Abstract
Natural killer (NK) cells are innate immune cells that mediate resistance against viruses and tumors. They express multiple activating receptors that couple to immunoreceptor tyrosine-based activation motif (ITAM)–containing signaling chains for downstream cell activation. Ligation of activating NK-cell receptors induces NK-cell cytotoxicity and cytokine release. How these distinct events are selectively controlled is not well defined. Here we report the identification of a specific signaling pathway that operates downstream of the ITAM-coupled NK-cell receptors NK1.1, Ly49D, Ly49H, and NKG2D. Using primary NK cells from Bcl10−/−, Malt1−/−, Carma1−/−, and Card9−/− mice, we demonstrate a key role for Bcl10 signalosomes in the activation of canonical NF-κB signaling as well as JNK and p38 MAPK upon NK-cell triggering. Bcl10 directly cooperates with Malt1 and depends on Carma1 (Card11) but not on Card9 for NK-cell activation. These Bcl10-dependent cascades selectively control cytokine and chemokine production but do not affect NK-cell differentiation or killing. Thus, we identify a molecular basis for the segregation of NK-cell receptor–induced signals for cytokine release and target cell killing and extend the previously recognized roles for CARD-protein/Bcl10/Malt1 complexes in ITAM receptor signaling in innate and adaptive immune cells.
Introduction
Natural killer (NK) cells are key components of the innate immune system that mediate resistance against pathogens, particularly against viruses and tumors.1–3 They comprise up to 15% of peripheral blood lymphocytes and are found in various peripheral tissues where they use perforin-containing cytolytic granules and cytokine production to mediate their effector function. NK-cell cytotoxicity and cytokine release are modulated by a wide variety of cell surface receptors that recognize and respond to transformed or infected cells. These NK-cell receptors can broadly be classified into inhibitory and activating receptors. How the distinct NK-cell receptors selectively mediate cytotoxicity and cytokine production remains an active area of research.3
The inhibitory NK-cell receptors monitor normal expression of cellular host proteins, particularly MHC class I molecules.1–3 In the absence of these molecules, for example on tumor cells, NK cells are released from their inhibition to kill their target. The activating NK-cell receptors in contrast recognize specific ligands that are induced by viral infection, cellular stress, or neoplastic transformation and directly activate NK cells for cytokine production and target cell killing. Prototypic examples of activating NK-cell receptors are NK1.1, Ly49D, Ly49H, and NKG2D, which possess charged transmembrane residues that mediate association with the immunoreceptor tyrosine-based activation motif (ITAM)–containing signaling chains DAP-12, FcRγ, or CD3ζ for downstream cell activation. NKG2D can also associate with the transmembrane adapter DAP-10.
Ligation of activating NK-cell receptors leads to a rapid tyrosine phosphorylation of the ITAM in the signaling chain mediated by Src family tyrosine kinases. This allows recruitment and activation of Syk or Zap70 kinases to the phosphorylated ITAM, which in turn recruits and activates phosphoinositide 3-kinases (PI3Ks) and other signaling mediators such as LAT, PLCγ1, PLCγ2, Vav2, and Vav3 and mobilizes free calcium.3,4 NKG2D signaling can in addition lead to a direct recruitment of PI3Ks to the phosphorylated YINM motif of DAP10. These proximal signaling events result in a cytoskeletal remodeling causing NK-cell degranulation necessary for rapid target cell killing, and they induce a reprogramming of gene transcription for the production of proinflammatory cytokines and chemokines. In addition, NK cells can also be activated by certain cytokines such as IL-12 and IL-18, which signal via ITAM-independent mechanisms.5
One key transcription factor that controls the expression of proinflammatory gene products in immune cells of many different lineages is nuclear factor κB (NF-κB).6 In resting cells, the activity of NF-κB is tightly controlled by inhibitory κB (IκB) proteins that can bind to NF-κB dimers and retain these in an inactive state in the cytoplasm. A conserved canonical NF-κB activation pathway operates downstream of most NF-κB–inducing stimuli to rapidly activate NF-κB. This signaling cascade depends on the activation of a multisubunit IκB kinase (IKK) that phosphorylates IκB proteins on conserved serine residues to target them for ubiquitin-dependent degradation. This process frees NF-κB and allows its translocation into the nucleus for the transactivation of target genes. Many of the proinflammatory cytokine genes that are induced by NK-cell activation, including TNF-α, GM-CSF, and IFN-γ are regulated by NF-κB.6–8 Yet, the exact molecular mechanisms that couple activating NK-cell receptors to the canonical NF-κB pathway are undefined.
The caspase recruitment domain (CARD) protein Bcl10 is a key regulator of ITAM-dependent antigen receptor signaling in both T and B lymphocytes.9 Recent work has revealed additional roles for Bcl10 downstream of the ITAM-coupled Fc∈RI in mast cells10,11 and downstream of various ITAM-coupled receptors on dendritic cells (DCs) and macrophages.12,13 For cell activation, Bcl10 can directly bind to the paracaspase Malt1 to assemble signalosomes that control the context-specific activation of IKK and NF-κB and in addition regulate the JNK and p38 MAP kinase pathways. However, Bcl10 also functions independently of Malt1, for example during neurodevelopment and in the control of B-cell proliferation.14–18 Individual receptor systems use distinct scaffold molecules that contain CARDs and coiled-coil domains to couple to Bcl10 signaling modules.19 In the immune system, the CARD-coiled-coil protein Carma1 (or Card11) is essential to relay TCR, BCR, and Fc∈RI signaling to Bcl10,20–22 whereas the related molecule Card9 is used by the ITAM-containing C-type lectin receptor Dectin-1 or other ITAM-coupled receptors on DCs and macrophages.12,13 Moreover, certain G protein–coupled receptors in nonimmune cells use Carma3 (Card14) to engage Bcl10 signaling for NF-κB activation.23
Based on the established role for Bcl10 in ITAM receptor signaling in T, B, and myeloid cells, we hypothesized that Bcl10-containing signalosomes could also play critical roles in NK-cell activation. Here we use primary NK-cells from Bcl10-, Malt1-, Carma1-, and Card9-deficient animals to analyze the function of these proteins in NK-cell activation. We extend the recently shown function of Bcl10 in NK-cell signaling24 and demonstrate the existence of a Card9-independent Carma1/Bcl10/Malt1 pathway that is engaged by multiple activating NK-cell receptors. We directly show that this cascade couples to the canonical IκB-controlled NF-κB pathway and in addition to JNK and p38 MAPK activation and that it is specifically required for cytokine production but largely dispensable for NK-cell killing.
