Abstract
Members of the immunity-related GTPase (IRG) family are interferon-inducible resistance factors against a broad spectrum of intracellular pathogens including Toxoplasma gondii. The molecular mechanisms governing the function and regulation of the IRG resistance system are largely unknown. We find that IRG proteins function in a system of direct, nucleotide-dependent regulatory interactions between family members. After interferon induction but before infection, the three members of the GMS subfamily of IRG proteins, Irgm1, Irgm2 and Irgm3, which possess an atypical nucleotide-binding site, regulate the intracellular positioning of the conventional GKS subfamily members, Irga6 and Irgb6. Following infection, the normal accumulation of Irga6 protein at the parasitophorous vacuole membrane (PVM) is nucleotide dependent and also depends on the presence of all three GMS proteins. We present evidence that an essential role of the GMS proteins in this response is control of the nucleotide-bound state of the GKS proteins, preventing their GTP-dependent activation before infection. Accumulation of IRG proteins at the PVM has previously been shown to be associated with a block in pathogen replication: our results relate for the first time the enzymatic properties of IRG proteins to their role in pathogen resistance.
Keywords: cell-autonomous resistance, IIGP, IRG-47, p47 GTPase, TGTP
Introduction
Several families of interferon-inducible large GTPases (including Mx proteins, guanylate-binding proteins (GBP) and immunity-related GTPases (IRG)) are involved in cell-autonomous resistance mechanisms functional against intracellular pathogens. The powerful biological activity of the IRG proteins (immunity-related or p47 GTPases) against a range of pathogens has been extensively reviewed (e.g. Martens and Howard, 2006). There have, however, been no reports relating the GTP binding and hydrolysis cycles of the IRG proteins to their activity in pathogen defence. We address this issue in the context of the well-documented ability of IRG proteins to assemble rapidly on the parasitophorous vacuole membrane (PVM) of the intracellular protozoal parasite, Toxoplasma gondii, and to participate in its destruction (Martens et al, 2005; Ling et al, 2006).
The IRG family is represented by 25 IRG coding units of about 1.2 kb in the C57BL/6 mouse genome representing 21 genes, as 4 of these are transcribed as pairs to generate tandem ∼2.4 kb products (Bekpen et al, 2005 and unpublished data). Three IRG genes (Irgm1–3) encode proteins with the non-canonical sequence GX4GMS in place of the otherwise universally conserved GX4GKS in the first nucleotide-binding motif (G1), giving rise to the subfamily name IRGM (Bekpen et al, 2005). As a convenient shorthand, we designate the IRGM subfamily members and the IRG proteins with the conventional G1 motif as GMS and GKS, respectively. Of the IRG genes studied so far by genomic knockout, Irgm1 (LRG-47), Irgm3 (IGTP) and Irgd (IRG-47) have all been shown to be non-redundantly required for resistance to T. gondii (Taylor et al, 2000; Collazo et al, 2001). Irga6 (IIGP) and Irgb6 (TGTP) also participate in the mechanism of cellular resistance to this parasite as its replication is unrestricted following expression of functionally dominant-negative mutants (Irga6-K82A, Irgb6-K69A) of these two IRG proteins in IFN-induced primary mouse astrocytes (Martens et al, 2005 and unpublished data). Irga6, Irgb6, Irgd, Irgm2 (GTPI) and Irgm3 have been shown to accumulate at T. gondii parasitophorous vacuoles (PVs) (Martens et al, 2005; Ling et al, 2006). Accumulation of IRG proteins at the PVM is followed by vesiculation and disruption of the PVM and death of the parasite. The modes of action of individual IRG proteins in resistance to T. gondii may, however, not all be the same. The accumulation of several family members at the PVM and the morphological disruption of the vacuole suggest an effector action at this site. However, Irgm1 does not localise to the PVM (Butcher et al, 2005; Martens et al, 2005 and unpublished data) and must therefore contribute to resistance in another way. In the present paper, we show that an essential function of Irgm1 as well as of Irgm2 and Irgm3 is to regulate other IRG proteins through control of their GTPase cycle.
There is an early report that partially purified Irgm3, a GMS protein, can bind GTP despite its anomalous P-loop sequence (Taylor et al, 1996). However, the GKS protein Irga6 is the only IRG family member for which the biochemical properties have been examined in detail and the crystal structure has been solved (Uthaiah et al, 2003; Ghosh et al, 2004). Purified, bacterially expressed Irga6 protein forms oligomeric structures in a GTP-dependent manner in vitro and hydrolyses GTP to GDP cooperatively. Nothing is known about the function, timing or regulation of the Irga6 GTPase cycle in vivo by heterologous regulators, such as GTPase-activating proteins (GAPs), guanine nucleotide exchange factors or guanine nucleotide dissociation inhibitors (GDIs). Thus, there is so far no direct link between the biochemical properties of the IRG proteins and their biological activity in cell-autonomous resistance. The fact that all three reported genomic knockouts of IRG genes display a non-redundant loss of T. gondii resistance suggested that different IRG proteins might interact with each other to exercise their function. Furthermore, the strikingly suppressive effects of Irga6-K82A and Irgb6-K69A on T. gondii resistance and PVM localisation in IFN-induced cells, noted above, was also compatible with an interactive model of non-redundancy. Furthermore, these mutants were designed to interfere with the nucleotide-binding site, implicating the control of the GTPase cycle in Irga6 and Irgb6 function in vivo. Both Irga6-K82A and Irgb6-K69A contain mutations of the lysine in the universally conserved P-loop GKS motif homologous to p21 Ras K16. Here, we show that the biochemical characteristic of Irga6-K82A is not the expected failure to bind GTP but rather a failure to hydrolyse it once bound. Irga6-K82A is thus a constitutively active form of the protein. WT Irga6 behaves as if it too is constitutively active in vivo in the absence of the three GMS proteins Irgm1, Irgm2 and Irgm3, suggesting that these latter proteins normally suppress or attenuate the spontaneous conversion of Irga6 into the active GTP-bound state. In the absence of this regulatory control, Irga6, such as Irga6-K82A, accumulates in cytoplasmic aggregates and fails to reach the PVM of infecting T. gondii. Irgb6 shows properties largely similar but not identical to those of Irga6, and we find that the regulatory interactions of the GMS proteins with these two GKS proteins are associated with direct nucleotide-dependent contact between the GTPases. We conclude that assembly of IRG proteins at the T. gondii PVM depends on the timing and subcellular location of nucleotide exchange, which is in turn governed by specific, nucleotide-dependent regulatory interactions between the three GMS proteins and members of the GKS group.
Results
Irga6-K82A is constitutively active and Irga6-S83N is inert
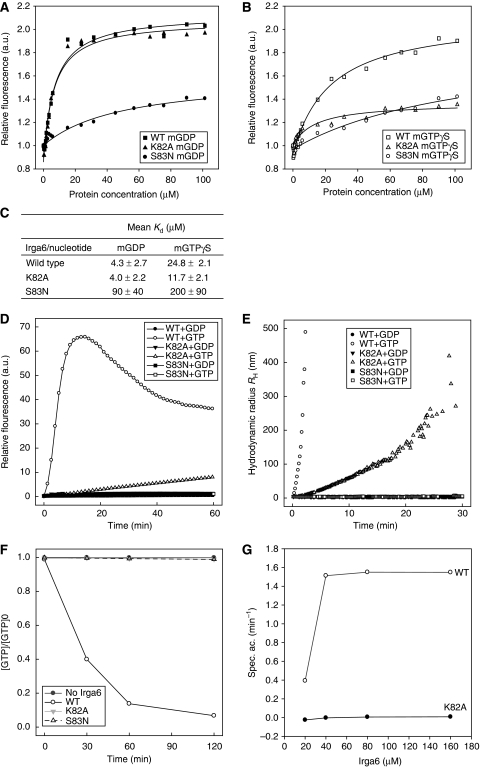
K82 of Irga6 is homologous to K16 of Ras; mutations at this P-loop residue have been shown to render several GTPases deficient in GTP binding (Sigal et al, 1986; Pitossi et al, 1993; Praefcke et al, 2004). Irga6-K82A has wild-type (WT) affinity for GDP (Uthaiah, 2002; Figure 1A and C) and also significant binding activity for methylanthraniloyl-(mant)-GTPγS (mGTPγS) (Figure 1B and C), with a calculated equilibrium affinity constant similar to WT Irga6. The absolute increase in fluorescence signal on mGTPγS binding was, however, unusually small (see Materials and methods), so this affinity estimate should be considered preliminary. Consistent with its affinity for GTP, Irga6-K82A, similar to WT Irga6 (Uthaiah et al, 2003), formed oligomers in vitro on addition of GTP (Figure 1D and E). In case of WT Irga6, the oligomers resolved as GTP was hydrolysed (Uthaiah et al, 2003), whereas Irga6-K82A oligomers formed slowly and continuously and did not resolve over the time course of the experiment. We further showed that Irga6-K82A shows no net GTP turnover activity over a wide protein concentration range (Figure 1F and G). Thus, Irga6-K82A, despite its dominant inhibitory effect on resistance to T. gondii (Martens et al, 2005), may become constitutively active in vivo, locked in the GTP-bound state.
Figure 1.
