAi in HeLa cells. Immunoblotting showed that by 48 h exposure of cells to a specific siRNA, CENP-V was significantly depleted (Figure 3A). At this time, >90% of cells showed an abnormal morphology when compared with cells exposed to control siRNA (Supplementary Figure 2A and A′). By 60 h, most cells exposed to CENP-V siRNA were dead (data not shown). Thus, CENP-V is essential for viability. Unfortunately, it was not possible to use siRNA rescue experiments to exclude possible off-target phenotypes, as all CENP-V constructs tested killed HeLa cells by 60 h after transfection.
CENP-V depletion caused a marked change in the structure of mitotic chromosomes in spreads prepared at 48 and 60 h after exposure to specific siRNA. Mitotic chromosomes from CENP-V-depleted cells appeared less compact overall than chromosomes from cells transfected with control siRNA (Figure 3B and C). In addition, the arms of CENP-V-depleted chromosomes frequently appeared to be less distinct from one another than the arms of chromosomes from control cells.
CENP-V depletion also had a remarkable effect on centromere structure in chromosome spreads. In CENP-V-depleted cells, the primary constriction of mitotic chromosomes appeared wider and less focused than that of chromosomes from control cells (Figure 3B and C). Measurement of DAPI-stained chromosomes revealed that the width of the primary constriction in chromosomes from cells exposed to control siRNA was ∼0.6 μm. After depletion of CENP-V, this distance increased to ∼1.5 μm (Figure 3E). Staining with anticentromere autoantibodies (ACA) revealed an increase in the distance between sister kinetochores to ∼0.8 μm, about twice the distance measured in cells exposed to control siRNA (0.45 μm; Figure 3D). This could not be explained by changes in tension, as these measurements were performed on spreads of chromosomes from nocodazole-blocked cells where tension is absent.
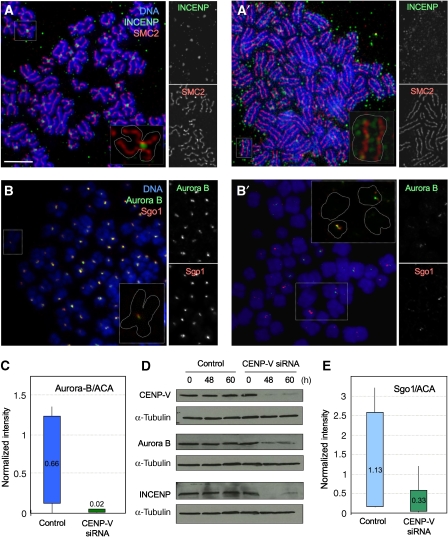
These changes in chromosomal morphology were not due to changes in the distribution of SMC2. Chromosome arms are slightly decondensed in cells lacking this key condensin subunit (Hudson et al, 2003). However, with the exception of the loss of a focus at the primary constriction, no differences in the distribution of SMC2 were observed in metaphase chromosomes from CENP-V-depleted cells (Figure 4A and A′).
Figure 4.
CENP-V depletion by RNAi affects the centromeric localization of chromosomal passenger proteins and Sgo1 but not SMC2. (A) At 48 h after transfection with control siRNA, HeLa cells show normal condensin and INCENP localization. (A′) At same time point, cells transfected with a specific-CENP-V siRNA show normal condensin localization but INCENP staining is decreased and appears delocalized to chromosome arms. (B, B′) Sgo1 and Aurora B localize to centromeres in cells exposed to a control siRNA (B), whereas the staining is reduced or absent in cells exposed to a specific CENP-V siRNA (B′′). Insets show higher magnification views of chromosomes. (C) Aurora B fluorescence intensity levels at centromeres (normalized relative to ACA staining for the same centromeres) are significantly decreased when CENP-V is depleted from cells. Average values are indicated for each condition. (D) Levels of CPC proteins Aurora B and INCENP decrease when cells are transfected with a CENP-V-specific siRNA. (E) Sgo1 fluorescence intensity levels at centromeres (normalized relative to ACA staining for the same centromeres) are significantly decreased when CENP-V is depleted from cells. Average values are indicated for each condition. Bar, 5 μm.
CENP-V depletion reduces the centromeric localization of chromosomal passenger proteins and Sgo1
The pronounced effects on the morphology of the primary constriction caused by CENP-V depletion were accompanied by a dramatic change in the distribution of several key centromeric components. In chromosome spreads prepared after 48 h exposure to CENP-V siRNA, INCENP was no longer concentrated in the inner centromere as in control cells, but instead was decreased in intensity and distributed all along the axis between the chromosome arms (Figure 4A′). A similar effect was observed on the distribution of Aurora B, the active kinase constituent of the CPC (Figure 4B and B′). In fixed samples, Aurora B is able to transfer to the central spindle and mid-body of CENP-V-depleted cells. However, its localization pattern was abnormal (Supplementary Figure 3B–B′′) when compared to the control experiment (Supplementary Figure 3A-A′′).
The apparent depletion of the CPC at inner centromeres was confirmed by quantification of the fluorescence intensity levels for Aurora B. In each case, the level of Aurora B fluorescence was normalized to the level of staining seen for the corresponding centromere using ACA (Figure 4C). Although there was a marked variability in the levels of Aurora B associated with individual centromeres, depletion of CENP-V caused a clear and dramatic decline in these levels. We further confirmed the decline of cellular Aurora B and INCENP by immunoblotting (Figure 4D).
To determine whether this decline in the levels of centromere-associated CPC proteins was functionally significant, we also stained chromosome spreads for the shugoshin protein Sgo1. The CPC is required for the stable localization of the Drosophila shugoshin MEI-S332 to centromeres in meiosis and mitosis (Resnick et al, 2006), and interactions between these proteins have been observed in a variety of cell types (Dai et al, 2006; Huang et al, 2007; Kawashima et al, 2007).