Methods
Mice
Bcl10, Malt1, Carma1, and Card9 knockout mice have been described previously.12,16,17,25 All mice were backcrossed into C57BL/6J genetic background for at least 5 generations. Animals were housed in accordance with standard protocols and German, European, and US laws and regulations.
Cytotoxicity assays
To preactivate NK cells, donor mice were infected with 5 × 105 pfu murine cytomegalovirus (MCMV salivary gland stock [Smith strain]) intraperitoneally 36 hours prior to splenectomy. NK cells were purified with DX5 (anti-CD49b) microbeads (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's recommendations. The percentage of DX5+ cells was determined by flow cytometry (60%-85%). Killing was quantified by 5-hour standard 51Cr release assays using 5000 target cells/well with different E/T ratios. Maximum release was determined from target cells lysed with 1% NP-40. YAC-1 and CHO cells bearing ligands to NKG2D and Ly49D, respectively, were used as target cells.
NK-cell culture and cytokine assays
NK cells were purified with DX5 microbeads (Miltenyi Biotec) and cultured for 7 days in RPMI 1640 with 10% FCS (Hyclone, Logan, UT) and 50 ng/mL rhIL-15 (R&D Systems, Minneapolis, MN), resulting in NK-cell proliferation and leading to a pure NK-cell culture. Identity and purity of NK cells were verified by fluorescence-activated cell sorting (FACS) analysis for NK1.1, Ly49D, and CD49b expression. These cells were stimulated for 24 hours with plate-bound anti-NK1.1 (PK136), anti-Ly49D (4e4), anti-Ly49H (3D10), and anti-NKG2D (A10) antibodies (5 μg/mL in PBS), 10 to 100 ng/mL phorbol myristate acetate (PMA) and ionomycin (Iono), or 10 ng/mL IL-12, and/or 25 ng/mL IL-18 at a density of 1 to 2 × 106 cells/mL on 96-well plates in the absence or presence of IL-15. Cytokine release into the supernatant was assayed using enzyme-linked immunosorbent assay (ELISA) reagents from BD Biosciences (San Jose, CA) for IFN-γ, TNF-α, and GM-CSF and R&D Systems reagents for MIP-1β. Quantification of cytokine mRNA transcripts by quantitative real-time polymerase chain reaction (PCR) was performed as previously described26 using RNA from 106 NK cells stimulated for 24 hours with plate-bound anti–NK-cell receptor antibodies or IL-18.
Western blots
IL-15–expanded NK cells were stimulated with plate-bound anti-NK1.1 and anti-Ly49D antibodies or PMA/Iono (50 ng/mL each). Six-well plates were coated with 5 μg/mL antibody in PBS for 3 hours at 37°C. Cells were stimulated for 20 minutes and 60 minutes at a density of 2 × 106 cells/mL. NK-cell lysates were analyzed by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and immunoblotting using antibodies against IκBα, P-ERK1/2 (Thr202/Tyr204), P-JNK (Thr183/Tyr185), and P-p38 (Thr180/Tyr182; New England Biolabs, Beverly, MA).
NF-κB activation
Nuclear extracts were prepared according to standard methods, and 1 μg nuclear protein was analyzed with a NF-κB p65 transcription factor assay kit (Pierce, Rockford, IL) as previously described.27
Flow cytometry and intracellular cytokine production
Splenocytes, bone marrow cells, and liver lymphocytes, purified by a 40%/80% percoll gradient, were stained with fluorescently labeled antibodies from eBioscience (San Diego, CA) and analyzed on a FACSCalibur cytometer (BD). For cytokine production assays, total splenocytes were cultivated for 2 days in RPMI 1640 with 10% FCS (Hyclone) and 250 U/mL rIL-2 in order to preactivate them. Cells were stimulated with plate-bound anti–NK-cell receptor antibodies for 5 hours with addition of 3 μM monensin after the first hour. Intracellular stain for cytokine production was performed with eBioscience reagents according to manufacturer's recommendations.
MCMV infection
Mice were infected intraperitoneally with 1.25 × 105 pfu murine cytomegalovirus (MCMV salivary gland stock [Smith strain]). On day 3 or day 6, mice were injected intraperitoneally with BrdU 3 hours prior to splenectomy. Proliferation of NK1.1+ NK cells was assayed by FACS using a BrdU staining kit (BD Biosciences) according to the manufacturer's recommendations. Viral DNA of MCMV-infected spleens and livers was extracted with the DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany) according to the manufacturer's recommendations. For absolute quantification of MCMV-genome copy numbers, a specific sequence of the M54 gene was amplified by PCR and cloned into a pCR2.1-TOPO-Vector (Invitrogen, Frederick, MD). Using a M54-plasmid standard curve, the number of MCMV genome copies in total DNA extracted from infected organs was quantified on an ABI 7700 PRISM TaqMan (Applied Biosystems, Foster City, CA) using SYBR Green and the M54 forward primer 5′-ATCATCCGTTGCATCTCGTTG-3′ and reverse primer 5′-CGCCAGTCTGTATCCGTCCAT-3′. Results are expressed as MCMV-DNA copies per 5 ng total organ DNA.28
Results
Regular development of NK cells without Bcl10
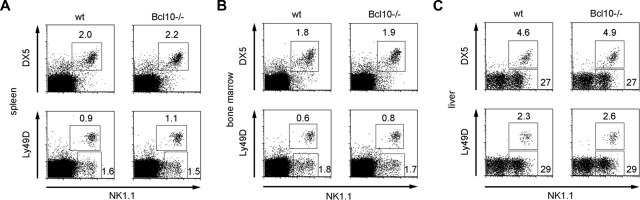
To study potential roles for Bcl10 signaling in NK-cell biology, we first determined whether Bcl10 is expressed in these cell types. Western blot analysis revealed that NK cells express substantial amounts of Bcl10 (data not shown, see also Figure 4A). Therefore, we first studied the effects of a Bcl10 deletion on NK-cell development in various organs. Using flow cytometry, we detected regular frequencies (Figure 1A-C) and numbers (not shown) of DX5-, Ly49D-, and NK1.1-positive NK cells in the bone marrow, spleen, and liver of Bcl10-deficient mice. Thus, although Bcl10 is expressed in normal NK cells, the absence of this signaling protein does not interfere with overall NK-cell development.