Nucleotide binding and hydrolysis by Irga6 and Irga6 mutants. (A–C) Nucleotide-binding affinities of recombinant WT Irga6, -K82A and -S83N measured by equilibrium titration with mant nucleotides: (A) mGDP, (B) mGTPγS and (C) average dissociation constants (Kd) from two independent experiments. (D, E) Nucleotide-dependent oligomerisation of WT Irga6 and mutant proteins, measured by (D) conventional and (E) dynamic light scattering. (F) Kinetics of α32P-GTP hydrolysis by Irga6 and Irga6 mutants displayed as ratio of the GTP concentration to the starting concentration against time. (G) Protein concentration dependence of the specific α32P-GTP hydrolysis activity of WT Irga6 and Irga6-K82A measured over 30 min.
The S17N mutation of Ras retains WT affinity for GDP but is unable to bind GTP, and is therefore dominant negative, locked in the inactive state (Feig and Cooper, 1988). The homologous mutation of Irga6, S83N, however, had greatly reduced binding affinity for both nucleotides (Figure 1A–C), did not hydrolyse GTP (Figure 1F) and did not form GTP-dependent oligomers (Figure 1D–E). Irga6-S83N, therefore, provided a negative control protein for the documentation of nucleotide-dependent processes mediated by Irga6.
Irga6 localisation to the T. gondii PVM is regulated by IFN
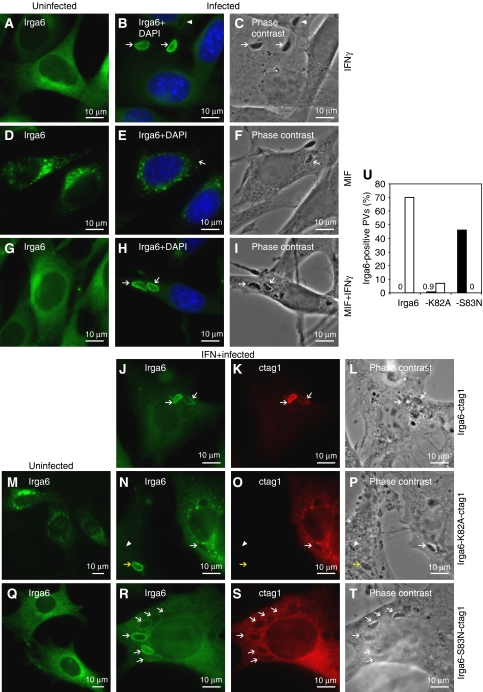
Irga6 transfected into unstimulated mouse L929 fibroblasts accumulated in cytoplasmic aggregates (Martens et al, 2004), instead of the smooth ER localisation characteristic of the endogenous, IFN-induced Irga6 protein (Figure 2A). Cells expressing such aggregates proliferated normally (Supplementary Figure S1) but showed striking distension of the ER lumen (Supplementary Figure S2). These results suggest that the correct cellular localisation of Irga6 requires the concomitant expression of additional IFNγ-inducible factors. To determine whether Irga6 accumulation at the PVM of infecting T. gondii also requires other IFNγ-inducible factors, we prepared cloned NIH3T3 fibroblast cell lines (gs3T3) stably expressing Irga6 under the control of a promoter inducible by the synthetic steroid, Mifepristone (MIF). On hormone induction, these cells expressed Irga6 at a level similar to that in IFNγ-induced gs3T3 cells (Supplementary Figure S3), and the protein accumulated in cytoplasmic aggregates (Figure 2D) similar to those reported previously in L929 cells transiently transfected with Irga6. When infected with T. gondii, the accumulation of Irga6 at the PVM seen in IFNγ-treated cells (Martens et al, 2005; Figure 2B and C) was not detected in MIF-induced cells (Figure 2E and F). However, when MIF-induced gs3T3-Irga6 cells were simultaneously induced with IFNγ, normal ER localisation of Irga6 was restored (Figure 2G) and the protein could also accumulate normally at the PVM (Figure 2H and I). Thus, cytoplasmic aggregation and failure to localise to the PVM are correlated properties of Irga6 when expressed in the absence of IFN, and both can be corrected by other IFN-inducible components.
Figure 2.
Subcellular localisation of Irga6 is dependent on IFNγ and nucleotide. (A–T) Localisation of Irga6 in uninfected (A, D, G, M, Q) and T. gondii-infected (B, C, E, F, H, I, J–L, N–P, R–T) fibroblasts. Arrows and arrowheads indicate intracellular T. gondii identified in phase contrast. Total Irga6 was detected with the mAb 10E7 (green). Transfected Irga6-ctag1 (1 μg DNA) was also detected by αctag1 AS (red). Nuclei were labelled with DAPI (blue). (A–I) gs3T3-Irga6 cells. (A–C) IFNγ-induced cells. (D–F) MIF-induced cells. The second T. gondii visible in (F) is probably still extracellular. (G–I) IFNγ and MIF double-induced cells. Note that not all PVs acquired Irga6 (arrowheads); 70–80% of PVs acquired Irga6 in IFNγ-induced gs3T3 cells (see Figure 4D). (J–L) Infected, IFNγ-induced WT MEFs transiently transfected with Irga6-ctag1. (M–P) Cells transiently transfected with Irga6-K82A-ctag1. (M) Uninduced gs3T3 cells. (N–P) Infected, IFNγ-induced WT MEFs. Irga6-K82A-ctag1 (O) did not transfer to the PVM and inhibited transfer of the endogenous WT Irga6 (N). The non-transfected cell shows a normal PVM stain with endogenous Irga6 (yellow arrow). (Q–T) Cells transiently transfected with Irga6-S83N-ctag1. (Q) Uninfected gs3T3 cells. (R–T) Infected, IFNγ-induced WT MEFs. (U) Quantification of the PVM localisation of Irga6 and its mutants in IFNγ-induced, T. gondii-infected gs3T3 cells transiently transfected with WT Irga6-ctag1, -K82A-ctag1 or -S83N-ctag1 (1 μg DNA). Transfected proteins were detected by αctag1 AS 2600. Total (i.e. transfected and induced) Irga6 was detected with mAb 10D7. The number of 10D7 singly (black bars) or 10D7 and 2600 double-positive PVs (white bars) per 100 intracellular parasites was determined in transfected cells. Around 100 PVs per data point were counted blind.
To distinguish between endogenous and ectopically expressed Irga6, we generated C-terminally epitope tagged Irga6 constructs (Irga6-ctag1). Irga6-ctag1 transiently transfected into IFNγ-induced mouse embryonic fibroblasts (MEFs) showed essentially normal resting localisation at the ER and accumulation at the T. gondii PVM (Figure 2J–L and U), indicating that this tag does not interfere with these aspects of normal Irga6 protein function. To test the possibility that aggregated WT Irga6 in cells not treated with IFNγ was trapped in the GTP-bound state, we examined the properties of ctag1-tagged Irga6-K82A, in transfected gs3T3 and MEFs. Irga6-K82A-ctag1 formed cytoplasmic aggregates in uninduced cells (Figure 2M) but unlike WT Irga6-ctag1, Irga6-K82A-ctag1 made similar aggregates in IFNγ-induced cells (Figure 2N). Furthermore, transfected Irga6-K82A-ctag1 almost completely prevented the endogenous IFNγ-induced Irga6 from accumulating at the T. gondii PVM (Figure 2N–P and U), thus acting functionally as a dominant negative, as previously reported for T. gondii-infected, IFNγ-induced astrocytes (Martens et al, 2005). The endogenous Irga6 apparently colocalised in aggregates with the transfected mutant (Figure 2N and O). These results suggested that cytoplasmic aggregation and failure to reach the T. gondii PVM are properties of GTP-bound Irga6, constitutively in the case of Irga6-K82A and, in the absence of IFNγ, also for the WT. The significance of nucleotide binding for the formation of Irga6 aggregates was indicated by the failure of transfected Irga6-S83N-ctag1 to form aggregates either in IFN-induced or uninduced cells (Figure 2Q and R). Irga6-S83N protein was distributed smoothly on cytoplasmic membranes but did not localise to the T. gondii PVM itself nor did it prevent WT Irga6 from doing so (Figure 2R–U).
These results suggest that WT Irga6 is normally kept in the cytoplasm in the GDP-bound state in IFNγ-induced cells, but accumulates at the T. gondii PVM in the active GTP-bound state. In the absence of IFNγ, WT Irga6 binds GTP and activates spontaneously in the cytoplasm to form ‘sterile' aggregates that cannot localise to the PVM. Irga6-K82A activates constitutively in the cytoplasm and forms ectopic GTP-bound aggregates that can also capture WT Irga6, thus acting as a functional dominant negative. Above all, these findings strongly suggest that the nucleotide-binding status of Irga6 must normally be regulated by other IFNγ-inducible components, and this regulation is essential for the ability of the protein to localise correctly to PVMs of infecting T. gondii.