Indeed, depletion of the CPC at centromeres was accompanied by a decrease in Sgo1 after 48 h exposure to CENP-V siRNA (Figure 4B′). In contrast, no loss of centromeric Sgo1 was seen in cells exposed to control siRNA (Figure 4B). Quantification of the fluorescence intensity values confirmed the decline in centromere-associated Sgo1 in CENP-V-depleted cells (Figure 4E). This drop in Sgo1, though significant, was not sufficient to cause premature sister chromatid separation.
Thus, CENP-V is required for the normal structure of the inner centromere in mitotic chromosomes and for the proper centromeric localization of Aurora B, INCENP and Sgo1.
CENP-V is required for proper alignment of metaphase chromosomes and for correct cytokinesis
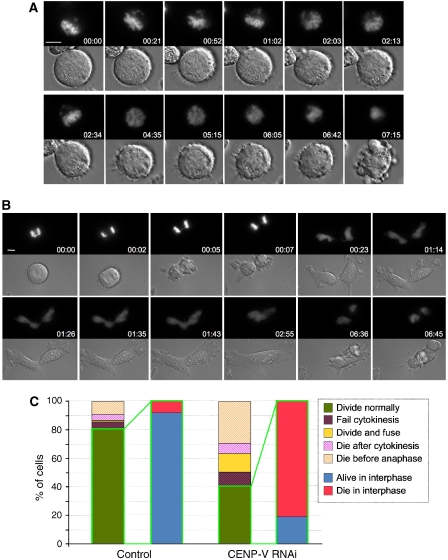
If CENP-V is required for CPC function as well as localization during mitosis, we would expect its depletion to produce strong phenotypes. To analyse the consequences of CENP-V depletion, we performed multi-site imaging by DIC and fluorescence microscopy of the chromosomes over 48 h with a 5 min time lapse, starting 24 h after transfection with fluorofore-labelled oligonucleotides. The analysis employed both control and siRNA oligonucleotides in three independent experiments, and more than 60 cells were assessed for each condition. This analysis enabled us to recognize three phenotypes following CENP-V depletion.
Approximately 30% of cells died without achieving a metaphase chromosome alignment or entering anaphase (Figure 5A and C; for a control cell, see Supplementary Figure 4).
Another ∼20% of cells either failed to complete cytokinesis or appeared to divide, but the furrow later regressed. Chromosome segregation in these cells was frequently characterized by lagging chromosomes. These cells generally underwent apoptosis soon after failing to complete cytokinesis (Figure 5B). This rapid cell death explains why the number of multinucleated cells in fixed samples (Supplementary Figure 2C) was much lower than that usually seen after perturbation of CPC function (Carvalho et al, 2003; Lens and Medema, 2003; Gassmann et al, 2004).
Finally, of the 40% of cells that appeared to complete mitosis normally, 80% underwent apoptosis without re-entering mitosis.
Figure 5.
Live imaging of CENP-V-depleted cells reveals defects in chromosome alignment and cytokinesis. (A, B) Selected fluorescence and DIC frames of live cell imaging experiments performed on a mitotic HeLa cell line stably expressing histone H2B–GFP treated with a specific CENP-V siRNA oligonucleotide. (A) Some cells are incapable of establishing a correct alignment in metaphase and fail to enter anaphase. (B) Other cells manage to enter anaphase with chromatin bridges and fail cytokinesis. (C) Quantification of the various phenotypes observed when CENP-V is depleted. Bar, 5 μm.
Thus, not only does CENP-V depletion produce an apparent perturbation of CPC localization and stability, the subsequent cell death appears to reflect a loss of CPC function during both chromosome bi-orientation and cytokinesis.
Mutations in the putative catalytic domain of CENP-V abolish its ability to induce hypercondensation of pericentromeric heterochromatin
In cells overexpressing CENP-V–GFP, the transfected protein was found in regions of dense chromatin reminiscent of the chromocentres seen in murine cell lines (Figure 6B). Centromeres were frequently associated with the surface of these condensed domains (Gassmann et al, 2005), and the domains contained the heterochromatin marker H3K9me3 (Supplementary Figure 5A). The CENP-V-induced chromatin hypercondensation was particularly prominent in HeLa cells in early G1 (e.g. in cells joined by a mature mid-body structure; Supplementary Figure 5B, D). Thus, CENP-V overexpression might either promote the condensation of pericentromeric chromatin or interfere with its decondensation during mitotic exit.
Figure 6.
Mutations in the putative catalytic domain of CENP-V abolish the chromatin hypercondensation phenotype of the wild-type protein but do not affect levels of histone modification H3K9me3. (A) Diagram depicting the cysteine residues mutated to alanine in CENP-VC174A (red) and CENP-VCC172/177AA (blue) mutants. (B) Chromatin morphology of non-transfected cells (NT) and cells transiently over expressing CENP-V–GFP (CENP-V) and the C-A mutants (C174A–GFP and CC172/177AA–GFP). The panel shows deconvolved images of DAPI staining of the DNA. (C) Dendrogram based on image similarity of the overexpression phenotype of non-transfected cells and cells overexpressing wild-type or mutant CENP-V–GFP. (D) Quantification of percentages of HeLa cells that undergo mitosis or enter apoptosis in non-transfected cells and in cells overexpressing CENP-V–GFP. (E) Immunoblots reveal that the levels of H3K9me3 are not affected when CENP-V–GFP, CENP-VC174A–GFP or CENP-VCC172/177AA–GFP are transiently overexpressed in HeLa cells. To enrich for transfected cells expressing the fusion proteins, at 24 h after transfection, cells were incubated 10 h with puromycin and extracts were prepared 14 h thereafter (nt, non-transfected cells). Bar, 5 μm.
CENP-V overexpression is highly toxic. Most cells transfected with CENP-V traverse mitosis apparently normally, but half of cells die in interphase 17–22 h later (Figure 6D). The mechanism of this death is not known, but it is unlikely to reflect interference with the function of the CPC as CENP-V overexpression had no obvious effect on the distribution of INCENP (Supplementary Figure 6) or Aurora B (Supplementary Figure 7). The timing of death suggests that CENP-V overexpression causes a fatal disruption in cell cycle progression in either S or G2.