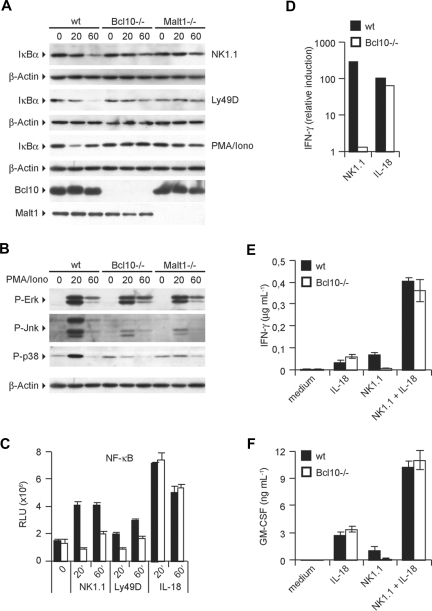
Figure 4.
ITAM-coupled NK-cell receptors activate NF-κB, JNK, and p38 via Bcl10 and Malt1. IL-15–expanded NK cells were stimulated with plate-bound anti-NK1.1 and anti-Ly49D antibodies or PMA/Iono for 20 and 60 minutes. Degradation of IκB-α (A) and phosphorylation of ERK, JNK, and p38 (B) was monitored by Western blot. (C) Activation of NF-κB was quantified with a NF-κB p65 transcription factor assay kit using nuclear protein lysates. (D) IFN-γ mRNA levels were quantified by real-time PCR after 24 hours of stimulation as indicated. (E,F) NK cells were stimulated as in Figure 2B,C with anti-NK1.1 and IL-18 either alone or in combination. Cytokine release was measured by ELISA. Data are means plus or minus SD of triplicate samples (where applicable) and representative of 3 independent experiments.
Figure 1.
Bcl10 deficiency does not interfere with NK cell development. Splenocytes (A), bone marrow cells (B), and liver lymphocytes (C) from Bcl10-deficient and wild-type control animals were stained for expression of CD49b (DX5), NK1.1, and Ly49D NK-cell surface markers. The percentages of NK cells with respect to total cell numbers are indicated. Data are representative of 4 mice per group.
Bcl10 and Malt1 signaling controls cytokine production in response to NK-cell activation
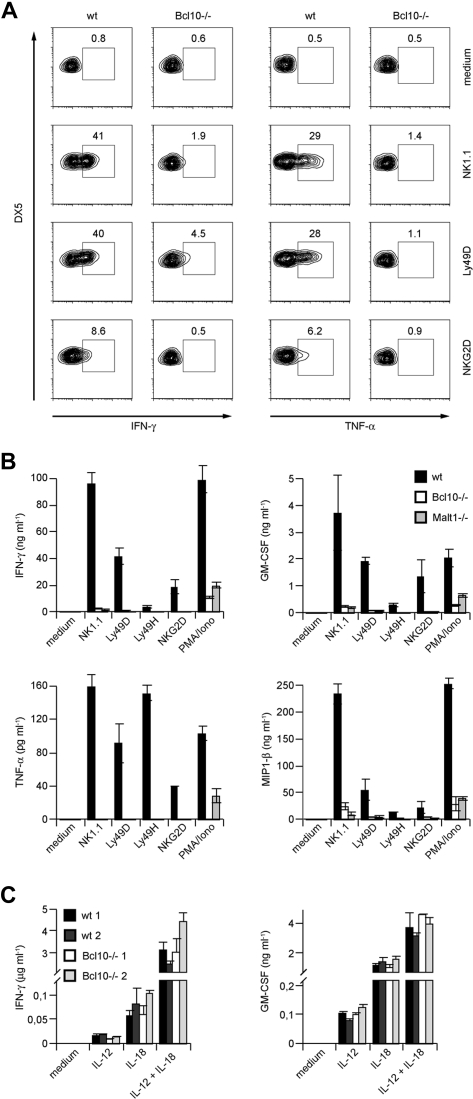
To investigate functions for Bcl10 signaling complexes during NK-cell activation, we next prepared primary splenic cell suspensions from wild-type mice and Bcl10-deficient animals and stimulated these cells with well-characterized agonistic antibodies directed against the NK-cell activating receptors NK1.1, Ly49D, and NKG2D (Figure 2A). Five hours later, we stained the cells with fluorescently labeled antibodies against the NK-cell surface marker DX5 and the T-cell receptor subunit CD3, permeabilized the cells, and investigated the production of IFN-γ and TNF-α specifically in the CD3−DX5+ NK-cell population using intracellular FACS. Whereas wild-type NK cells robustly synthesized both TNF-α and IFN-γ in response to NK1.1, Ly49D, or NKG2D triggering, the production of either cytokine was severely impaired in the absence of Bcl10.
Figure 2.
Impaired cytokine production in NK cells from Bcl10- and Malt1-deficient NK cells upon NK-cell receptor stimulation. (A) Total splenocytes from control or Bcl10-deficient mice were stimulated with plate-bound anti-NK1.1, anti-Ly49D, or anti-NKG2D, stained for CD49b (DX5) and CD3 and intracellular cytokine production, and analyzed by flow cytometry. Cells were gated for DX5+CD3− cells. The percentages of IFN-γ– or TNF-α–positive cells with respect to total NK cells are indicated. (B) IL-15–expanded splenic NK cells from Bcl10- and Malt1-deficient mice were stimulated with plate-bound antibodies directed against various activating NK- cell receptors or PMA and ionomycin (PMA/Iono) as indicated for 24 hours. (C) NK cells from 2 wild-type and 2 Bcl10-deficient mice were prepared as in panel B and stimulated with 10 ng/mL IL-12 and/or 25 ng/mL IL-18 for 24 hours. Cytokine release was measured by ELISA. Data are means plus or minus SD of triplicate samples and representative of 3 independent experiments.
We also expanded NK cells from Bcl10−/− and wild-type control animals in vitro with IL-15 prior to stimulation via NK1.1, Ly49D, Ly49H, or NKG2D and then tested for secretion of TNF-α and IFN-γ into the culture supernatant (Figure 2B). In addition, we analyzed the production of GM-CSF and MIP-1β. Consistent with the results presented in Figure 2A, we detected again severe defects in TNF-α and IFN-γ production and also in GM-CSF and MIP-1β secretion in the absence of Bcl10. Thus, Bcl10 plays a key role downstream of several activating NK-cell receptors to control the production of cytokines.