The GMS proteins are the IFN-inducible regulators of Irga6
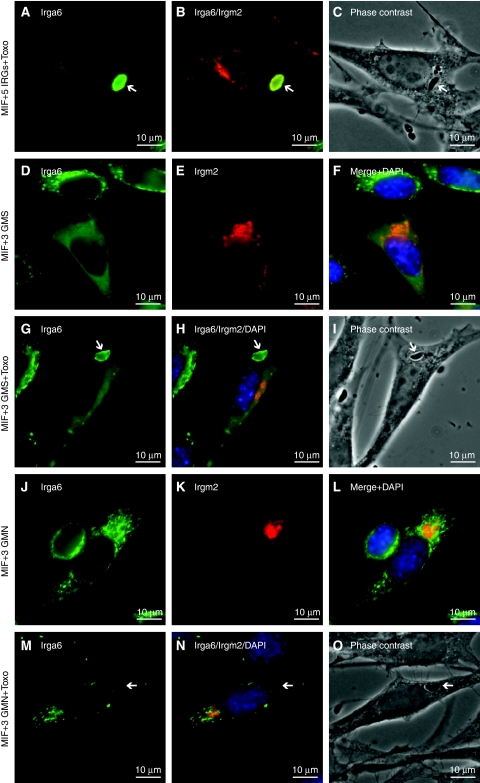
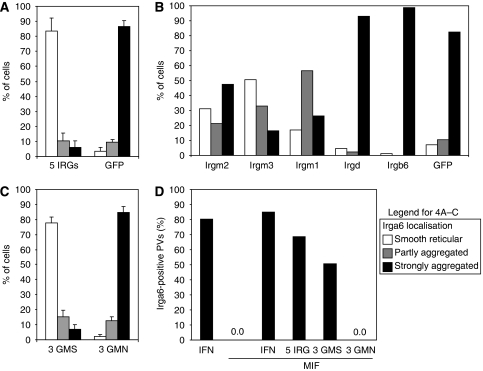
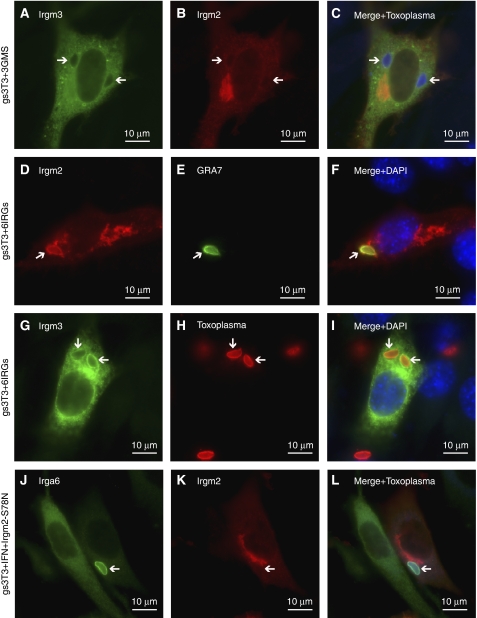
The non-redundancy documented for several IRG proteins in resistance to T. gondii suggested that the behaviour of Irga6 in IFNγ-induced cells may be regulated by the presence of other IRG proteins. We therefore attempted to reconstitute normal intracellular behaviour of WT Irga6 in the absence of IFNγ by transfecting MIF-induced gs3T3-Irga6 cells with a pool of expression vectors encoding Irgb6, Irgd, Irgm1, Irgm2 and Irgm3. Transfected cells identified by the expression of Irgm2 at the Golgi apparatus (Figure 3B; Martens and Howard, 2006) showed essentially complete restoration of the normal smooth distribution of Irga6 on the ER, and Irga6 also accumulated on the T. gondii PVM (Figures 3A–C, 4A and D). In control cells transfected with EGFP alone, Irga6 showed the usual cytoplasmic aggregates (Figure 4A). Only the GMS proteins proved to be necessary for this reconstitution, but surprisingly all three were required as transfection with any one or any two of the GMS proteins gave incomplete phenotypes independent of DNA dose (Figures 3D–I, and 4B and C and data not shown). Thus, the regulation of Irga6 behaviour in cells is controlled by the three anomalous GMS GTPases.
Figure 3.
Subcellular Irga6 localisation is dependent on the three GMS proteins. gs3T3-Irga6 cells were induced with IFNγ or MIF or both, or induced with MIF and transfected transiently with other IRGs (2 μg DNA total). Subsequently cells were infected with T. gondii or fixed immediately for analysis. Irga6 was identified by mAb 10E7 (green) and Irgm2 with H53 AS (red). Nuclei were stained with DAPI (blue). Intracellular T. gondii (arrows) were identified in phase contrast. Note the diffuse localisation of Irga6 only in cells transfected with the three WT GMS proteins (A, D, G) and the accumulation of Irga6 to the PVM under these conditions (A, G). (A–C) MIF-induced, infected cells transiently transfected with Irgm1, -m2, -m3, -b6 and -d (five IRGs). (D–I) MIF-induced cells transiently transfected with Irgm1, -m2 and -m3 (three GMS): (D–F) uninfected and (G–I) infected cells. (J–O) MIF-induced cells transiently transfected with Irgm1-S90N, Irgm2-S78N and Irgm3-S98N (three GMN). (J–L) uninfected and (M–O) infected cells.
Figure 4.
Quantification of the effect of GMS proteins on Irga6 localisation. Irga6 localisation in MIF-induced gs3T3-Irga6 cells transiently transfected with the indicated expression constructs (2 μg DNA total; see also Figure 3). (A–C) Effect of other IRG proteins on the resting localisation of Irga6, recorded as smooth reticular (WT) (white), partly aggregated (grey) and strongly aggregated (black). 150 cells were counted blind per data point. (A) Cells transfected with Irgm1, -m2, -m3, -b6 and -d (five IRGs) compared with cells transfected with EGFP. (B) Cells transfected with single IRG proteins. Each GMS protein alone showed a significant beneficial but incomplete effect on Irga6 localisation. Similar effects were also seen when pairs of GMS proteins were transfected (data not shown). Irgd was inactive. Cells transfected with the Irgb6 showed striking Irgb6 aggregates that largely colocalised with aggregated Irga6 (see also Figure 7A). (C) Cells transfected with a pool of the three WT GMS proteins were compared with cells transfected with the three inactive GMN mutants. (D) Accumulation of Irga6 at the T. gondii PV following induction with IFNγ, MIF or both, or MIF induction and transfection with Irgm1, -m2, -m3, -b6 and -d, the three GMS proteins or the three GMN mutants. Data are recorded as the percentage of Irga6-positive PVs per 100 intracellular T. gondii. Around 100 PVs per data point were counted blind.
It was reported that the mutation S98N in Irgm3, homologous to S83N of Irga6, prevents GTP binding (Taylor et al, 1997). We therefore prepared Irgm3-S98N and the homologous mutations Irgm1-S90N and Irgm2-S78N to ask whether the ability of GMS proteins to regulate Irga6 was dependent on their nucleotide-bound states. The co-transfection of all three mutant GMS sequences together did not reconstitute normal Irga6 behaviour in MIF-induced gs3T3-Irga6 cells (Figures 3J–O, and 4C and D). Thus, regulation of the nucleotide-bound state of Irga6 by GMS proteins is itself a nucleotide-dependent process.
Nucleotide-dependent direct interactions between IRGs in yeast two-hybrid assays
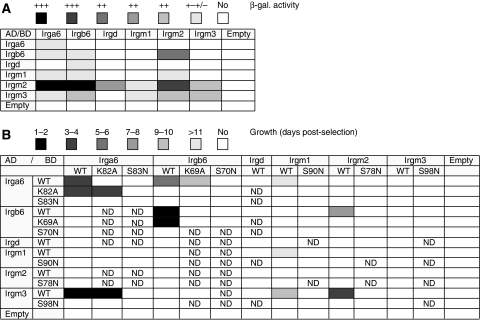
To determine whether nucleotide-dependent interactions between Irga6 and the GMS proteins involved physical contact, two yeast two-hybrid (Y2H) systems employing different DNA-binding (BD) and activation domain (AD) fusion partners for the IRG proteins and different expression properties were used (see Materials and methods). The IRG proteins showed multiple interactions in these assay systems and each family member displayed a distinct pattern of behaviour (Figure 5A and B). The strong homotypic interaction of Irga6 observed in the Gal4-based and weak interaction in the LexA Y2H system reproduced the in vitro oligomerisation of Irga6 in the presence of GTP (Uthaiah et al, 2003), strengthening the potential in vivo relevance of this interaction. Very strong homotypic interaction was also observed for Irgb6. As frequently seen in Y2H, the two assay systems highlighted distinct interactions and some of the interactions in both systems were unidirectional, probably as a result of the different structural impacts of the AD and BD on the fused IRGs (Estojak et al, 1995). Interactions involving Irgm1 were all weak, though reproducible.
Figure 5.
Nucleotide-dependent interactions of IRG proteins in Y2H. Graphic representation of the interaction behaviour of WT and mutant IRGs observed in Y2H. Interactions were measured by β-galactosidase activity in the LexA system (A) and growth on selective medium in the Gal4-based system (B). The strengths of the interactions are indicated in shades of grey. ND, not determined.
To confirm the specificity of the interactions seen and to determine their relevance to the in vivo findings, the Y2H assay in the Gal4-based system was repeated with nucleotide-binding site mutants of the individual proteins. All Y2H interactions of GMS proteins with other family members were completely abolished by the GMS to GMN mutants, Irgm3-S98N, Irgm2-S78N and Irgm1-S90N (Figure 5B). Likewise, Irga6-S83N was also completely negative for all interactions displayed by WT Irga6 (Figure 5B). This result is consistent with the essentially passive behaviour of Irga6-S83N in fibroblasts (see above). The corresponding mutant of Irgb6, S70N, similarly failed to show any of the interactions observed with Irgb6WT. The constitutively active mutant Irga6-K82A retained many interactions in Y2H (Figure 5B) including, perhaps surprisingly, the interaction with Irgm3 (see next section) but lost bi-directionality in its interaction with WT Irga6 and interaction with Irgb6 was completely abolished. The corresponding mutant of Irgb6, K69A, preserved its interactions with WT Irgb6 and Irga6, though the latter was weakened significantly, whereas the interactions with Irgm2 and Irgm3 were lost (Figure 5B).