To test whether the chromatin-condensing activity of CENP-V requires its putative catalytic site, we used a site-directed mutagenesis approach to specifically inactivate the putative enzyme function of CENP-V. Mutant CENP-VC174A removes a cysteine involved in binding of the cofactor glutathione in GFA-Pn. Mutant CENP-VCC172/177AA removes the two remaining cysteines of the putative catalytic site (Figure 6A). For subsequent analysis, cells overexpressing GFP-tagged proteins were identified using a × 20 objective—a magnification at which the detailed chromatin morphology could not be distinguished. Images were then captured using the × 100 objective, deconvolved and analysed.
The appearance of the nuclear DNA in HeLa cells transiently transfected and overexpressing CENP-VC174A–GFP or CENP-VCC172/177AA–GFP differed from that of cells overexpressing CENP-V–GFP. Although some regions of condensed chromatin were observed in cells overexpressing the mutant proteins, these were smaller and less abundant than those seen in cells overexpressing wild-type CENP-V (Figure 6B; Supplementary Figure 5B–B′′, C, D).
We used a computational pattern recognition approach to provide a quantitative assessment of the differences between wild-type and mutant CENP-V overexpression phenotypes. Deconvolved images of DAPI-stained cells overexpressing CENP-V, the two mutants and untransfected cells were subjected to an analysis of image similarity using WND-CHARM (weighted neighbour distance using a compound hierarchy of algorithms representing morphology) (Orlov et al, 2006). Briefly, in this analysis a four-way classifier was trained to distinguish images of CENP-V–GFP (GFP), mutant A (CENP-VC174A–GFP), mutant B (CENP-VCC172/177AA–GFP), and untransfected cells (NT). The classifier output for each image is a set of marginal probabilities corresponding to its similarity to the four training classes. Relative similarities between images can thus be computed from their respective Euclidian distances in a ‘marginal probability space'. The pairwise similarities between all images were used to construct the dendrogram in Figure 6C.
This analysis readily differentiated between the nuclear morphologies of non-transfected cells and those induced by CENP-V and the two mutants. The phenotypes induced by the two mutants were most closely related, whereas those of the non-transfected and CENP-V-expressing cells were readily distinguished. Close examination of the dendrogram reveals some of the heterogeneity of the raw data (e.g. mutant C174A nucleus A11 had a condensed phenotype similar to that seen with wild-type CENP-V). Overall, this analysis clearly reveals that mutation of the putative catalytic site of CENP-V causes a marked alteration in its ability to reorganize the nuclear chromatin in transfected cells.
Depletion of CENP-V by RNAi affects the distribution of pericentromeric heterochromatin in interphase nuclei
CENP-V overexpression or knockdown by RNAi cause either a hypercondensation of pericentromeric heterochromatin or expansion of the centromere of mitotic chromosomes, respectively. As chromatin states are regulated at least in part by post-translational modifications of the core histones, we therefore asked whether alteration of CENP-V levels has an effect on the levels or distribution of certain key modifications.
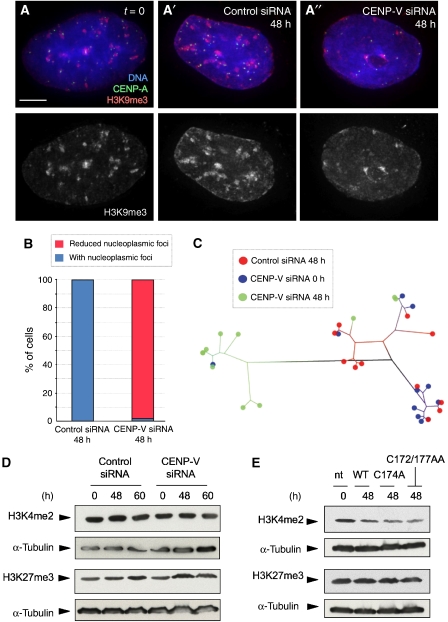
Alteration of CENP-V levels had no detectable effect on overall levels of three key histone modifications. H3K9me3 (a marker for heterochromatin), H3K4me2 (a marker for ‘open' and centromeric chromatin) and H3K27me3 (a marker for silenced genes regulated by EZH2) levels did not change following knockdown of CENP-V by siRNA (Figures 3A and 7D) or its overexpression (Figures 6E and 7E).
Figure 7.
Depletion of CENP-V by RNA interference affects the distribution of pericentromeric heterochromatin. (A, A′) Distribution of heterochromatin stained with anti-H3K9me3 in nucleoplasmic foci in HeLa cells at time point zero (A) and at 48 h after transfection with a control siRNA (A′). (A′′) At 48 h after transfection with a specific CENP-V siRNA, the distribution of H3K9me3 was more diffuse in the nucleoplasm. (B) Quantification of the distribution of H3K9me3 in the nucleoplasm of cells depleted of CENP-V. (C) Dendrogram based on image similarity of the H3K9me3 distribution between time point zero (blue), 48 h after transfection with a CENP-V-specific siRNA (green) and 48 h transfection with a control siRNA (red). (D) Immunoblots reveal that the levels of the two characteristic chromatin modifications H3K27me3 and H3K4me2 are not affected when CENP-V is depleted at 48 and 60 h after transfection with specific siRNA. (E) Levels of these modifications also do not change when either CENP-V–GFP or the putative catalytic mutants are overexpressed. Bar, 5 μm.