To test whether Malt1 is required for these Bcl10-mediated effects, we additionally studied NK cells from Malt1-deficient mice (Figure 2B). Similar to Bcl10-deficient cells, Malt1−/− NK cells also exhibit a severe impairment in TNF-α, IFN-γ, GM-CSF, and MIP-1β production upon stimulation with anti-NK1.1, -Ly49D, -Ly49H, and -NKG2D antibodies indicating that Bcl10 and Malt1 cooperate in NK-cell activation. Finally, we activated the NK cells from Bcl10−/− and Malt1−/− mice with phorbol myristate acetate (PMA) and the calcium ionophore ionomycin (Iono) to bypass receptor proximal signaling and to stimulate the cells directly by mobilizing free calcium ions and activating PKC enzymes. Whereas wild-type NK cells robustly produced IFN-γ, TNF-α, GM-CSF, and MIP-1β in response to PMA/Iono treatment, the production of these cytokines was defective in the absence of Bcl10 or Malt1. Thus, Bcl10 and Malt1 operate downstream of several ITAM-coupled NK-cell receptors and downstream of PKCs for NK-cell activation.
To study ITAM-independent NK-cell activation, we next stimulated NK cells from Bcl10−/− mice with IL-12 or IL-18 or a combination of IL-12 + IL-18 (Figure 2C). All these stimuli induced a similar production of IFN-γ and GM-CSF in the presence or absence of Bcl10, indicating that Bcl10-deficient cells are in principle able to synthesize cytokines and that certain non–ITAM receptor signals can induce NK-cell activation in a Bcl10-independent manner.
The Bcl10 complex is dispensable for NK cell–mediated target cell killing
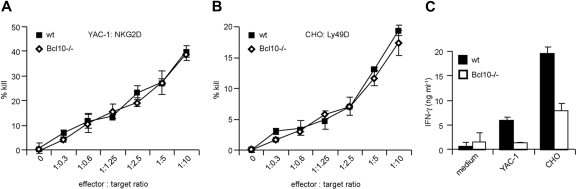
NK-cell activation not only induces the production of proinflammatory cytokines and interferons, but also efficiently kills target cells that express NK-cell activation ligands. To investigate potential roles for Bcl10 signaling complexes in NK-cell killing, we used YAC-1 cells expressing NKG2D ligands or CHO cells expressing Ly49D ligands as target cells in standard 51Cr release assays.29 The target cells were coincubated with anti-CD49b (DX5)–purified splenic NK cells from wild-type and Bcl10-deficient mice in various effector to target ratios. NK cells were preactivated in vivo by an injection of 5 × 105 pfu MCMV 36 hours prior to killing. After 5 hours of coincubation, we measured target cell killing by studying 51Cr release into the culture supernatant (Figure 3A,B). In a dose-dependent manner, wild-type and Bcl10−/− NK cells induced target cell killing similarly, indicating that Bcl10 disruption does not interfere with the killing machinery of NK cells.
Figure 3.
Bcl10 is dispensable for NK-cell receptor–induced killing. NK cells were isolated from the spleens of WT and Bcl10−/− mice 36 hours after infection with 5 × 105 pfu MCMV. NK-cell cytotoxicity was assessed in a 5-hour 51Cr release assay using YAC-1 (A) and CHO (B) cells as target cells at the indicated effector to target ratios. Data are representative of 3 independent experiments with a total of 6 mice per group. (C) IL-15–expanded NK cells were stimulated with YAC-1 or CHO cells at a 1:1 ratio and IFN-γ production was quantified by ELISA. Data are representative of 2 independent experiments.
In addition, we coincubated wild-type and Bcl10−/− NK cells in a 1:1 ratio with target cells and measured 24 hours later the production of IFN-γ in the culture supernatant (Figure 3C). Consistent with the results reported in Figure 2, we again found defects in IFN-γ production in Bcl10−/− NK cells.
Bcl10 and Malt1 couple NK-cell activation to NF-κB and MAPK pathways
The production of cytokines such as TNF-α, GM-CSF, and IFN-γ is controlled by transcription factors of the NF-κB family and by signals through MAPK kinase pathways.7,8,30 Based on known roles of Bcl10 and Malt1 in activating canonical NF-κB signaling as well as JNK and p38 MAP kinase activation in T cells, B cells, and myeloid cells, we hypothesized that the Bcl10/Malt1 signaling module could couple activating NK-cell receptors to NF-κB and MAPK activation. To test these hypotheses, we investigated the activation of the NF-κB pathway in NK cells by first studying the degradation of the IκBα inhibitory protein (Figure 4A). Whereas a triggering of the NK-cell activating receptors NK1.1 and Ly49D induces a progressive degradation of IκBα in wild-type cells, the degradation of the NF-κB inhibitor was impaired in the absence of either Bcl10 or Malt1. Similar effects were observed after a stimulation of NK cells with PMA/Iono, suggesting that PKCs engage Bcl10 and Malt1 for downstream signaling to NF-κB (Figure 4A). Consistent with an intact negative feedback loop, the strong cell activator PMA/Iono induces a regular NF-κB–dependent resynthesis of IκB-α already after 60 minutes in wild-type cells.
We then studied the NK cell–specific roles of Bcl10 and Malt1 in MAPK activation (Figure 4B). Stimulation of wild-type NK cells with PMA/Iono resulted in a robust activation of ERK, JNK, and p38 MAPKs as determined by Western blotting using activation-specific antibodies that detect the phosphorylated forms of the respective kinases. We also observed a regular activation of ERK in the absence of Bcl10 or Malt1. However, the activation of JNK as well as the activation of p38 were defective in the absence of either Bcl10 or Malt1, indicating a key role for both proteins in regulating JNK and p38 signaling in NK cells.
Together, these signaling experiments indicate that the Bcl10/Malt1 module couples activating NK-cell receptors to the canonical NF-κB pathway and in addition controls the activation of JNK and p38 MAP kinases in these cells.
To directly study nuclear translocation and DNA binding of NF-κB, we then prepared nuclear extracts from NK1.1-, Ly49D-, or IL-18–stimulated cells and used those to perform oligonucleotide-binding assays for the NF-κB subunit p65. In line with a defective IκB degradation in NK1.1- or Ly49D-stimulated Bcl10−/− cells, NF-κB DNA binding induced by these ITAM-coupled receptors was also severely impaired (Figure 4C). However, the ITAM-independent NK-cell activator IL-18 is able to induce regular NF-κB activation in the absence of Bcl10. To test whether this differential activation of the NF-κB pathway by ITAM and non-ITAM signals results in a differential activation of gene transcription, we focused on NK1.1- or IL-18–mediated NK-cell activation and studied the synthesis of IFN-γ mRNA (Figure 4D). Consistent with a regular IL-18–mediated NF-κB activation and with our results presented in Figure 2, IL-18 stimulation induced similar amounts of IFN-γ mRNA in wild-type and in Bcl10−/− cells. However, NK1.1 signaling induced an up-regulation of IFN-γ mRNA only in wild-type cells but not in those that lack Bcl10 (Figure 4D). Since IL-18 signaling can synergize with signals from ITAM-coupled receptors,31 we also costimulated the cells with agonistic antibodies against NK1.1 and with IL-18 (Figure 4E,F). Intriguingly, the combination of NK1.1 triggering and IL-18 stimulation induced similar amounts IFN-γ and GM-CSF in wild-type and in Bcl10−/− cells, suggesting that the IL-18 signal can complement some of the defects in Bcl10-deficient cells. Similar rescue effects were also observed for a synergism of IL-18 signaling with Ly49D, NKG2D, or Ly49H stimulation for the production of IFN-γ or GM-CSF (data not shown).