Direct interaction between Irga6 and Irgm3 is dependent on GDP
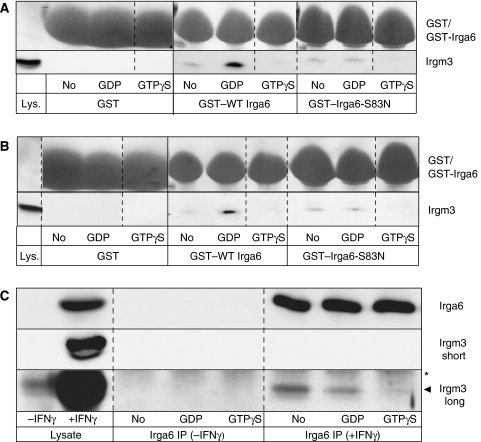
Functionally significant nucleotide-dependent interactions between Irga6 and the GMS proteins occur in IFNγ-induced cells, and the Y2H data suggested that at least some of these interactions were likely to be direct. The strongest interaction of Irga6 with a GMS protein in Y2H was with Irgm2 (Figure 5A) and Irgm3 (Figure 5B). Irgm3 localises, like Irga6, to the ER membrane in IFN-stimulated cells (Taylor et al, 1997) (Supplementary Figure S4). We accordingly used purified recombinant GST-tagged Irga6 bound to glutathione Sepharose beads to pull down Irgm3 from lysates of IFN-treated gs3T3 fibroblasts. In the absence of nucleotide, a very weak Irgm3 signal was detected. However, Irgm3 was efficiently pulled down upon addition of GDP to the binding reaction, whereas GTPγS inhibited the interaction completely (Figure 6A). When the nucleotide-binding deficient mutant Irga6-S83N was employed in the same assay, no increment in Irgm3 pull-down above background was seen on addition of GDP. Thus, Irga6 probably interacts with Irgm3 in the IFNγ-treated cell in the GDP-bound state. That the interaction between Irga6 and Irgm3 does not depend on other IFNγ-induced intermediaries was shown by a pull down of Irgm3 by Irga6 from MIF-induced gs3T3-Irgm3 cell lysates (Figure 6B). Again, strong interaction with WT Irga6 required GDP and was not observed with Irga6-S83N. The identity of the nucleotide-bound in Irgm3 when interacting with Irga6 is not clear but may also be GDP in view of the high concentration (0.5 mM) of the nucleotide maintained throughout the experiment. An interaction between Irga6 and Irgm3 was also shown by coimmunoprecipitation of Irgm3 with Irga6 from IFN-induced gs3T3 cells in the presence of exogenous GDP but not GTPγS (Figure 6C). The interaction was substoichiometric and was detected most strongly in the absence of exogenous nucleotide, presumably reflecting complexes formed in vivo containing trapped endogenous nucleotides of unknown identity. These experiments confirm the potential for physical interaction between Irga6 and Irgm3 suggested by the Y2H results. This interaction depends on the structural integrity of the nucleotide-binding site of Irga6, and both the pull-down and co-precipitation experiments suggest that the interaction may be favoured when Irga6 is in the GDP-bound state. The interaction of Irga6-K82A with Irgm3 in Y2H noted above might suggest that GTP binding by Irga6 does not necessarily inhibit the interaction, at least under the conditions of the Y2H experiments. However, it is unknown which nucleotide is bound to Irga6-K82A in the interaction with Irgm3 seen in Y2H. Furthermore, the apparently altered conformation of Irga6-K82A, which hinders interaction with WT Irga6 in Y2H (see above), may permit interaction with Irgm3 even in the presence of GTP.
Figure 6.
Nucleotide-dependent interaction of Irga6 with cellular Irgm3. Pull down of Irgm3 with recombinant GST–WT Irga6 and –Irga6-S83N protein from IFNγ-induced gs3T3 cells (A) and from MIF-induced gs3T3-Irgm3 cells (B) lysed in the presence or absence of guanine nucleotides. GST alone served as a negative control. Recombinant protein was visualised by Ponceau S staining of WB membranes (top row). Irgm3 was detected with αIrgm3 mAb (bottom row). Lys: 4% of lysate input. Dotted lines indicate positions where irrelevant lanes were removed from the image. (C) Coimmunoprecipitation of Irgm3 with Irga6 from IFNγ and non-induced gs3T3 cells in the absence or presence of exogenous nucleotides. Irga6 was detected on WB membranes with 10D7, Irgm3 with αIGTP mAb. The Irgm3 blot was exposed longer to visualise the substoichiometric interaction with Irga6 (bottom row 5 min, middle and top rows 10 s). Lysate: 5% of the input. Arrowhead: Irgm3; star: heavy chain of the immunoprecipitating AS (165); IP, immunoprecipitated.
Regulatory interactions between Irgb6 and the GMS proteins
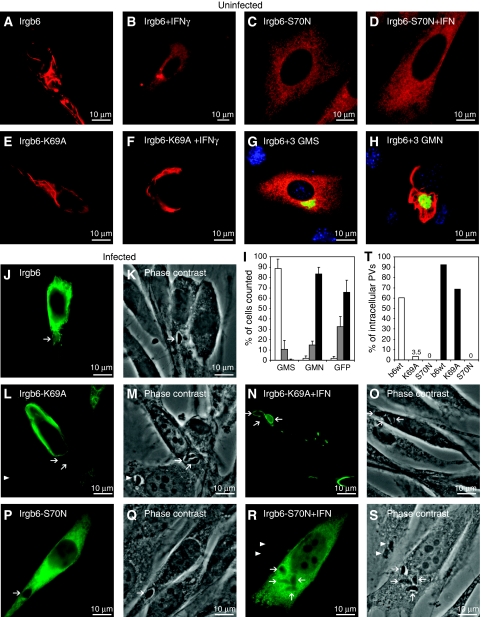
Most cell biological properties of Irgb6 resembled those described for Irga6. Native and C-terminally FLAG-tagged Irgb6 expressed by transfection in unstimulated cells formed distinctive aggregates (Figure 7A and data not shown). In IFNγ-induced cells, the aggregates were almost eliminated (Figure 7B). Irgb6-S70N formed no aggregates (Figure 7C and D), whereas Irgb6-K69A formed aggregates both in IFNγ-induced and uninduced cells, though some reduction of aggregates in IFNγ-induced cells was observed (Figure 7E and F). Aggregate formation by transfected Irgb6WT in the absence of IFNγ was eliminated by co-transfection with the three GMS proteins (Figure 7G and I) but not by co-transfection of the three GMN mutants (Figure 7H and I). However, unlike Irga6, WT Irgb6 could accumulate somewhat less efficiently (Figure 7T) on T. gondii PVM in cells not induced with IFNγ, despite the concomitant presence of Irgb6 cytoplasmic aggregates (Figure 7J and K). Irgb6-K69A could not accumulate on the PVM in uninduced cells (Figure7L and M), but could accumulate in IFNγ-induced cells, again in the presence of cytoplasmic aggregates (Figure 7N and O). Like Irga6-S83N, Irgb6-S70N was unable to accumulate on the PVM either in the absence (Figure 7P and Q) or presence (Figure 7R and S) of IFNγ.
Figure 7.
Irgb6 localisation in uninfected and T. gondii-infected fibroblasts. Localisation of transiently transfected (0.5 μg per construct), FLAG-tagged Irgb6 and G1-motif mutants in gs3T3 cells detected with the αFLAG mAb (red in A–H, green in J, L, N, P, R). Irgb6WT in uninduced (A) or IFNγ-induced (B) cells. Irgb6-S70N in uninduced (C) or IFNγ-induced (D) cells. Irgb6-K69A in uninduced (E) or IFNγ-induced (F) cells. Irgb6WT plus the three GMS WT (G) or GMN mutant (H) proteins in uninduced cells. Irgm2 (green) was detected with the H53 AS. (I) Quantification of Irgb6 localisation data illustrated in (A, G, H) recorded as smooth reticular WT staining (white), partly aggregated (grey) and strongly aggregated (black). A total of 150 cells were counted blind per data point. (J–S) Cells infected with T. gondii. Arrows indicate intracellular T. gondii identified in phase contrast. (J, K) Irgb6WT in uninduced cells. Irgb6-K69A in uninduced (L, M) or IFN-induced (N, O) cells. Irgb6-S70N in uninduced (P, Q) or IFN-induced cells (R, S). (T) The percentage of αFLAG M2 mAb-positive PVs was quantified in gs3T3 cells transiently transfected with WT Irgb6-FLAG and its mutants in the presence (black bars) and absence (white bars) of IFNγ.
