Abstract
Introns may affect gene expression by increasing the time required to transcribe the gene. One way for extended transcription times to affect the behavior of a gene expression program is through a negative feedback loop. Here, we show that a logically engineered negative feedback loop in animal cells produces expression pulses, which have a broad time distribution that increases with intron length. These results in combination with mathematical models provide insight into what may produce the intron-dependent pulse distributions. We conclude that the long production time required for large intron-containing genes is significant for the behavior of gene expression programs.
Keywords: In vivo quantitation, intron, negative feedback, oscillations, synthetic biology, transcription elongation
Introns contribute significant length to genes, break the protein coding information so that it can be alternatively processed into different splice isoforms, and introduce unique levels for regulation during the coordination of transcription and processing. In humans, intron lengths contribute 95% of the average gene’s sequence (Venter et al. 2001). One simple mechanism by which introns may affect the dynamics of gene expression is by increasing the time required to transcribe the gene (Shermoen and O’Farrell 1991; Rothe et al. 1992; Tennyson et al. 1995; Swinburne and Silver 2008). The self-evident transcriptional time delay that introns make much longer can potentially impact four types of expression programs: the activation of multiple genes by the same cue, the repression of multiple genes by the same cue, gene expression at cell division when transcription elongation is interrupted because of chromatin condensation during mitosis, and negative feedback loops where the product of the gene reduces the gene’s expression (Swinburne and Silver 2008). While only a few specific instances where intron delays impact biology have been recognized, their presence in developmentally expressed genes could alter the accuracy of timing and dynamics of developmental events. Functional genomic studies in Drosophila melanogaster suggest that introns may function as modular time delays through the use of very distal alternative promoters by many genes during development (Manak et al. 2006). While the presence of introns is well appreciated, the consequences of increased transcription times on a gene’s activation kinetics and expression dynamics in individual cells are unclear.
Transcriptional negative feedback loops are common throughout nature; introducing a time delay in such a feedback loop can theoretically produce oscillatory pulses of gene expression (Goodwin 1965; Lewis 2003; Monk 2003). In prokaryotes, where delays during gene expression are shorter because of the absence of introns and because translation begins cotranscriptionally, autoinhibition by transcription factors includes much shorter time delays. As such, autoinhibition has been shown to reduce the distribution of protein expression levels and the time needed to reach a steady state after activation (Becskei and Serrano 2000; Rosenfeld et al. 2002; Dublanche et al. 2006). In eukaryotes, the behavior of negative feedback loops may be fundamentally different because of elevated transcriptional delays and compartmentalization by the nucleus that separates both physically and temporally the processes of transcription and translation.
In this study we pursued the potential impact introns have as transcriptional time delays by engineering a gene network and modifying only intron length between clonal variants. Specifically, we asked how intron length in a negative feedback loop affects gene expression. We found that gene length affects quantitative aspects of dynamic expression from an engineered negative feedback loop. Additionally, we show how quantitative dynamics exhibited by many cells in a clonal population are consistent with transcriptional bursting that increases with gene length. These results are of relevance to our understanding of developmentally regulated networks where transcriptional delays may alter both timing during early gene activation programs and expression dynamics during patterning such as those underlying vertebrate somitogenesis.
Results and Discussion
Endogenous intron-containing genes are not amenable to study the impact intron lengths have on gene expression in a natural context. Orthologous introns vary in length from species to species, and mutants with large intron insertions or deletions are either not found or not properly characterized for timing defects in genetic screens. To study the affect of transcriptional time delays by introns, we engineered a negative feedback loop (Fig. 1A) that expresses a humanized Tet repressor (TetR) fused to the fast-maturing Venus variant of yellow fluorescent protein (YFP) under the strong β-actin promoter (Anastassiadis et al. 2002; Nagai et al. 2002). The TetR fusion contains a nuclear localization signal from SV40 that sends the protein to the nucleus, where it inhibits transcription initiation of its own gene by binding tet-operators (tetO) (Fig. 1A, gray boxes) in the promoter region; repression can be relieved by the addition of doxycycline. We varied the size of the introns in the reporter gene by introducing into the first intron of β-actin (1 kb), either a 7- or 16-kb intron cassette. Since the coding region is 2 kb long, this yields three variants of different lengths (3 kb, 10 kb, and 19 kb), all with the same coding region.