Carma1-dependent, Card9-independent engagement of Bcl10 signaling in NK cells
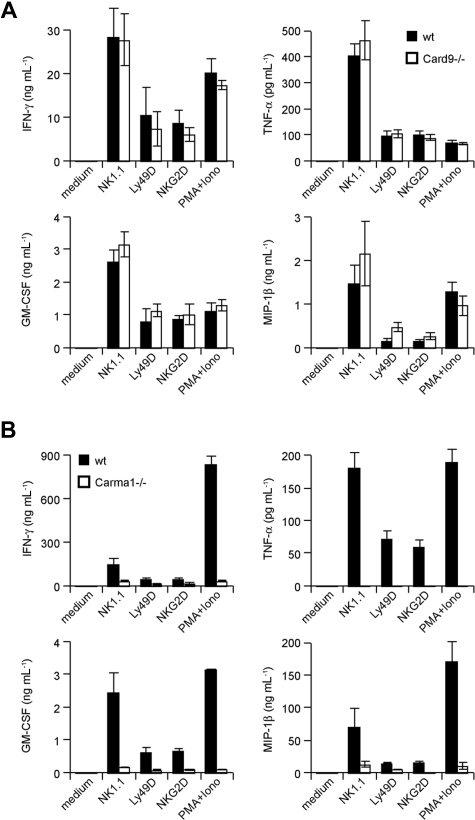
The CARD-coiled-coil proteins Carma1 and Card9 can couple immunoreceptors to Bcl10 signalosomes in a context- and cell lineage–specific manner.12,13,19 How activating NK-cell receptors engage Bcl10-regulated signalosomes is unclear. To test whether Carma1 or Card9 might be involved in Bcl10-dependent NK-cell activation, we isolated NK cells from Carma1- and Card9-deficient mice. Subsequently, we stimulated the cells in vitro with agonistic antibodies against NK1.1, Ly49D, and NKG2D and measured the production of the Bcl10- and Malt1-controlled cytokines TNF-α, IFN-γ, GM-CSF, and MIP-1β in the culture supernatant (Figure 5A,B). Whereas Card9-deficient NK cells produce regular amounts of these cytokines in response to NK-cell activation, the production of all measured cytokines was severely impaired in the absence of Carma1. These results indicate a Carma1-dependent, Card9-independent coupling of activating NK-cell receptors to Bcl10/Malt1 complexes for NK-cell activation.
Figure 5.
Carma1, but not Card9, is required for NK-cell receptor signaling. IL-15–expanded NK cells from Card9-deficient (A) or IL-15–expanded NK cells from Carma1-deficient (B) or control mice were stimulated with plate-bound anti–NK-cell receptor antibodies or PMA/Iono as indicated for 24 hours. Cytokine release was measured by ELISA. Data are means plus or minus SD of triplicate samples and representative of 3 independent experiments.
Lack of Bcl10 signaling does not affect early immunity against murine cytomegalovirus in vivo
Our results indicate a segregation of ITAM receptor–mediated NK-cell killing and NK-cell cytokine production at the level of Bcl10 signaling complexes. This allows us to selectively investigate the role of NK-cell receptor–mediated cytokine production during early antiviral immunity in vivo without considering killing. To this end, we used the herpes virus murine cytomegalovirus (MCMV) as a model, since it is well established that mice that are NK cell–depleted32 are highly susceptible to MCMV infection, as manifested by high viral titers in the spleen, and that the NK cell–activating receptor Ly49H, which recognizes the MCMV protein m157,33 is critically involved in resistance to MCMV in vivo.34
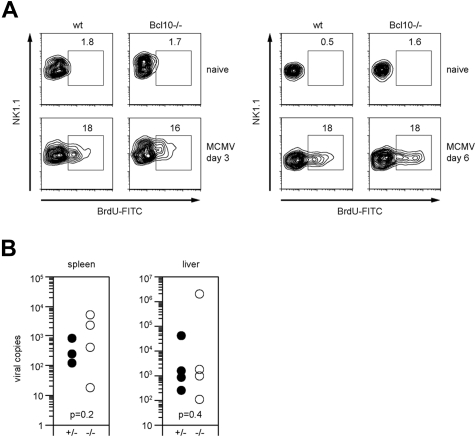
To test the role of Bcl10 in antiviral immunity, we infected wild-type and Bcl10-deficient animals with 1.25 × 105 pfu MCMV intravenously. As MCMV infection induces an expansion of NK cells in vivo that is at least in part dependent on the ITAM adapter DAP12,35,36 we first investigated the role of Bcl10 in NK-cell proliferation signaling (Figure 6A). Three days after infection, we applied a pulse of BrdU by intraperitoneal injection and measured the incorporation of BrdU into NK cells in vivo using FACS. Both wild-type and Bcl10−/− mice exhibited similar frequencies of BrdU-positive NK cells in the spleen, indicating that NK-cell proliferation does not depend on Bcl10. In a second set of in vivo experiments, we killed the animals at day 6 after infection and, in addition to proliferation, determined MCMV titers in the liver and the spleens (Figure 6A,B). No significant differences in the viral titers were detected between wild-type and Bcl10−/− animals in either organ. Thus, although Bcl10 signaling couples multiple activating NK-cell receptors to cytokine production, this pathway is not essential for early antiviral immunity.
Figure 6.
Bcl10 deficiency does not affect NK-cell proliferation and early control of viral replication in MCMV infection. (A) Proliferation of NK1.1+ NK cells upon intraperitoneal infection with 1.25 × 105 pfu MCMV was assessed on day 3 and day 6 after infection by BrdU staining. Percentages of proliferating BrdU+ NK cells are indicated. Data are representative of 4 mice per group. (B) Viral load of spleen and liver was quantified 6 days after intraperitoneal infection with 1.25 × 105 pfu MCMV using quantitative real-time PCR. Results are indicated as MCMV-DNA copies per 5 ng total purified organ DNA. Shown are values of single mice and of 4 mice per group.