The ability of transfected Irgb6WT to accumulate on the T. gondii PVM in uninduced cells was initially surprising. However, the results may suggest that in the absence of GMS proteins, WT Irga6 and Irgb6 establish equilibria between GTP-bound, aggregated and GDP-bound, free forms, which are more in favour of GDP in the case of Irgb6 than in the case of Irga6. In IFNγ-induced cells, the biochemically dominant-positive Irgb6-K69A can perhaps also be partially repaired by co-expressed GMS proteins, which further push the equilibrium towards the GDP-bound state and release monomers that can activate correctly at the PVM, associated with some reduction of aggregates (Figure 7N).
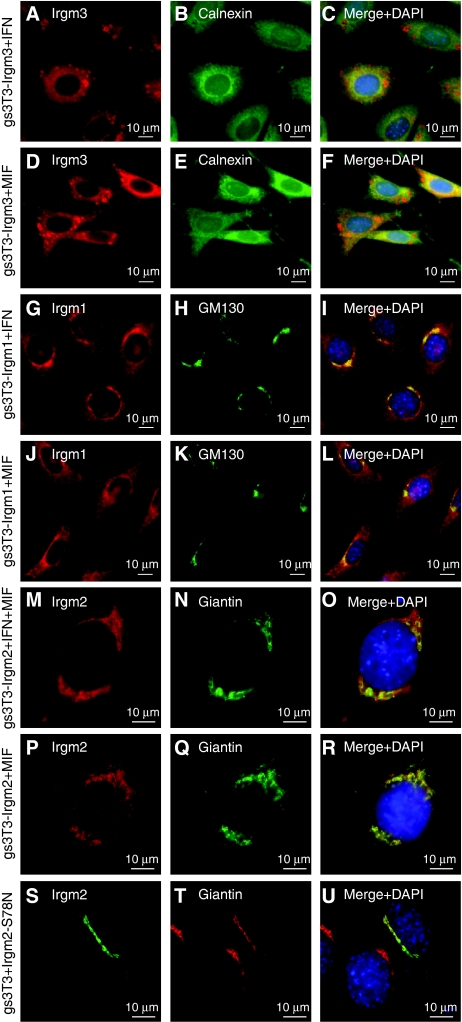
Localisation of GMS proteins in the absence of IFN
We have shown that GMS proteins are essential regulators of Irga6 and Irgb6. Unlike the GKS proteins, which have large cytosolic pools, the GMS proteins are largely or completely membrane-associated (Martens et al, 2004). Each GMS protein resides in a distinctive compartment: Irgm1 predominantly on Golgi (Martens et al, 2004) but also on endolysosomal membranes (Zhao et al, in preparation), Irgm2 on Golgi membranes (Martens and Howard, 2006) and Irgm3 on ER membranes and unidentified globular structures (Taylor et al, 1997; Martens et al, 2004) (Figure 8A–C, G–I and M–O; Supplementary Figure S4). These localisations were faithfully reproduced in uninduced gs3T3 cells transfected with single expression constructs (data not shown) and in MIF-induced stable gs3T3 cells (Figure 8D–F, J–L and P–R). As we show here for Irgm2 and as already shown for Irgm3 (Taylor et al, 1997) and Irgm1 (Martens et al, 2004), these localisations were unaffected by the inactivating GMS to GMN mutations in the G1 motif (Figure 8S–U).
Figure 8.
Irgm1, Irgm2 and Irgm3 localisation in uninfected fibroblasts is independent of IFNγ and nucleotide. gs3T3 cells expressing single GMS proteins were induced with IFNγ (A–C, G, H), MIF (D–F, J–L, P–R) or both (M–O). GMS proteins were colocalised with markers for defined cellular organelles: (A–F) gs3T3-Irgm3 cells, (A, D) αIrgm3 mAb; (B, E) αCalnexin AS; (C, F) merge with DAPI. (G–L) gs3T3-Irgm1 cells, (G, J) αIrgm1 AS; (H, K) αGM130 mAb; (I, L) merge with DAPI. (M–R) gs3T3-Irgm2 cells; (S–U) gs3T3 cells transiently transfected with Irgm2-S78N (1 μg), (M, P, S) αIrgm2 AS H53; (N, Q, T) αGiantin mAb; (O, R, U) merge with DAPI. Endogenous Irgm2 in gs3T3 cells is not detected by H53 AS due to a polymorphism between C57BL/6 and NIH Swiss mouse strains (unpublished data).
Both Irgm2 and Irgm3 have been shown to accumulate on the T. gondii PVM in IFNγ-induced, infected cells (Martens et al, 2005). We found that this accumulation depended on the presence of other IRG proteins. Thus, it did not occur in MIF-inducible gs3T3 cell lines expressing only Irgm2 or Irgm3 (data not shown). PVM localisation was also not seen in unstimulated gs3T3 cells transiently transfected with the three GMS proteins (Figure 9A–C). However, normal localisation of Irgm2 and Irgm3 to the T. gondii PVM was restored in cells transfected with the GKS proteins Irga6, Irgb6 and Irgd in addition to the three GMS proteins (Figure 9D–I) and when cells transfected with either Irgm2 or Irgm3 alone were co-induced with IFNγ (data not shown). Under these latter conditions, the ability of Irgm2 and Irgm3 to accumulate on the PVM was dependent on the integrity of the G1 motif (Figure 9J–L and data not shown).
Figure 9.
PVM localisation of Irgm2 and Irgm3 is dependent on other IRG proteins and nucleotide. gs3T3 cells infected with T. gondii. Parasites were detected with αGRA7 mAb (E, green), αT. gondii rabbit (H, red) or goat (C, L, blue) AS, Irga6 with mAb 10E7 (J, green), Irgm2 with H53 AS (B, D, K, red) and Irgm3 with αIrgm3 mAb (A, G, green). Nuclei were stained with DAPI (blue). Arrows indicate intracellular T. gondii. (A–C) Cells transiently transfected with expression constructs for Irgm1, -m2 and -m3 (666 ng each). (D–I) Accumulation of Irgm2 (D–F, red) and Irgm3 (G–I, green) at the PVM in uninduced cells transiently transfected with six IRGs (Irgm1, -m2, -m3, -a6, -b6 and -d; total 2 μg DNA). (J–L) IFNγ-induced cells transiently transfected with Irgm2-S78N (1 μg).
Discussion
We have shown that the cell-autonomous resistance mechanism mediated by the family of IFN-inducible IRG proteins is underlain by a complex pattern of nucleotide-dependent interactions between members of the family that determine both the positioning of the IRG proteins in the cell before infection and their subsequent ability to target the PV of the intracellular protozoal pathogen, T. gondii (see Supplementary Table SI). The three GMS proteins, Irgm1, Irgm2 and Irgm3, are necessary and sufficient to determine the normal intracellular localisation, including accumulation at the T. gondii PVM, of at least two of the GKS proteins, Irga6 (Figures 3 and 4) and Irgb6 (Figure 7). The regulatory function of the GMS proteins is dependent on the integrity of their nucleotide-binding sites (Figures 3, 4 and 7), as is the ability of the GKS proteins to relocate to the PVM (Figures 2 and 7). We also demonstrate direct nucleotide-dependent interactions between GMS and GKS proteins in Y2H systems (Figure 5). Furthermore, CD-immunoprecipitation and pull-down experiments employing purified Irga6 to capture cellular Irgm3, suggest that the nucleotide required for binding, at least in the case of Irga6, is GDP (Figure 6).
On the basis of these results, we can formulate a tentative model (Figure 10) of the regulatory regimen that governs Irga6 (and possibly also Irgb6) in the IFNγ-induced cell and mediates its rapid assembly on the PVM of infecting T. gondii. In the uninfected cell, we propose that the GMS proteins interact with GKS proteins in the GDP-bound state and inhibit uptake of GTP, acting as GDIs (Vetter and Wittinghofer, 2001) and functionally as attenuators of activation. The GMS proteins are largely or completely membrane-bound so these interactions presumably occur at cytoplasmic membranes rather than in the cytosol. Irga6 appears to exist in equilibrium between cytosol and ER membrane. Cytosolic Irga6 will be predominantly in the GDP-bound state on the basis of free cellular nucleotide concentrations (327 μM GTP/91 μM GDP) (Kleineke et al, 1979) and single-site equilibrium affinity constants of Irga6 for GTP of 15 μM and for GDP of 1 μM (Uthaiah et al, 2003). GMS proteins should attenuate GTP uptake by Irga6 primarily at ER membranes because the much higher local concentrations of Irga6 molecules will favour local GTP-dependent oligomerisation events at these endomembranes. The need to inhibit Irga6 (and other GKS proteins) activation on multiple endogenous membrane systems may explain why all three GMS proteins are required for full control of Irga6 and Irgb6 activation. Irgm1 localises to the Golgi (Martens et al, 2004) and endolysosomal membranes (Zhao et al, in preparation), Irgm2 to the Golgi (Martens and Howard, 2006; Figures 3 and 8) and Irgm3 to the ER (Taylor et al, 1997). We suggest that when GMS proteins are absent, as in resting cells transfected with Irga6 or when Irga6 is induced from a synthetic promoter, GTP-dependent oligomerisation initiates at cellular membranes and is uncontrolled, resulting in cytoplasmic aggregates (Figures 2D and 7A) and deformation of the ER (Supplementary Figure S2). Consistent with this interpretation is the observation that Irga6-S83N, which is unable to bind nucleotides (Figure 1), fails to form aggregates in uninduced cells (Figure 2Q), whereas Irga6-K82A that binds GTP, but does not measurably hydrolyse it (Figure 1), develops aggregates even in the IFNγ-induced cell (Figure 2N), either because GMS proteins are unable to interact normally with the mutant, as indicated by the modified interactions in Y2H assays (Figure 5B), and/or because incipient oligomerisation events cannot be terminated by GTP turnover. Finally, we have shown in a separate study that Irga6 aggregated in the cytoplasm or accumulated at the T. gondii PV in IFNγ-induced cells expresses an epitope detected by a monoclonal antibody (mAB), 10D7, that is dependent on the GTP-bound state of Irga6 (Papic et al, in preparation). Similar considerations apply to Irgb6. In support of the presented model, both Irga6 and Irgb6 form cytoplasmic aggregates in IFN-induced bone marrow-derived macrophages from Irgm1 and Irgm3 single knockouts as well in double-deficient mice (Taylor et al, in preparation).