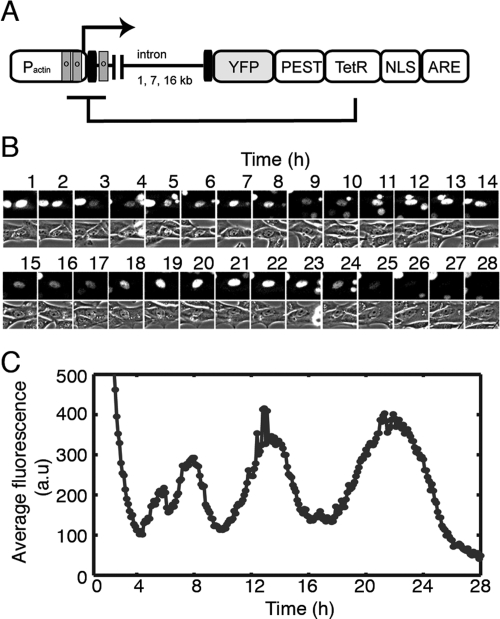
Figure 1.
An engineered network with delayed autoinhibition exhibits pulses of protein expression. (A) Schematic diagram of the reporter used for the study of intron delays in a gene network with delayed autoinhibition (sequences and assembly strategy are detailed in Supplemental Table 1). The mouse β-actin promoter (Pactin) that includes its first intron drives the expression of the reporter. Fluorescence detection is possible because of YFP, the Venus fast-maturing variant of yellow fluorescent protein. TetR is the tetracycline repressor that, when delivered to the nucleus because of a fusion with a nuclear localization signal (NLS), binds to operator sequences (o) that have been engineered into the promoter to drive repression by steric hindrance and promoter looping-out. The PEST sequence (rich in proline, glutamic acid, serine, and threonine) is a protein degradation element originating in the mouse ornithine decarboxylase gene. AU-rich elements present in the 3′-untranslated region of our gene were included to reduce the stability of the mRNA. Destabilization of both the protein and mRNA increases the responsiveness of the negative feedback loop. (B) Fluorescence micrographs of YFP signal and phase contrast micrographs from a 3T3 cell of a clone that contains a Flp-In, 3-kb version of the gene depicted in A. Images were taken every 6 min, but only images for every 10th exposure are presented. (C) Time-lapse trajectory of the quantified average fluorescence intensity in arbitrary units (a.u.) from the cell in B. The data presented were acquired in a 28-h window that encompasses a single cell division.
Oscillations in gene expression are characteristics of the biological networks underlying vertebrate somitogenesis, the cell cycle, hormonal signaling, and circadian rhythms (Kondo 1993; Goldbeter 2002; Lewis 2003; Monk 2003; Lahav et al. 2004). We focused on several factors that make negative feedback loops more likely to show the dynamic behavior of oscillations, including reduced protein and RNA stability and cooperative promoter binding. We therefore engineered all three factors into our artificial feedback loop (for a full description, see the Supplemental Material). Three islands of tetO were added to the β-actin promoter with the goal of increasing the cooperativity of TetR repression through chromatin looping. We also added a PEST sequence to the repressor protein and AU-rich elements (ARE) to the messenger RNA to reduce their respective stabilities (Zubiaga et al. 1995; Li et al. 1998). We generated clonal populations of 3T3 mouse fibroblast cell lines containing each of the three length variants of the negative feedback loop by using the Flp-In system (Invitrogen) for controlled, site-specific integration. We then studied the expression of the reporter protein using time-lapse fluorescence microscopy.