Depletion of CENP-V did, however, cause a significant alteration in the nuclear distribution of H3K9me3. Control HeLa cell nuclei exhibited dense patches stained with antibody specific for H3K9me3 that were distributed throughout the nuclear interior and adjacent to the inner nuclear membrane and periphery of the nucleolus (Figure 7A and A′). In contrast, after 48 h of exposure to CENP-V siRNA, these nucleoplasmic patches of H3K9me3 were less frequent (Figure 7A′′ and B). Some patches did remain adjacent to the inner nuclear membrane and nucleolus. Pattern analysis using WND-CHARM confirmed this difference in the distribution of H3K9me3 in most CENP-V-depleted nuclei (Figure 7C). Thus, although CENP-V siRNA did not cause a drop in the overall cellular levels of H3K9me3, it did cause a redistribution of the heterochromatin, but only in interphase nuclei: no difference in the distribution of H3K9me3 was observed in mitotic chromosomes (Supplementary Figure 8G and G′).
Discussion
CENP-V is a putative enzyme and novel centromeric protein identified in a proteomic screen of mitotic chromosome scaffolds (Gassmann et al, 2005) that has the ability to induce hypercondensation of pericentromeric chromatin in interphase nuclei and is required for the normal morphology of the primary constriction of mitotic chromosomes. CENP-V is required for the stability and targeting of the CPC to centromeres and when it is depleted by RNAi, cells exhibit classical CPC phenotypes including failure to complete chromosome bi-orientation and failure to complete cytokinesis. CENP-V levels are critical for cell viability, and either lowering or increasing levels of the protein leads to cell death. CENP-V RNAi also induces dramatic changes in cell shape prior to death, so the protein may have other functions in addition to its role(s) at centromeres.
The role of CENP-V in the architecture of the primary constriction of mitotic chromosomes may involve its ability to induce chromatin condensation. We showed previously that CENP-V overexpression causes hypercondensation of chromatin in interphase nuclei, and that the condensed chromatin domains are proximal to the kinetochore (Gassmann et al, 2005). Here, we confirmed that the condensed domains largely colocalize with H3K9me3, a marker for heterochromatin. This suggests that CENP-V overexpression can promote the condensation of interphase pericentromeric heterochromatin.
In a recent study examining the spring-like behaviour of centromeres, it was shown that altering chromatin higher order structure by lowering levels of a core histone causes an increase in the ‘rest length' of the spring (i.e. the distance between sister kinetochores when the centromere is not under load) (Bouck and Bloom, 2007). In previous work from our laboratory, we showed that although the condensin complex is required for complete compaction of the arms of mitotic chromosomes (Vagnarelli et al, 2006), it is not required to set the rest length of the centromere spring (Hudson et al, 2003). Here, we find that depletion of CENP-V also causes an increase in the centromeric rest length by a factor of ∼1.7, an effect greater than that seen following depletion of histone H4 in yeast (Bouck and Bloom, 2007). CENP-V depletion has no apparent effect on the distribution of condensin, and furthermore, when CENP-V-depleted cells enter anaphase, they do not exhibit the dramatic loss of compact architecture seen in condensin-depleted cells (Hudson et al, 2003; Vagnarelli et al, 2006).
One important functional consequence of the alteration of the primary constriction caused by CENP-V depletion is destabilization and delocalization of the CPC. This CPC delocalization is reminiscent of the phenotype seen following depletion of the essential cohesin subunit Rad21/Scc1 and the key mitotic regulator Bub1 from chicken centromeres (Sonoda et al, 2001; Kitajima et al, 2005; Boyarchuk et al, 2007) and is accompanied by partial delocalization of the cohesin regulator Sgo1. The CPC is required for stable localization of shugoshins such as MEI-S332 to centromeres (Resnick et al, 2006) and indeed heterochromatin itself also has an important role in promoting cohesin binding (Bernard et al, 2001; Nonaka et al, 2002). Interestingly, Bub1 and CENP-50 depletion has been reported to cause an increase in the spacing between sister kinetochores, although this was interpreted as a decrease in cohesion/adhesion rather than an increase in centromeric ‘rest length' (Kitajima et al, 2005; Minoshima et al, 2005).
The CPC can regulate the chromosomal association of chromatin remodelling complexes in Xenopus (MacCallum et al, 2002) and can also interact with histone and non-histone proteins in pericentromeric chromatin (Rangasamy et al, 2003). Whether CENP-V exerts its effect on centromeric architecture indirectly through effects on the chromosomal passenger proteins or more directly by modulating the formation or organization of pericentromeric heterochromatin remains to be determined.
Importantly, CENP-V depletion causes no detectable effect on sister chromatid cohesion. Instead, the dominant phenotypes seen after CENP-V depletion resemble those observed following disruption of the CPC. Chromosomes are perturbed in their ability to undergo normal bi-orientation, and those cells that do enter anaphase and undergo mitotic exit typically fail to complete cytokinesis. Where the CENP-V phenotype differs, however, is in the aftermath of this mitotic failure. Typically, interference with CPC function results in the accumulation of multinucleated cells and eventual cell death by apoptosis. In the case of CENP-V depletion, the consequences are more severe—the mitotic failures are followed by apoptotic death with little or no delay. The reason for this more severe phenotype is not known, but one possibility is that CENP-V also has other essential functions during interphase.
The cellular distribution of CENP-V is consistent with its apparent role(s) both as an architectural factor in the primary constriction and as a factor influencing CPC stability and function. The distribution of endogenous CENP-V in mitosis is most similar to that of the kinesin CENP-E, although the two proteins do not exactly colocalize in the kinetochore. Both proteins are found in the outer kinetochore (with CENP-V showing most overlap with Hec1), but then transfer to the central spindle and mid-body after anaphase onset. How CENP-V influences centromere architecture from this position is not known. However, it is worth pointing out that PLK1, which also influences the architecture of the inner centromere, has a similar localization in the outer kinetochore (Nishino et al, 2006).
The suggestion that CENP-V might be a factor preferentially involved in the architecture of centromeric chromatin is particularly interesting, as examination of its sequence suggests that the protein may be an enzyme functionally related to GFA of P. denitrificans. A structural model for CENP-V based on a target-to-template alignment shared many similarities with GFA-Pn, raising the possibility that CENP-V could have a similar biochemical function. Importantly, two catalytic mutants of CENP-V designed based on this predicted structure were shown by a novel pattern recognition analysis algorithm to be unable to induce chromatin hypercondensation in a pattern similar to the wild-type protein when overexpressed in HeLa cells.