Discussion
Together, our results identify a novel signal transduction pathway that is specifically engaged by multiple ITAM-coupled activating NK-cell receptors. This pathway depends on Carma1, Bcl10, and Malt1 and operates independently of Card9 to activate NF-κB, JNK, and p38 upon activating NK-cell receptor triggering to segregate the signals for cytokine production from NK-cell proliferation and NK-cell killing.
Previous work from us and others has revealed essential functions for Bcl10 in the assembly of multiprotein complexes that relay ITAM-dependent signals in T cells, B cells, mast cells, dendritic cells, and macrophages to the canonical NF-κB pathway and to the activation of JNK and p38.9–13 Distinct CARD-coiled-coil proteins of the Carma family couple receptor-proximal signals to Bcl10, and Bcl10 can cooperate with Malt1 in a context-dependent manner for downstream signaling.19 Here we broaden the concept that ITAM receptors in the immune system engage Bcl10 signaling modules for cell activation by showing a key function for Bcl10 complexes in NK-cell activation. In line with our results, a parallel study using an independently generated Bcl10-deficient mouse has also recently reported essential roles for Bcl10 in NK-cell activation and reported that NK-cell development was unaffected in these animals.24 We extend the findings of Malarkannan et al by additionally demonstrating that the NK-cell activation signal depends on Carma1 but not on Card9 and by showing that Malt1 is also essential for the NK-cell activation pathways. This latter finding is not trivial, as Bcl10 is known to signal independently of Malt1 in certain cells, for example during neurodevelopment or for B-cell proliferation.14–18 In addition, we demonstrate directly on a genetic basis that Bcl10 complexes control the activation the NF-κB, JNK, and p38 pathways in primary NK cells. Although ERK signaling is intact in Bcl10- or in Malt1-deficient cells and is required for IFN-γ production,31 our findings indicate that ITAM receptor–induced ERK activation by itself is not sufficient to drive cytokine gene transcription and cytokine production.
Bcl10 is not only required for the activation of NK cells via agonistic antibodies directed against specific activating NK-cell receptors but also for the direct NK-cell activation via PMA/Iono. These results suggest that PKC enzymes are involved in NK-cell activation signaling. This suggestion is in line with the essential role of Carma1 in NK-cell signaling and the known function of Carma1 but not Card9 downstream of PKCs in immune cell activation. Recent work has identified conserved serine residues in Carma1 in a region that is now referred to as the linker region.37,38 These serines can directly be phosphorylated by PKCβ upon BCR signaling or by PKCθ upon TCR signaling.37,38 This linker region is not present in Card9, and Card9-dependent engagement of Bcl10 in DCs does not appear to involve PKCs.12 During antigen receptor signaling, the PKC-mediated phosphorylation of Carma1 is thought to release an intramolecular autoinhibition in Carma1 resulting in the recruitment of Bcl10 to Carma1 and the assembly and stabilization of large signalosomes for signal propagation. Based on the apparent conservation between the antigen receptor and the NK-cell receptor pathways, we suggest that one or several PKCs in NK cells might directly phosphorylate Carma1 in the linker region to couple to Bcl10/Malt1 signalosomes. Future work with NK cells that lack specific PKC isoforms is required to define the precise PKC enzymes that control NK-cell activation.
The downstream effectors that mediate Bcl10/Malt1-dependent NF-κB and MAPK activation upon activating NK-cell receptor stimulation are also not defined. However, these are likely TRAF proteins, presumably TRAF2 or TRAF6, ubiquitin modulators such as UBC13 and UEV1A, as well as the kinase TAK1 together with TAB2 and TAB3. All of these proteins are known to regulate TCR signaling together with Carma1 and Bcl1039 and are implicated in the BCR pathway. Notably, a conditional genetic disruption of TAK1 specifically in T or in B cells indicates a differential requirement for this signal transducer in TCR and BCR signaling to NF-κB, JNK, and p38.40–42 Thus the Bcl10 downstream effectors are also assembled in a context-specific fashion. Comprehensive studies investigating the roles of TRAFs, UBC13, UEV1A, TAB2, and TAB3 in NK cells are therefore necessary to precisely define the molecular mechanisms of Bcl10-mediated NF-κB, JNK, and p38 activation in response to activating NK-cell receptors.
Intriguingly, although our findings indicate essential functions for Carma1/Bcl10/Malt1 signaling in NK-cell cytokine and chemokine production, a genetic disruption of this pathway does not result in elevated viral loads early after MCMV infections. NK cells are known to mediate the innate resistance of mammals to herpes virus,1,32,34 and signals from activating NK-cell receptors are essential for this defense.33,34 However, a deficiency in Bcl10 impairs only the specific pathway for ITAM receptor–mediated cytokine production without affecting NK-cell killing. Thus, in vivo the NK-cell killing function seems to play a predominant role in the early control of MCMV infection. This assumption is in line with highly elevated MCMV titers that are observed early upon infection of perforin-deficient mice, which have specific defects in NK-cell killing.43 In addition, cytokines such as IL-12 and IL-18 have been demonstrated to activate NK cells via p38, JNK, and NF-κB,44,45 and we show here that IL-12 and IL-18 signaling is independent of Bcl10. A costimulation of Bcl10−/− cells through activating NK-cell receptors and a cytokine such as IL-18 can rescue cytokine production in vitro. It is therefore conceivable that during a viral infection in vivo, similar rescue effects could contribute to the normal early course of MCMV infection. Moreover, NK cell–produced cytokines that are released during the innate antiviral immune phase are thought to couple to the activation of the adaptive immune response by modulating and enhancing dendritic cell activation and T-cell function. It is therefore also possible that a later adaptive immune response to viral infections might be impaired in the absence of Bcl10 signaling in NK cells. However, we cannot directly test this hypothesis with our current models, as the mutant mice that we use lack Carma1, Bcl10, or Malt1 in all immune cells and therefore have also deficiencies in DC and T-cell responses. Conditional mouse mutants with a specific disruption of Bcl10 only in NK cells are required to address this question.
In conclusion, we report that Carma1 specifically links multiple ITAM-coupled receptors in NK cells to Bcl10 and Malt1 for the activation of the canonical NF-κB, p38, and JNK signaling pathways. Together with previous results that were obtained in T cells, B cells, mast cells, DCs, and macrophages,9–13 we propose the common principle that potentially all ITAM receptors in the immune system engage Bcl10/Malt1 signaling. Whereas the innate immune receptors in DCs and macrophages use the Card9 protein for Bcl10/Malt1-dependent cell activation, the PKC-coupled ITAM receptors in T cells, B cell, mast cells, and presumably NK cells appear to signal via phosphorylation of Carma1 for Bcl10/Malt1 engagement.