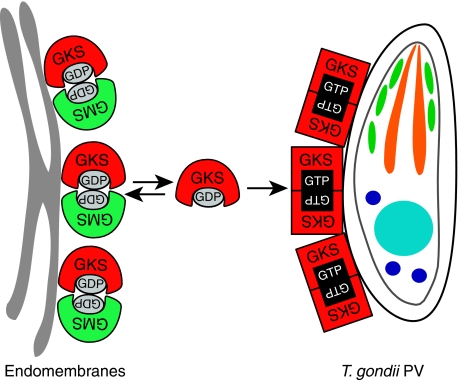
Figure 10.
Model of IRG protein regulation and function in resistance to T. gondii. In the IFN-induced, uninfected cell, GKS IRGs (red) are held in the inactive, GDP-bound form by transient GDP-dependent interactions with GMS proteins (green) on endomembranes. This membrane-bound form of GKS proteins is in equilibrium with free monomeric, GDP-bound molecules in the cytosol. Upon infection with T. gondii, cytosolic GKS molecules reach the PVM by diffusion. Because of the initial absence of inhibitory GMS proteins on the PVM, GKS proteins activate by GTP-dependent homo- and probably also hetero-oligomerisation promoting further IRG protein accumulation at the PV and ultimately vacuole rupture.
The access of cytosolic, presumably GDP-bound Irga6 or Irgb6 to the PVM, which is detectable within a few minutes after infection, may be by simple diffusion, as depolymerisation of microtubules by nocodazole has no effect on PVM localisation of Irga6 or Irgb6 (Khaminets et al, in preparation). We propose that activation of IRG proteins by GTP binding at the PVM may occur either through a local, perhaps transient, deficiency of GMS proteins at this membrane or through a specific, activating interaction with a factor presumably derived from the T. gondii and expressed on the cytosolic face of the PVM. Proteins derived from rhoptry secretions (Dubremetz, 2007) would be strong candidates for this role in view of their established association with T. gondii invasion and virulence. As Irga6 forms GTP-dependent oligomers with accelerated GTPase activity in vitro, the Irga6 observed at the PVM may be in the form of GTP-bound oligomers, possibly both homo- and hetero-oligomers in view of the strong nucleotide-dependent interaction in Y2H between Irga6 and Irgb6 (Figure 5). The membranes of some PVs also carry Irgm2 and Irgm3, but interestingly only those that also carry Irga6 and Irgb6 (Khaminets et al, in preparation). It is possible that these two regulatory GMS proteins bind secondarily if GDP-bound Irga6 is generated significantly at these sites following GTP hydrolysis.
Several non-Ras GTPases have recently been shown to accelerate GTP hydrolysis by G-domain–G-domain dimerisation (Sun et al, 2002; Egea et al, 2004; Ghosh et al, 2006; Scrima and Wittinghofer, 2006), the two G-domains functioning effectively as GAP proteins for each other. Such mutually activating GTPase pairs may be homodimeric (GBP1) or heterodimeric (SRP, TOC, MnmE). We now know that accelerated hydrolysis of GTP in Irga6 oligomers in vitro also occurs through reciprocal G-domain–G-domain interactions (Pawlowski et al, in preparation). As noted above, hetero-oligomeric interactions, as between Irga6 and Irgb6, may also contribute to events occurring at the PVM in vivo. By analogy, in the resting state, Irga6 and Irgb6 may be negatively regulated by heterodimeric G-domain interactions with GMS members of the IRG family, perhaps by successful competition with Irga6 and Irgb6 for the G-domain interaction site. The enhancement of Irgm3 pull-down by the addition of GDP suggests that GDP is an essential cofactor in this regulation. In the model (Figure 10), we have indicated that both Irga6 and its GMS attenuator are in the GDP-bound state but this view, though plausible, needs verification by an in vitro reconstruction of the interaction since Taylor and colleagues reported some years ago that the guanine nucleotides co-precipitated with Irgm3 from IFNγ-induced cells were 96% GTP to 4% GDP (Taylor et al, 1997). Unfortunately, nothing is yet known about the hydrolysis, binding specificity and affinity of any of the GMS proteins for nucleotide so it is difficult to put these earlier observations into context. At present, in the absence of information to the contrary, the action of the GMS proteins in limiting GDP to GTP nucleotide exchange, and thus preventing activation of Irga6 and Irgb6, seems analogous to the action of GDI proteins in controlling the activation of small GTPases (Vetter and Wittinghofer, 2001). Interestingly, many GDI proteins also regulate the shuttling of their target GTPases between intracellular compartments (DerMardirossian and Bokoch, 2005), as GMS proteins regulate access of Irga6 to the T. gondii PVM.
The finding that the IRG GTPases are involved in complex regulatory interactions with each other may account for some obscure observations in the IRG field. Bernstein-Hanley et al (2006) observed that overexpression of Irgm3 resulted in impaired resistance to Chlamydia trachomatis in MEFs, consistent with the here proposed role of Irgm3 as an attenuator of GKS activation. Second, our data show that GMS proteins are required for normal regulation of the nucleotide status and subcellular localisation of Irga6 and Irgb6. Thus, the susceptibility phenotypes shown by mice with disrupted Irgm1 or Irgm3 genes may not be due to non-redundant activity of GMS gene products against the pathogens, but rather to dysregulation of other IRG proteins. Furthermore, absence of GMS proteins leads to formation of ectopic aggregates of the GKS proteins (Figures 2 and 7) that may have cytopathic consequences. This hints at an explanation for the lymphomyeloid failure repeatedly observed in Irgm1-deficient mice infected with strongly immunostimulatory pathogens such as Mycobacteria (Feng et al, 2004, 2008) and Trypanosoma (Santiago et al, 2005), if such cytopathic aggregates form in lymphomyeloid stem cells (Howard, 2007). In this context, Carlow et al (1998) reported problems recovering transfected clones stably expressing Irgb6 in vitro, and in those few clones recovered, Irgb6 expression was low and unstable, again suggesting cytopathic effects of expressing GKS proteins ‘unprotected' by their GMS regulators. Finally, regulatory interactions between GMS and GKS GTPases have been invoked recently, in the absence of a specific model, to account for the unexpected phenotype of mice deficient for both Irgm1 and Irgm3. The severe deficiency in resistance against Salmonella seen in the Irgm1 single-deficient mouse (Henry et al, 2007) was largely reversed in the doubly deficient mouse (Taylor et al, in preparation).
Here, we show that IRG proteins function in a system of nucleotide-dependent interactions that regulate the behaviour of these powerful resistance proteins in response to intracellular infection. Detailed insight into mechanistic aspects of these effects will have to await the purification of more IRG family members, in particular of the GMS proteins. At a functional level, the next challenge is to understand how the binding and hydrolysis of GTP by IRG proteins localised to the T. gondii PVM contributes to the destruction of the vacuole and the subsequent demise of the pathogen.
Materials and methods
Expression constructs
Expression constructs were generated as described in the Supplementary data.
Cell culture
gs3T3 cells (Invitrogen) and C57BL/6 MEFs (Boehm et al, 1998) were cultured in DMEM, high glucose (Invitrogen), 10% FCS (Biochrom), 2 mM L-glutamine, 1 mM sodium pyruvate, 1 × MEM non-essential amino acids, 100 U/ml penicillin, 100 μg/ml streptomycin (all PAA), transiently transfected using FuGENE6 (Roche) according to the manufacturer's instructions, induced with 200 U/ml of mouse IFNγ (Cell Concepts) or 10−9 M MIF (Invitrogen) for 24 h.
Generation of inducible cell lines
gs3T3 cells were transfected with linearised pGene-IRG constructs by calcium phosphate precipitation (Graham and van der Eb, 1973) and selected for stable integration with 200 μg/ml Zeocin (Invivogen). The integrated pSwitch inducer plasmid was maintained with 50 μg/ml hygromycin (Invivogen). Clones expressing comparable protein amounts following MIF and IFNγ induction were identified by SDS–PAGE and WB (Supplementary Figure S3; see Supplementary data).