Consideration of the function of GFA-Pn suggests a possible mechanism for CENP-V action in chromatin. Pericentromeric chromatin is characterized by having high concentrations of H3K9me3 and H4K20me3 (Bannister et al, 2001; Peters et al, 2003; Rice et al, 2003; Schotta et al, 2004), whereas transcribed bodies of genes are enriched in H3K4me3 (Santos-Rosa et al, 2002). In recent years, the ‘histone code' hypothesis has attracted interest in part because of the discovery of the enzymes that catalyse these methylation reactions. Histone methylation was originally considered a static modification, but this changed with the identification of histone demethylases. The first of these, LSD1, specifically demethylates lysine 4 of histone H3 (Shi et al, 2004), releasing formaldehyde as a by-product.
Formaldehyde is toxic and is usually metabolized by cells through a pathway that involves glutathione. Although this reaction occurs spontaneously, bacteria use enzymes such as GFA-Pn to catalyse the formation of S-hydroxymethylglutathione. This suggests that CENP-V could be an enzyme that scavenges the formaldehyde produced in histone demethylation reactions. If removal of formaldehyde helped to drive the forward reaction, then depending on the histone demethylation events involved, this could be one mechanism by which CENP-V could promote chromatin compaction.
Overexpression of wild-type CENP-V or depletion of the protein by siRNA did not affect global levels of H3K4me2, H3K27me3, or global levels and localization of H3K9me3 (Figures 3A, 6E, and 7D and E; Supplementary Figure 8). However, CENP-V downregulation did alter the distribution of H3K9me3 foci in interphase nucleoplasm. Thus, CENP-V appears to be an architectural factor that can influence the compaction of heterochromatin in mitosis and its distribution in interphase, rather than adjusting global levels of particular histone modifications per se.
CENP-V has provided an intriguing link between the CPC, the primary constriction of mitotic chromosomes and pericentromeric heterochromatin. Unfortunately, very preliminary attempts to observe a catalytic effect of CENP-V on formaldehyde produced during histone demethylation assays in vitro were negative (R Klose, personal communication). However, this initial effort was hampered by our lack of information about the cofactors and auxiliary subunits, if any, required for CENP-V activity. Determining the detailed mechanism of CENP-V action remains an exciting challenge for future studies.
Materials and methods
Oligonucleotides
A 21-mer oligonucleotide end labelled with Alexa555 fluorophore (GCAGCAUUUGCAAGAAGAAdTdT) covering bases 519–539 downstream of the translational start codon of human CENP-V cDNA (Uniprot/SWISSPROT Q7Z7K6) was selected as the targeting sequence among four oligonucleotides generated according to ‘HP OnGuard siRNA Design' from Qiagen. This sequence is unique to human CENP-V. A 21-mer oligonucleotide end labelled with Alexa555 fluorophore (CGUACGCGGAAUACUUCGAdTdT) with no significant homology to any known human mRNA in the databases was used as a control (Elbashir et al, 2001). The double-stranded RNA oligomers (siRNA) were supplied as annealed duplexes (Qiagen).
Cell culture and RNAi
HeLa cells in exponential growth were seeded onto polylysine-coated glass coverslips and grown overnight in RPMI/10% FBS (Gibco-BRL), 5% CO2 and at 37°C. RNAi was performed as previously described (Elbashir et al, 2001). A single pulse of 60 pmol of siRNA was administered to the cells at 30–40% confluence by transfection with OligoFectamine (Invitrogen) in complete medium without antibiotics. Cells were maintained in this medium for the duration of the experiment and assayed for CENP-V silencing, for CPC localization and cellular levels and for heterochromatin marker levels by indirect immunofluorescence and immunoblotting.
Unfortunately, we were unable to perform siRNA rescue experiments to confirm the specificity of the RNAi phenotype, as expression of wild-type or mutant CENP-V was highly toxic, killing transfected HeLa cells within ∼48–60 h. Similar toxicity has been seen with the expression of the centromere-associated helicase PICH (Baumann et al, 2007). However, we could demonstrate that the siRNA causes a specific depletion of CENP-V, and the resulting centromeric phenotypes are entirely consistent with the localization of the endogenous protein.
Indirect immunofluorescence microscopy and immunoblotting
HeLa cells grown on polylysine-coated coverslips were fixed for 5 min with 4% (v/v) paraformaldehyde (Electron Microscopy Services) in CB buffer (137 mM NaCl, 5 mM KCl, 1.1 mM Na2HPO4, 0.4 mM KH2PO4, 2 mM MgCl2, 2 mM EGTA, 5.5 mM glucose, 5 mM PIPES, pH 6.1). After permeabilization with 0.15 % (v/v) Triton X-100 in CB, coverslips were blocked with 1% (v/v) BSA in CB buffer. Cells were probed with antibodies against H3(S10)p (1/200, mouse; Cell Signaling). Alternatively, HeLa cells grown on polylysine-coated coverslips were fixed with cold MeOH at −20°C and blocked with 0.5% (v/v) BSA, 0.1% Tween 20 in PBS. Later, cells were probed with antibodies against CENP-V (Ra4552, 1:100, rabbit), Hec1 (1:500, mouse; Abcam), α-tubulin (B512, 1:2000, mouse; Sigma), CENP-B (2D7, 1:20, monoclonal; William C Earnshaw), CENP-E (1:200, monoclonal; William C Earnshaw), CENP-A (1:300; Hiroshi Masumodo, Nagoya University, Japan), H3K9me3 (1:2000, rabbit; Upstate); INCENP (1:50, mouse; Upstate), Aurora B (AIM-11:500, monoclonal; BD Transduction Laboratories), Sgo1 (1:1000, rabbit; Abcam), EB1 (1:100, monoclonal; BD Transduction Laboratories) and ACA were used to stain the centromeres (1/300, human).