Acknowledgments
We thank K. Strasser and M. Schweneker for critical reading of the paper, and K. Heinrich, B. Grohs, and W. Ballhorn for excellent technical assistance.
This work was supported by a Max-Eder-Program grant from Deutsche Krebshilfe and by SFB grants from Deutsche Forschungsgemeinschaft (J.R.).
Footnotes
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: O.G. and J.R. designed the research; O.G., C.G., C.S., S.Z., D.S., N.H., and W.R. performed experiments; O.G., C.G., C.P., A.K., and J.R. analyzed results; H.J., W.M.Y., H.H., and D.R.L. provided critical reagents; O.G. prepared the figures; and O.G. and J.R. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jürgen Ruland, Klinikum rechts der Isar, III. Medizinische Klinik, Ismaninger Str 22, 81675 Munich, Germany; e-mail: jruland@lrz.tu-muenchen.de.
References
- 1.Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3:304–316. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- 2.Cerwenka A, Lanier LL. Natural killer cells, viruses and cancer. Nat Rev Immunol. 2001;1:41–49. doi: 10.1038/35095564. [DOI] [PubMed] [Google Scholar]
- 3.Kirwan SE, Burshtyn DN. Regulation of natural killer cell activity. Curr Opin Immunol. 2007;19:46–54. doi: 10.1016/j.coi.2006.11.012. [DOI] [PubMed] [Google Scholar]
- 4.Lanier LL. Natural killer cell receptor signaling. Curr Opin Immunol. 2003;15:308–314. doi: 10.1016/s0952-7915(03)00039-6. [DOI] [PubMed] [Google Scholar]
- 5.Carson WE, Dierksheide JE, Jabbour S, et al. Coadministration of interleukin-18 and interleukin-12 induces a fatal inflammatory response in mice: critical role of natural killer cell interferon-gamma production and STAT-mediated signal transduction. Blood. 2000;96:1465–1473. [PubMed] [Google Scholar]
- 6.Hayden MS, Ghosh S. Signaling to NF-kappaB. Genes Dev. 2004;18:2195–2224. doi: 10.1101/gad.1228704. [DOI] [PubMed] [Google Scholar]
- 7.Holloway AF, Rao S, Chen X, Shannon MF. Changes in chromatin accessibility across the GM-CSF promoter upon T cell activation are dependent on nuclear factor kappaB proteins. J Exp Med. 2003;197:413–423. doi: 10.1084/jem.20021039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tato CM, Mason N, Artis D, et al. Opposing roles of NF-kappaB family members in the regulation of NK cell proliferation and production of IFN-gamma. Int Immunol. 2006;18:505–513. doi: 10.1093/intimm/dxh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thome M. CARMA1, BCL-10 and MALT1 in lymphocyte development and activation. Nat Rev Immunol. 2004;4:348–359. doi: 10.1038/nri1352. [DOI] [PubMed] [Google Scholar]
- 10.Klemm S, Gutermuth J, Hultner L, et al. The Bcl10-Malt1 complex segregates Fc epsilon RI-mediated nuclear factor kappa B activation and cytokine production from mast cell degranulation. J Exp Med. 2006;203:337–347. doi: 10.1084/jem.20051982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen Y, Pappu BP, Zeng H, et al. B cell lymphoma 10 is essential for FcepsilonR-mediated degranulation and IL-6 production in mast cells. J Immunol. 2007;178:49–57. doi: 10.4049/jimmunol.178.1.49. [DOI] [PubMed] [Google Scholar]
- 12.Gross O, Gewies A, Finger K, et al. Card9 controls a non-TLR signalling pathway for innate anti-fungal immunity. Nature. 2006;442:651–656. doi: 10.1038/nature04926. [DOI] [PubMed] [Google Scholar]
- 13.Hara H, Ishihara C, Takeuchi A, et al. The adaptor protein CARD9 is essential for the activation of myeloid cells through ITAM-associated and Toll-like receptors. Nat Immunol. 2007;8:619–629. doi: 10.1038/ni1466. [DOI] [PubMed] [Google Scholar]
- 14.Ferch U, Buschenfelde CM, Gewies A, et al. MALT1 directs B cell receptor-induced canonical nuclear factor-kappaB signaling selectively to the c-Rel subunit. Nat Immunol. 2007;8:984–991. doi: 10.1038/ni1493. [DOI] [PubMed] [Google Scholar]
- 15.Ruefli-Brasse AA, French DM, Dixit VM. Regulation of NF-kappaB-dependent lymphocyte activation and development by paracaspase. Science. 2003;302:1581–1584. doi: 10.1126/science.1090769. [DOI] [PubMed] [Google Scholar]
- 16.Ruland J, Duncan GS, Elia A, et al. Bcl10 is a positive regulator of antigen receptor-induced activation of NF-kappaB and neural tube closure. Cell. 2001;104:33–42. doi: 10.1016/s0092-8674(01)00189-1. [DOI] [PubMed] [Google Scholar]
- 17.Ruland J, Duncan GS, Wakeham A, Mak TW. Differential requirement for Malt1 in T and B cell antigen receptor signaling. Immunity. 2003;19:749–758. doi: 10.1016/s1074-7613(03)00293-0. [DOI] [PubMed] [Google Scholar]
- 18.Xue L, Morris SW, Orihuela C, et al. Defective development and function of Bcl10-deficient follicular, marginal zone and B1 B cells. Nat Immunol. 2003;4:857–865. doi: 10.1038/ni963. [DOI] [PubMed] [Google Scholar]
- 19.Wegener E, Krappmann D. CARD-Bcl10-Malt1 signalosomes: missing link to NF-kappaB. Sci STKE. 2007;2007:pe21. doi: 10.1126/stke.3842007pe21. [DOI] [PubMed] [Google Scholar]
- 20.Blonska M, Pappu BP, Matsumoto R, et al. The CARMA1-Bcl10 signaling complex selectively regulates JNK2 kinase in the T cell receptor-signaling pathway. Immunity. 2007;26:55–66. doi: 10.1016/j.immuni.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newton K, Dixit VM. Mice lacking the CARD of CARMA1 exhibit defective B lymphocyte development and impaired proliferation of their B and T lymphocytes. Curr Biol. 2003;13:1247–1251. doi: 10.1016/s0960-9822(03)00458-5. [DOI] [PubMed] [Google Scholar]
- 22.Hara H, Wada T, Bakal C, et al. The MAGUK family protein CARD11 is essential for lymphocyte activation. Immunity. 2003;18:763–775. doi: 10.1016/s1074-7613(03)00148-1. [DOI] [PubMed] [Google Scholar]