Immunoreagents
Immunoreagents used were αIrga6 165 rabbit antiserum (AS) (Martens et al, 2004), 10E7 and 10D7 mouse mAbs, αIGTP (Irgm3) mAb (BD Biosciences), αIrgb6 A20 goat AS and αIrgm1 A19 goat AS (Santa Cruz Biotechnology), αIrgm2 H53 rabbit AS (Martens et al, 2005), αIrgd 2078 rabbit AS (Martens et al, 2004), αT. gondii rabbit (BioGenex) and goat AS (Abcam), αGRA7 5-241-178 mouse mAb (Bonhomme et al, 1998), αctag1 2600 rabbit AS (Martens et al, 2005), αGM130 mAb (BD Biosciences), αGiantin mAb (Linstedt and Hauri, 1993), αCalnexin AS (StressGene), Alexa 350/488/555 labelled donkey αmouse, -rabbit and -goat sera (Molecular Probes), donkey αrabbit- (GE Healthcare), donkey αgoat- (Santa Cruz Biotechnology) and goat αmouse-HRP (Pierce) AS.
Immunocytochemistry
Immunocytochemistry was performed as described earlier (Martens et al, 2005), analysed using an Axioplan II fluorescence microscope and AxioCam MRm camera and images were processed with Axiovision4 (Zeiss).
Infection of fibroblasts
ME49 strain T. gondii tachyzoites were passaged in vitro and used for infection of untreated, transiently transfected, IFNγ- and/or MIF-induced fibroblasts at a multiplicity of infection of six for 2 h as described by Martens et al (2005). Intracellular parasites were identified by immunostaining for the T. gondii protein GRA7 or in phase contrast.
Y2H
In the GAL4-based Y2H system, the IRGs were expressed constitutively at low level as N-terminal fusions with the Gal4 BD and Gal4 AD (James et al, 1996). pGAD- and pGBD-IRG constructs were lithium acetate transformed (Gietz et al, 1995) into PJ69-4a-a and -α yeast cells, respectively, followed by selection on synthetic complete (SC) medium lacking Leu (SC-L) or Trp (SC-T) (James et al, 1996). Co-expression was achieved by mating on YPD plates (Sherman, 2002) and selection for diploid cells on SC-L-T. Protein–protein interaction was determined by growth on SC-L-T also lacking Ade and His. At least two independent crossings were performed in each case. The Y2H analysis using the Matchmaker LexA-system (Clontech) based on inducible expression of the IRGs as N-terminal fusions with the LexA BD and B42 AD was performed as described by Kaiser et al (2004) and Kaiser (2005).
Expression and purification of recombinant protein
WT Irga6 and mutant proteins were expressed as N-terminal GST fusions from pGEX-4T-2 constructs in Escherichia coli BL-21 upon overnight (ON) induction with 0.1 mM IPTG at 18°C. The cells were lysed in B1 buffer (PBS/2 mM DTT)/Complete Mini Protease Inhibitor Cocktail, EDTA free (Roche) using a microfluidiser (EmulsiFlex-C5; Avestin). Cleared lysates were purified on a GSTrap FF glutathione Sepharose affinity column (GE Healthcare) in B1 buffer. GST was cleaved off by ON incubation of the resin with thrombin (Serva) at 4°C. Free Irga6 was eluted with B1 buffer. Irga6 containing fractions were subjected to size exclusion chromatography (Superdex 75; GE Healthcare) in B2 buffer (50 mM Tris–HCl pH 7.4, 5 mM MgCl2 and 2 mM DTT). Pure Irga6 proteins were concentrated with Vivaspin 20 centrifugal concentrators (Sartorius).
Pull down
IFNγ- and MIF-induced gs3T3 cells were lysed for 1 h at 4°C in lysis buffer (PBS, 0.1% Thesit (Sigma-Aldrich), 3 mM MgCl2, Complete Mini Protease Inhibitor Cocktail, EDTA free (Roche)) with or without 0.5 mM GDP or GTPγS (Sigma-Aldrich). Postnuclear supernatants were incubated at 4°C ON with glutathione Sepharose-bound (high performance; GE Healthcare) recombinant GST–Irga6 and –Irga6-S83N that was pre-incubated for 1 h with or without 1 mM GDP or GTPγS in PBS, 5 mM MgCl2 and 1 mM DTT. Bound cellular proteins were eluted from the washed beads with elution buffer (30 min RT) and subjected to SDS–PAGE and WB. Input of recombinant Irga6 was monitored by Ponceau S staining.
Coimmunoprecipitation
IFNγ-induced and untreated gs3T3 cells were lysed in lysis buffer in the absence or presence of 0.5 mM GDP and GTPγS (both Sigma-Aldrich). Irga6-specific AS 165 was bound to protein A Sepharose CL-4B (GE Healthcare) and crosslinked using 20 mM dimethyl pimelimidate (Sigma-Aldrich) (Harlow and Lane, 1988). Coupled beads were incubated with the postnuclear cell lysates for 2 h at 4°C. Bound proteins were eluted from washed beads with elution buffer (100 mM Tris pH 8.5 and 0.5% SDS) for 30 min at room temperature and subjected to SDS–PAGE and WB. One-quarter of the eluate was used for detection with αIrga6 mAb 10D7 and three-quarters for detection with the αIrgm3 mAb.
Guanine nucleotide-binding parameters
The nucleotide-binding affinities of WT Irga6, -K82A and -S83N for mGDP and mGTPγS (Jena Bioscience) were determined by equilibrium titration of 0–100 μM protein against 0.5 μM mant nucleotides in B2 buffer at 20°C. The mant nucleotides were excited at 355 nm, and monitored at 448 nm (Aminco-Bowman 2 Luminescence Spectrometer; SLM Instruments). Equilibrium dissociation constants were obtained as described by Herrmann and Nassar (1996). The fluorescence emission intensity increases upon binding of mant nucleotides to GTPases due to exclusion of solvent that otherwise quenches the fluorophore in solution (Rojas et al, 2003). Thus, mGTPγS seemed to be more solvent exposed when bound to Irga6-K82A than to WT Irga6 (Figure 1B).
GTP hydrolysis assay
Here, 80 μM WT Irga6 and mutant recombinant proteins were incubated with 10 mM GTP (Sigma-Aldrich) containing traces of α32P-labelled GTP (GE Healthcare) at 37°C in B2 buffer for up to 3 h. Reactions were separated on PEI Cellulose F TLC plates (Merck) in 1 M acetic acid and 0.8 M LiCl. Signals were detected with the BAS 1000 phosphoimager analysis system (Fujifilm) and quantified with the AIDA Image Analyser v3 software (Raytest). Similar results were obtained with 1 mM GTP and 50 μM protein (data not shown).
Oligomerisation assays
Oligomerisation of 80 μM Irga6 in the presence of 10 mM GDP and GTP, respectively, in B2 buffer at 37°C was determined by conventional and dynamic light scattering. Conventional light scattering was performed at 350 nm in a DM45 Spectrofluorimeter (Olis) and dynamic light scattering at 650 nm with a DynaPro molecular sizing instrument (Protein Solutions; Wyatt Technologies). Data were obtained and analysed using the DYNAMICS software (v.5).
Supplementary Material
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Information
Acknowledgments
We thank G Taylor, J Coers and G Praefcke for critical commentary on earlier versions of this paper and for valuable discussions, the Centre for Ultrastructural Imaging, King's College London, for generation of the electron microscopy images, G Reichmann and H-P Hauri for the gift of αGRA7 and αGiantin mAb, respectively, and J Dohmen for the Y2H system. C Poschner assisted with cell culture. This study was supported by the DFG grants SPP1110, SFB635 and SFB670, by the University of Cologne and by a stipend from the Cologne Graduate School in Genetics and Functional Genomics (JPH).