To check CENP-V localization in the absence of microtubules, HeLa JW cells were treated with colcemid (100 ng/ml) for 2 h prior to MeOH fixation as described above.
Chromosome spreads were made by hypotonically swelling colcemid-treated cells with 75 mM KCl prior to fixation. Fixation was done with cold methanol. Washes with 0.1% (v/v) TEEN (1 mM triethanolamine HCl pH 8.5, 0.2 mM Na EDTA, 25 mM NaCl), 0.1% (v/v) BSA and 0.1% (v/v) Triton X-100 followed the fixation protocol. Later, cells were incubated with primary antibodies against Sgo1, ACA, SMC2 (1:200, rabbit; William C Earnshaw), Aurora B and INCENP, diluted in 0.1% (v/v) TEEN, 0.1% (v/v) BSA and 0.1% (v/v) Triton X-100. Coverslips were washed with KB− (10 mM Tris HCl pH 7.7, 0.15 M NaCl, 1% (v/v) BSA). In all cases, we used secondary antibodies coupled to FITC, Texas red, rhodamine or Cy5, and DNA was stained with DAPI (Sigma).
Total extracts of HeLa cells treated with control or CENP-V-specific oligonucleotides, HeLa cells transiently transfected with the constructs: CENP-V–GFP, C174A–GFP or CC174/177AA, and HeLa cells stably expressing CENP-V–GFP–TrAP were harvested in Laemmli buffer containing β-mercaptoethanol. The samples were run in a SDS–PAGE gel and blotted onto nitrocellulose membranes (Amersham). The membranes were blocked with 5% skimmed milk in PBS/0.05% Tween 20 and incubated later with either an anti-CENP-V antibody (Ra4553 affinity purified, diluted 1/250) in 3% milk/PBS/0.1% Tween 20 with Aurora B, INCENP, α-tubulin or with anti-H3K9me3, H4K9me2 and H3K27me3, according to the manufacturer's instructions. The membranes were incubated with anti-rabbit horseradish peroxidase-linked secondary antibody (diluted 1/10 000; Amersham) in 1% milk/PBS/0.05% Tween 20 and results were visualized using the enhanced chemiluminescence protocol (Amersham).
Time-lapse imaging
Cells were seeded onto coverslips, and treated with siRNA or transfected with CENP-V–GFP construct (see Supplementary data). For the RNAi experiment, the coverslip was transferred to a Rose Chamber 24 h after transfection with the siRNA oligonucleotide. DIC and FITC 3D image datasets were collected every minute using an inverted Nikon Eclipse TE2000-E fluorescence microscope heated to 37°C and equipped with × 40 and × 60 objectives. At least 30 cells were followed for 48 h in each experiment. For determining the fate of cells overexpressing CENP-V–GFP, the coverslip was transferred to a Rose Chamber, 24 h after transfection. DIC 3D image datasets were collected every 5 min using the Nikon Eclipse TE2000-E microscope heated to 37°C and equipped with a × 40 objective. At least 50 mitotic and interphase transfected cells as well as non-transfected cells were followed for 48 h.
Bioinformatic and pattern recognition analysis
The CENP-V structural model was constructed based on a target-to-template sequence alignment of CENP-V and GFA-Pn. The automated PFAM alignment (pfam04828) was manually adjusted taking into account the known 3D structure of GFA-Pn (PDB:1xa8) and coincidences of predicted (CENP-V) and known (GFA-Pn) secondary structure (Figure 1). Although the wide set of ‘seed' representatives of the Gfa domain family of proteins was considered in this alignment, particular attention was given to the two relevant subfamilies by including the close homologues of the target and the templates as well as a multiple sequence alignment of the GFA-Pn and its homologues. Only minor adjustments were necessary due to the noticeable sequence similarity (22% identity between template and target sequence over the Gfa domain) and the anchoring effect of the conserved cysteines.
The 3D structure and secondary structure of GFA-Pn (PDB: 1xa8) were also used for the modelling of the CENP-V structure. The atomic coordinates of the structural model were generated using the software SYBYL (Tripos™: www.tripos.com, Tripos Inc.).
The difference between the overexpression phenotypes of CENP-V–GFP and the mutants C174A–GFP and CC172/177AA–GFP was quantified using WND-CHARM (Orlov et al, 2006). WND-CHARM is a general purpose pattern analysis and classification tool for images.
Given a set of training images segregated into classes, WND-CHARM reports similarity statistics between images or classifies unknown images into one of the defined classes. Four classes were defined for WND-CHARM (A, B, GFP and NT) corresponding to the two mutants, CENP-V–GFP and untransfected cells. Twenty maximum-intensity projected images were used to train each class. For each input image, WND-CHARM reports a set of marginal probabilities corresponding to the image's similarity to each of the classes used in training. The four values reported for each image in this case were used as coordinates in a ‘marginal probability space' to compute similarities between images. The set of pairwise distances between all images were used as inputs to the ‘fitch' program from the PHYLIP package to compute a dendrogram. The dendrogram was rendered by ‘drawtree' from PHYLIP, and was further colourized based on each image's class membership and relative distance to other images in the dendrogram.