- 23.McAllister-Lucas LM, Ruland J, Siu K, et al. CARMA3/Bcl10/MALT1-dependent NF-kappaB activation mediates angiotensin II-responsive inflammatory signaling in nonimmune cells. Proc Natl Acad Sci U S A. 2007;104:139–144. doi: 10.1073/pnas.0601947103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Malarkannan S, Regunathan J, Chu H, et al. Bcl10 plays a divergent role in NK cell-mediated cytotoxicity and cytokine generation. J Immunol. 2007;179:3752–3762. doi: 10.4049/jimmunol.179.6.3752. [DOI] [PubMed] [Google Scholar]
- 25.Egawa T, Albrecht B, Favier B, et al. Requirement for CARMA1 in antigen receptor-induced NF-kappa B activation and lymphocyte proliferation. Curr Biol. 2003;13:1252–1258. doi: 10.1016/s0960-9822(03)00491-3. [DOI] [PubMed] [Google Scholar]
- 26.Woo AL, Gildea LA, Tack LM, et al. In vivo evidence for interferon-gamma-mediated homeostatic mechanisms in small intestine of the NHE3 Na+/H+ exchanger knockout model of congenital diarrhea. J Biol Chem. 2002;277:49036–49046. doi: 10.1074/jbc.M205288200. [DOI] [PubMed] [Google Scholar]
- 27.LeibundGut-Landmann S, Gross O, Robinson MJ, et al. Syk- and CARD9-dependent coupling of innate immunity to the induction of T helper cells that produce interleukin 17. Nat Immunol. 2007;8:630–638. doi: 10.1038/ni1460. [DOI] [PubMed] [Google Scholar]
- 28.Tang-Feldman YJ, Wojtowicz A, Lochhead GR, Hale MA, Li Y, Pomeroy C. Use of quantitative real-time PCR (qRT-PCR) to measure cytokine transcription and viral load in murine cytomegalovirus infection. J Virol Methods. 2006;131:122–129. doi: 10.1016/j.jviromet.2005.07.013. [DOI] [PubMed] [Google Scholar]
- 29.Cella M, Fujikawa K, Tassi I, et al. Differential requirements for Vav proteins in DAP10- and ITAM-mediated NK cell cytotoxicity. J Exp Med. 2004;200:817–823. doi: 10.1084/jem.20031847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ishizuka T, Terada N, Gerwins P, et al. Mast cell tumor necrosis factor alpha production is regulated by MEK kinases. Proc Natl Acad Sci U S A. 1997;94:6358–6363. doi: 10.1073/pnas.94.12.6358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ortaldo JR, Winkler-Pickett R, Wigginton J, et al. Regulation of ITAM-positive receptors: role of IL-12 and IL-18. Blood. 2006;107:1468–1475. doi: 10.1182/blood-2005-04-1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bukowski JF, Woda BA, Habu S, Okumura K, Welsh RM. Natural killer cell depletion enhances virus synthesis and virus-induced hepatitis in vivo. J Immunol. 1983;131:1531–1538. [PubMed] [Google Scholar]
- 33.Arase H, Mocarski ES, Campbell AE, Hill AB, Lanier LL. Direct recognition of cytomegalovirus by activating and inhibitory NK cell receptors. Science. 2002;296:1323–1326. doi: 10.1126/science.1070884. [DOI] [PubMed] [Google Scholar]
- 34.Brown MG, Dokun AO, Heusel JW, et al. Vital involvement of a natural killer cell activation receptor in resistance to viral infection. Science. 2001;292:934–937. doi: 10.1126/science.1060042. [DOI] [PubMed] [Google Scholar]
- 35.Dokun AO, Kim S, Smith HR, Kang HS, Chu DT, Yokoyama WM. Specific and nonspecific NK cell activation during virus infection. Nat Immunol. 2001;2:951–956. doi: 10.1038/ni714. [DOI] [PubMed] [Google Scholar]
- 36.French AR, Sjolin H, Kim S, et al. DAP12 signaling directly augments proproliferative cytokine stimulation of NK cells during viral infections. J Immunol. 2006;177:4981–4990. doi: 10.4049/jimmunol.177.8.4981. [DOI] [PubMed] [Google Scholar]
- 37.Matsumoto R, Wang D, Blonska M, et al. Phosphorylation of CARMA1 plays a critical role in T cell receptor-mediated NF-kappaB activation. Immunity. 2005;23:575–585. doi: 10.1016/j.immuni.2005.10.007. [DOI] [PubMed] [Google Scholar]
- 38.Sommer K, Guo B, Pomerantz JL, et al. Phosphorylation of the CARMA1 linker controls NF-kappaB activation. Immunity. 2005;23:561–574. doi: 10.1016/j.immuni.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 39.Rawlings DJ, Sommer K, Moreno-Garcia ME. The CARMA1 signalosome links the signalling machinery of adaptive and innate immunity in lymphocytes. Nat Rev Immunol. 2006;6:799–812. doi: 10.1038/nri1944. [DOI] [PubMed] [Google Scholar]
- 40.Sato S, Sanjo H, Takeda K, et al. Essential function for the kinase TAK1 in innate and adaptive immune responses. Nat Immunol. 2005;6:1087–1095. doi: 10.1038/ni1255. [DOI] [PubMed] [Google Scholar]
- 41.Wan YY, Chi H, Xie M, Schneider MD, Flavell RA. The kinase TAK1 integrates antigen and cytokine receptor signaling for T cell development, survival and function. Nat Immunol. 2006;7:851–858. doi: 10.1038/ni1355. [DOI] [PubMed] [Google Scholar]
- 42.Liu HH, Xie M, Schneider MD, Chen ZJ. Essential role of TAK1 in thymocyte development and activation. Proc Natl Acad Sci U S A. 2006;103:11677–11682. doi: 10.1073/pnas.0603089103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tay CH, Welsh RM. Distinct organ-dependent mechanisms for the control of murine cytomegalovirus infection by natural killer cells. J Virol. 1997;71:267–275. doi: 10.1128/jvi.71.1.267-275.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mavropoulos A, Sully G, Cope AP, Clark AR. Stabilization of IFN-gamma mRNA by MAPK p38 in IL-12- and IL-18-stimulated human NK cells. Blood. 2005;105:282–288. doi: 10.1182/blood-2004-07-2782. [DOI] [PubMed] [Google Scholar]
- 45.Hoshino K, Tsutsui H, Kawai T, et al. Cutting edge: generation of IL-18 receptor-deficient mice: evidence for IL-1 receptor-related protein as an essential IL-18 binding receptor. J Immunol. 1999;162:5041–5044. [PubMed] [Google Scholar]