References
- Bekpen C, Hunn JP, Rohde C, Parvanova I, Guethlein L, Dunn DM, Glowalla E, Leptin M, Howard JC (2005) The interferon-inducible p47 (IRG) GTPases in vertebrates: loss of the cell autonomous resistance mechanism in the human lineage. Genome Biol 6: R92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein-Hanley I, Coers J, Balsara ZR, Taylor GA, Starnbach MN, Dietrich WF (2006) The p47 GTPases Igtp and Irgb10 map to the Chlamydia trachomatis susceptibility locus Ctrq-3 and mediate cellular resistance in mice. Proc Natl Acad Sci USA 103: 14092–14097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm U, Guethlein L, Klamp T, Ozbek K, Schaub A, Futterer A, Pfeffer K, Howard JC (1998) Two families of GTPases dominate the complex cellular response to IFN-gamma. J Immunol 161: 6715–6723 [PubMed] [Google Scholar]
- Bonhomme A, Maine GT, Beorchia A, Burlet H, Aubert D, Villena I, Hunt J, Chovan L, Howard L, Brojanac S, Sheu M, Tyner J, Pluot M, Pinon JM (1998) Quantitative immunolocalization of a P29 protein (GRA7), a new antigen of Toxoplasma gondii. J Histochem Cytochem 46: 1411–1422 [DOI] [PubMed] [Google Scholar]
- Butcher BA, Greene RI, Henry SC, Annecharico KL, Weinberg JB, Denkers EY, Sher A, Taylor GA (2005) p47 GTPases regulate Toxoplasma gondii survival in activated macrophages. Infect Immun 73: 3278–3286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlow DA, Teh SJ, Teh HS (1998) Specific antiviral activity demonstrated by TGTP, a member of a new family of interferon-induced GTPases. J Immunol 161: 2348–2355 [PubMed] [Google Scholar]
- Collazo CM, Yap GS, Sempowski GD, Lusby KC, Tessarollo L, Woude GF, Sher A, Taylor GA (2001) Inactivation of LRG-47 and IRG-47 reveals a family of interferon gamma-inducible genes with essential, pathogen-specific roles in resistance to infection. J Exp Med 194: 181–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DerMardirossian C, Bokoch GM (2005) GDIs: central regulatory molecules in Rho GTPase activation. Trends Cell Biol 15: 356–363 [DOI] [PubMed] [Google Scholar]
- Dubremetz JF (2007) Rhoptries are major players in Toxoplasma gondii invasion and host cell interaction. Cell Microbiol 9: 841–848 [DOI] [PubMed] [Google Scholar]
- Egea PF, Shan SO, Napetschnig J, Savage DF, Walter P, Stroud RM (2004) Substrate twinning activates the signal recognition particle and its receptor. Nature 427: 215–221 [DOI] [PubMed] [Google Scholar]
- Estojak J, Brent R, Golemis EA (1995) Correlation of two-hybrid affinity data with in vitro measurements. Mol Cell Biol 15: 5820–5829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feig LA, Cooper GM (1988) Inhibition of NIH 3T3 cell proliferation by a mutant ras protein with preferential affinity for GDP. Mol Cell Biol 8: 3235–3243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng CG, Collazo-Custodio CM, Eckhaus M, Hieny S, Belkaid Y, Elkins K, Jankovic D, Taylor GA, Sher A (2004) Mice deficient in LRG-47 display increased susceptibility to mycobacterial infection associated with the induction of lymphopenia. J Immunol 172: 1163–1168 [DOI] [PubMed] [Google Scholar]
- Feng CG, Weksberg DC, Taylor GA, Sher A, Goodell MA (2008) The p47 GTPase Lrg-47 (Irgm1) links host defense and hematopoietic stem cell proliferation. Cell Stem Cell 2: 83–89 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A, Praefcke GJ, Renault L, Wittinghofer A, Herrmann C (2006) How guanylate-binding proteins achieve assembly-stimulated processive cleavage of GTP to GMP. Nature 440: 101–104 [DOI] [PubMed] [Google Scholar]
- Ghosh A, Uthaiah R, Howard J, Herrmann C, Wolf E (2004) Crystal structure of IIGP1; a paradigm for interferon-inducible p47 resistance GTPases. Mol Cell 15: 727–739 [DOI] [PubMed] [Google Scholar]
- Gietz RD, Schiestl RH, Willems AR, Woods RA (1995) Studies on the transformation of intact yeast cells by the LiAc/SS-DNA/PEG procedure. Yeast 11: 355–360 [DOI] [PubMed] [Google Scholar]
- Graham FL, van der Eb AJ (1973) A new technique for the assay of infectivity of human adenovirus 5 DNA. Virology 52: 456–467 [DOI] [PubMed] [Google Scholar]
- Harlow E, Lane D (1988) Antibodies. A laboratory manual. Cold Spring Harbor, NY, USA: Cold Spring Harbor Laboratory [Google Scholar]
- Henry SC, Daniell X, Indaram M, Whitesides JF, Sempowski GD, Howell D, Oliver T, Taylor GA (2007) Impaired macrophage function underscores susceptibility to Salmonella in mice lacking Irgm1 (LRG-47). J Immunol 179: 6963–6972 [DOI] [PubMed] [Google Scholar]
- Herrmann C, Nassar N (1996) Ras and its effectors. Prog Biophys Mol Biol 66: 1–41 [DOI] [PubMed] [Google Scholar]
- Howard JC (2007) Introduction: cell-autonomous immunity. Microbes Infect 9: 1633–1635 [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA (1996) Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144: 1425–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaiser F (2005) Molekulare Charakterisierung und funktionelle Analyse der Interferon-induzierten 47 kDa GTPase IIGP. PhD Thesis, Institut für Biotechnologie, Technische Universität Berlin, Germany
- Kaiser F, Kaufmann SH, Zerrahn J (2004) IIGP, a member of the IFN inducible and microbial defense mediating 47 kDa GTPase family, interacts with the microtubule binding protein hook3. J Cell Sci 117: 1747–1756 [DOI] [PubMed] [Google Scholar]
- Kleineke J, Duls C, Soling HD (1979) Subcellular compartmentation of guanine nucleotides and functional relationships between the adenine and guanine nucleotide systems in isolated hepatocytes. FEBS Lett 107: 198–202 [DOI] [PubMed] [Google Scholar]
- Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, Yap GS (2006) Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J Exp Med 203: 2063–2071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linstedt AD, Hauri HP (1993) Giantin, a novel conserved Golgi membrane protein containing a cytoplasmic domain of at least 350 kDa. Mol Biol Cell 4: 679–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Howard J (2006) The interferon-inducible GTPases. Annu Rev Cell Dev Biol 22: 559–589 [DOI] [PubMed] [Google Scholar]
- Martens S, Parvanova I, Zerrahn J, Griffiths G, Schell G, Reichmann G, Howard JC (2005) Disruption of Toxoplasma gondii parasitophorous vacuoles by the mouse p47-resistance GTPases. PLoS Pathog 1: e24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martens S, Sabel K, Lange R, Uthaiah R, Wolf E, Howard JC (2004) Mechanisms regulating the positioning of mouse p47 resistance GTPases LRG-47 and IIGP1 on cellular membranes: retargeting to plasma membrane induced by phagocytosis. J Immunol 173: 2594–2606 [DOI] [PubMed] [Google Scholar]
- Pitossi F, Blank A, Schroder A, Schwarz A, Hussi P, Schwemmle M, Pavlovic J, Staeheli P (1993) A functional GTP-binding motif is necessary for antiviral activity of Mx proteins. J Virol 67: 6726–6732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Praefcke GJ, Kloep S, Benscheid U, Lilie H, Prakash B, Herrmann C (2004) Identification of residues in the human guanylate-binding protein 1 critical for nucleotide binding and cooperative GTP hydrolysis. J Mol Biol 344: 257–269 [DOI] [PubMed] [Google Scholar]
- Rojas RJ, Kimple RJ, Rossman KL, Siderovski DP, Sondek J (2003) Established and emerging fluorescence-based assays for G-protein function: Ras-superfamily GTPases. Comb Chem High Throughput Screen 6: 409–418 [DOI] [PubMed] [Google Scholar]
- Santiago HC, Feng CG, Bafica A, Roffe E, Arantes RM, Cheever A, Taylor G, Vieira LQ, Aliberti J, Gazzinelli RT, Sher A (2005) Mice deficient in LRG-47 display enhanced susceptibility to Trypanosoma cruzi infection associated with defective hemopoiesis and intracellular control of parasite growth. J Immunol 175: 8165–8172 [DOI] [PubMed] [Google Scholar]
- Scrima A, Wittinghofer A (2006) Dimerisation-dependent GTPase reaction of MnmE: how potassium acts as GTPase-activating element. EMBO J 25: 2940–2951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F (2002) Getting started with yeast. Methods Enzymol 350: 3–41 [DOI] [PubMed] [Google Scholar]
- Sigal IS, Gibbs JB, D'Alonzo JS, Temeles GL, Wolanski BS, Socher SH, Scolnick EM (1986) Mutant ras-encoded proteins with altered nucleotide binding exert dominant biological effects. Proc Natl Acad Sci USA 83: 952–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun YJ, Forouhar F, Li Hm HM, Tu SL, Yeh YH, Kao S, Shr HL, Chou CC, Chen C, Hsiao CD (2002) Crystal structure of pea Toc34, a novel GTPase of the chloroplast protein translocon. Nat Struct Biol 9: 95–100 [DOI] [PubMed] [Google Scholar]
- Taylor GA, Collazo CM, Yap GS, Nguyen K, Gregorio TA, Taylor LS, Eagleson B, Secrest L, Southon EA, Reid SW, Tessarollo L, Bray M, McVicar DW, Komschlies KL, Young HA, Biron CA, Sher A, Vande Woude GF (2000) Pathogen-specific loss of host resistance in mice lacking the IFN-gamma-inducible gene IGTP. Proc Natl Acad Sci USA 97: 751–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor GA, Jeffers M, Largaespada DA, Jenkins NA, Copeland NG, Woude GF (1996) Identification of a novel GTPase, the inducibly expressed GTPase, that accumulates in response to interferon gamma. J Biol Chem 271: 20399–20405 [DOI] [PubMed] [Google Scholar]
- Taylor GA, Stauber R, Rulong S, Hudson E, Pei V, Pavlakis GN, Resau JH, Vande Woude GF (1997) The inducibly expressed GTPase localizes to the endoplasmic reticulum, independently of GTP binding. J Biol Chem 272: 10639–10645 [DOI] [PubMed] [Google Scholar]
- Uthaiah RC (2002) Biochemical, structural and cellular studies on IIGP1, a member of the p47 family of GTPases. PhD Thesis, Institute for Genetics, Cologne, Germany
- Uthaiah RC, Praefcke GJ, Howard JC, Herrmann C (2003) IIGP1, an interferon-gamma-inducible 47-kDa GTPase of the mouse, showing cooperative enzymatic activity and GTP-dependent multimerization. J Biol Chem 278: 29336–29343 [DOI] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A (2001) The guanine nucleotide-binding switch in three dimensions. Science 294: 1299–1304 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Figure S1
Supplementary Figure S2
Supplementary Figure S3
Supplementary Figure S4
Supplementary Information