Supplementary Material
Supplementary data
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Supplementary Movie 6
Supplementary Movie 7
Supplementary Movie 8
Supplementary Movie 9
Supplementary Figures
Acknowledgments
We thank R Klose (Oxford, UK) for performing histone de-methylase assays, H Masumoto (Nagoya, Japan) for anti-CENP-A. In Edinburgh, we thank D Kelly for help with image analysis, P Vagnarelli for advice and all members of the Earnshaw laboratory for comments on the paper. AMB Tadeu is a PhD student at the Programa Doutoral em Biologia Experimental e Biomedicina of the Universidade de Coimbra supported by a PhD fellowship from FCT-Fundação para a Ciência e Tecnologia of Portugal. The laboratory of WC Earnshaw is funded by the Wellcome Trust, of which he is a Principal Research Fellow.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215: 403–410 [DOI] [PubMed] [Google Scholar]
- Bannister AJ, Zegerman P, Partridge JF, Miska EA, Thomas JO, Allshire RC, Kouzarides T (2001) Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature 410: 120–124 [DOI] [PubMed] [Google Scholar]
- Baumann C, Korner R, Hofmann K, Nigg EA (2007) PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 128: 101--114 [DOI] [PubMed] [Google Scholar]
- Bernard P, Maure JF, Partridge JF, Genier S, Javerzat JP, Allshire RC (2001) Requirement of heterochromatin for cohesion at centromeres. Science 11: 11. [DOI] [PubMed] [Google Scholar]
- Bjerling P, Ekwall K (2002) Centromere domain organization and histone modifications. Braz J Med Biol Res 35: 499–507 [DOI] [PubMed] [Google Scholar]
- Bouck DC, Bloom K (2007) Pericentric chromatin is an elastic component of the mitotic spindle. Curr Biol 17: 741–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyarchuk Y, Salic A, Dasso M, Arnaoutov A (2007) Bub1 is essential for assembly of the functional inner centromere. J Cell Biol 176: 919–928 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y (2002) Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 298: 1039–1043 [DOI] [PubMed] [Google Scholar]
- Carroll CW, Straight AF (2006) Centromere formation: from epigenetics to self-assembly. Trends Cell Biol 16: 70–78 [DOI] [PubMed] [Google Scholar]
- Carvalho A, Carmena M, Sambade C, Earnshaw WC, Wheatley SP (2003) Survivin is required for stable checkpoint activation in Taxol-treated HeLa cells. J Cell Sci 116: 2987–2998 [DOI] [PubMed] [Google Scholar]
- Chan GK, Liu ST, Yen TJ (2005) Kinetochore structure and function. Trends Cell Biol 15: 589–598 [DOI] [PubMed] [Google Scholar]
- Cheeseman IM, Desai A (2008) Molecular architecture of the kinetochore–microtubule interface. Nat Rev Mol Cell Biol 9: 33–46 [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ (2004) The Jalview Java alignment editor. Bioinformatics 20: 426–427 [DOI] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ (2002) Proteomic analysis of the mammalian nuclear pore complex. J Cell Biol 158: 915–927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Sullivan BA, Higgins JM (2006) Regulation of mitotic chromosome cohesion by Haspin and Aurora B. Dev Cell 11: 741–750 [DOI] [PubMed] [Google Scholar]
- Dunleavy E, Pidoux A, Allshire R (2005) Centromeric chromatin makes its mark. Trends Biochem Sci 30: 172–175 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Cooke CA (1991) Analysis of the distribution of the INCENPs throughout mitosis reveals the existence of three distinct substages of metaphase and early events in cleavage furrow formation. J Cell Sci 98: 443–461 [DOI] [PubMed] [Google Scholar]
- Earnshaw WC, Rothfield N (1985) Identification of a family of human centromere proteins using autoimmune sera from patients with scleroderma. Chromosoma 91: 313–321 [DOI] [PubMed] [Google Scholar]
- Elbashir SM, Harborth J, Lendeckel W, Yalcin A, Weber K, Tuschl T (2001) Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411: 494--498 [DOI] [PubMed] [Google Scholar]
- Gassmann R, Carvalho A, Henzing AJ, Ruchaud S, Hudson DF, Honda R, Nigg EA, Gerloff DL, Earnshaw WC (2004) Borealin: a novel chromosomal passenger required for stability of the bipolar mitotic spindle. J Cell Biol 166: 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gassmann R, Henzing AJ, Earnshaw WC (2005) Novel components of human mitotic chromosomes identified by proteomic analysis of the chromosome scaffold fraction. Chromosoma 113: 385–397 [DOI] [PubMed] [Google Scholar]
- Heitz E (1928) Das heterochromatin der moose. I Jb wiss Bot 69: 762–818 [Google Scholar]
- Huang H, Feng J, Famulski J, Rattner JB, Liu ST, Kao GD, Muschel R, Chan GK, Yen TJ (2007) Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol 177: 413–424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson DF, Vagnarelli P, Gassmann R, Earnshaw WC (2003) Condensin is required for nonhistone protein assembly and structural integrity of vertebrate mitotic chromosomes. Dev Cell 5: 323–336 [DOI] [PubMed] [Google Scholar]
- Hyman AA, Sorger PK (1995) Structure and function of kinetochores in budding yeast. Ann Rev Cell Dev Biol 11: 471–495 [DOI] [PubMed] [Google Scholar]
- Jenuwein T (2001) Re-SET-ting heterochromatin by histone methyltransferases. Trends Cell Biol 11: 266–273 [DOI] [PubMed] [Google Scholar]
- Kawashima SA, Tsukahara T, Langegger M, Hauf S, Kitajima TS, Watanabe Y (2007) Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes Dev 21: 420–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima TS, Hauf S, Ohsugi M, Yamamoto T, Watanabe Y (2005) Human Bub1 defines the persistent cohesion site along the mitotic chromosome by affecting Shugoshin localization. Curr Biol 15: 353–359 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128: 693–705 [DOI] [PubMed] [Google Scholar]
- Lens SM, Medema RH (2003) The survivin/Aurora B complex: its role in coordinating tension and attachment. Cell Cycle 2: 507–510 [DOI] [PubMed] [Google Scholar]
- MacCallum DE, Losada A, Kobayashi R, Hirano T (2002) ISWI remodeling complexes in Xenopus egg extracts: identification as major chromosomal components that are regulated by INCENP-aurora B. Mol Biol Cell 13: 25–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Deluca J, Salmon ED, Earnshaw WC (2004) The dynamic kinetochore–microtubule interface. J Cell Sci 117: 5461–5477 [DOI] [PubMed] [Google Scholar]
- Minoshima Y, Hori T, Okada M, Kimura H, Haraguchi T, Hiraoka Y, Bao YC, Kawashima T, Kitamura T, Fukagawa T (2005) The constitutive centromere component CENP-50 is required for recovery from spindle damage. Mol Cell Biol 25: 10315–10328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Musacchio A, Salmon ED (2007) The spindle-assembly checkpoint in space and time. Nat Rev Mol Cell Biol 8: 379–393 [DOI] [PubMed] [Google Scholar]
- Neculai AM, Neculai D, Griesinger C, Vorholt JA, Becker S (2005) A dynamic zinc redox switch. J Biol Chem 280: 2826–2830 [DOI] [PubMed] [Google Scholar]
- Nightingale KP, O'Neill LP, Turner BM (2006) Histone modifications: signalling receptors and potential elements of a heritable epigenetic code. Curr Opin Genet Dev 16: 125–136 [DOI] [PubMed] [Google Scholar]
- Nishino M, Kurasawa Y, Evans R, Lin SH, Brinkley BR, Yu-Lee LY (2006) NudC is required for Plk1 targeting to the kinetochore and chromosome congression. Curr Biol 16: 1414–1421 [DOI] [PubMed] [Google Scholar]
- Nonaka N, Kitajima T, Yokobayashi S, Xiao G, Yamamoto M, Grewal SI, Watanabe Y (2002) Recruitment of cohesin to heterochromatic regions by Swi6/HP1 in fission yeast. Nat Cell Biol 4: 89–93 [DOI] [PubMed] [Google Scholar]
- Notredame C, Higgins DG, Heringa J (2000) T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol 302: 205–217 [DOI] [PubMed] [Google Scholar]
- Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA (2001) Functional analysis of kinetochore assembly in Caenorhabditis elegans. J Cell Biol 153: 1209–1226 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orlov N, Johnston J, Macura T, Wolkow C, Goldberg I (2006) Pattern recognition approaches to compute image similarities: application to age related morphological change. In Third IEEE International Symposium on Biomedical Imaging: from Macro to Nano, pp 1152–1155 [Google Scholar]
- Peters AH, Kubicek S, Mechtler K, O'Sullivan RJ, Derijck AA, Perez-Burgos L, Kohlmaier A, Opravil S, Tachibana M, Shinkai Y, Martens JH, Jenuwein T (2003) Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell 12: 1577–1589 [DOI] [PubMed] [Google Scholar]
- Rangasamy D, Berven L, Ridgway P, Tremethick DJ (2003) Pericentric heterochromatin becomes enriched with H2A.Z during early mammalian development. EMBO J 22: 1599–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resnick TD, Satinover DL, MacIsaac F, Stukenberg PT, Earnshaw WC, Orr-Weaver TL, Carmena M (2006) INCENP and Aurora B promote meiotic sister chromatid cohesion through localization of the Shugoshin MEI-S332 in Drosophila. Dev Cell 11: 57–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JC, Briggs SD, Ueberheide B, Barber CM, Shabanowitz J, Hunt DF, Shinkai Y, Allis CD (2003) Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell 12: 1591–1598 [DOI] [PubMed] [Google Scholar]
- Rieder CL, Maiato H (2004) Stuck in division or passing through: what happens when cells cannot satisfy the spindle assembly checkpoint. Dev Cell 7: 637–651 [DOI] [PubMed] [Google Scholar]
- Ruchaud S, Carmena M, Earnshaw WC (2007) Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol 8: 798–812 [DOI] [PubMed] [Google Scholar]
- Santos-Rosa H, Schneider R, Bannister AJ, Sherriff J, Bernstein BE, Emre NC, Schreiber SL, Mellor J, Kouzarides T (2002) Active genes are tri-methylated at K4 of histone H3. Nature 419: 407–411 [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T (2004) A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 18: 1251–1262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schueler MG, Sullivan BA (2006) Structural and functional dynamics of human centromeric chromatin. Annu Rev Genomics Hum Genet 7: 301–313 [DOI] [PubMed] [Google Scholar]
- Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, Casero RA, Shi Y (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119: 941–953 [DOI] [PubMed] [Google Scholar]
- Sonnhammer EL, Eddy SR, Durbin R (1997) Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins 28: 405–420 [DOI] [PubMed] [Google Scholar]
- Sonoda E, Matsusaka T, Morrison C, Vagnarelli P, Hoshi O, Ushiki T, Nojima K, Fukagawa T, Waizenegger IC, Peters JM, Earnshaw WC, Takeda S (2001) Scc1/Rad21/Mcd1 is required for sister chromatid cohesion and kinetochore function in vertebrate cells. Dev Cell 1: 759–770 [DOI] [PubMed] [Google Scholar]
- Strahl BD, Allis CD (2000) The language of covalent histone modifications. Nature 403: 41–45 [DOI] [PubMed] [Google Scholar]
- Sullivan BA, Karpen GH (2004) Centromeric chromatin exhibits a histone modification pattern that is distinct from both euchromatin and heterochromatin. Nat Struct Mol Biol 11: 1076–1083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vader G, Medema RH, Lens SM (2006) The chromosomal passenger complex: guiding Aurora-B through mitosis. J Cell Biol 173: 833–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vagnarelli P, Earnshaw WC (2004) Chromosomal passengers: the four-dimensional regulation of mitotic events. Chromosoma 113: 211–222 [DOI] [PubMed] [Google Scholar]
- Vagnarelli P, Hudson DF, Ribeiro SA, Trinkle-Mulcahy L, Spence JM, Lai F, Farr CJ, Lamond AI, Earnshaw WC (2006) Condensin and Repo-Man-PP1 co-operate in the regulation of chromosome architecture during mitosis. Nat Cell Biol 8: 1133–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yen TJ, Compton DA, Earnshaw WC, Cleveland DW (1991) CENP-E, a human centromere associated protein released from chromosomes at the onset of anaphase. EMBO J 10: 1245–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary data
Supplementary Movie 1
Supplementary Movie 2
Supplementary Movie 3
Supplementary Movie 4
Supplementary Movie 5
Supplementary Movie 6
Supplementary Movie 7
Supplementary Movie 8
Supplementary Movie 9
Supplementary Figures