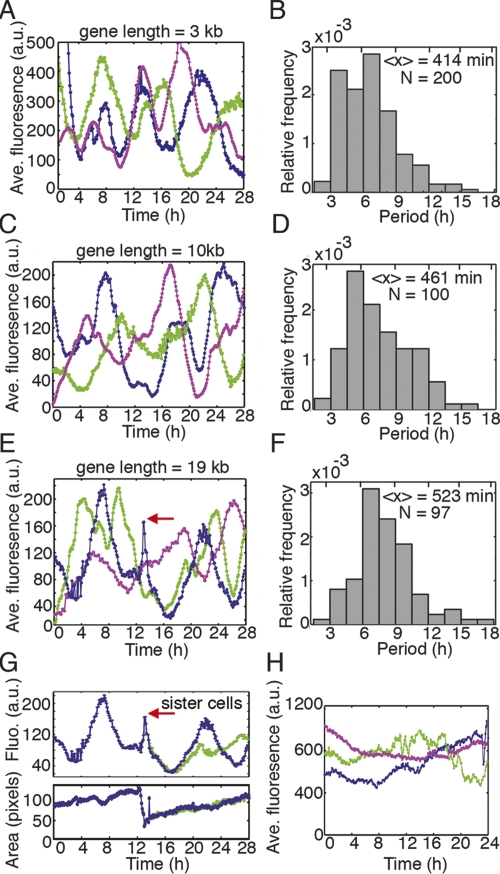
Negative feedback loops studied with short bacterial genes show that autoinhibition can reinforce homeostasis and reduce gene activation times (Becskei and Serrano 2000; Rosenfeld et al. 2002; Dublanche et al. 2006). In contrast, we found that in animal cells our engineered autoinhibitory network can indeed produce pulses of protein expression (Fig. 1B,C; Supplemental Movie 1). Time-lapse imaging of a single cell containing the 3-kb variant of the gene (Fig. 1B) and quantification of the images obtained from this cell (Fig. 1C) show that the shortest pulse length this cell exhibits is 5 h, although the length of the pulse varies widely. Varying pulse length is also seen in other cells expressing this construct (Fig. 2A), and in cells expressing the longer intron constructs (Fig. 2C,E). Data from 152 individual cells are summarized in Figure 2, B, D and F. Although the distribution of pulse length is broad, both the median value (3 kb: 408 min, 10 kb: 435 min, 19 kb: 480 min) and the mean value (3 kb: 414 min, 10 kb: 461 min, 19 kb: 523 min) of the pulse length distribution increase with gene length (Fig. 2B,D,F). The increase in period from the 3-kb to 19-kb gene is significant (P < 0.0001, Mann-Whitney U-test). The cause of this increase could be due to either transcription elongation alone or the combined influence of transcription times and altered splicing rates. Either mechanism yields a greater time delay with longer introns. Additionally, both transcription and negative feedback are required for oscillations. In the presence of doxycycline, which prevents the Tet repressor from binding to tetO, all three cell types showed slow fluctuations instead of clear oscillations (Fig. 2H). Inhibition of transcriptional elongation using α-amanitin also abrogated pulsing (data not shown), indicating that post-transcriptional events such as nuclear transport were not sufficient to cause oscillations.
Figure 2.
Time between pulses of protein expression increases with intron length. (A,C,E) Representative expression trajectories from single cells of clones (each color is a different cell) containing either a 3-kb (A), 10-kb (C), or 19-kb (E) version of the gene from Figure 1. (B,D,F) One-hundred-fifty-two cells were examined and presented are histograms of pulse lengths (n = 97–200) from single cells within the clones represented in A–C. Pulses are interrupted by the mitotic constraint as in E and G (red arrow highlights the mitotic event). (G) After a cell division, sister cells behave similarly for a window of time (here ∼8 h) before the reporter’s behavior begins to diverge in the sibling cells. In the bottom panel, the area of the dividing cell is present to illustrate several hallmarks that indicate a mitotic event: rounding (sharp decrease in area) and division (area becomes about half of predivision area). (H) Over longer periods of time, cells without autoinhibition exhibit relatively constant expression that fluctuates slowly.
We also observed that mitosis affects expression of the delayed negative feedback loop; the trajectories of expression pulses are interrupted after cell divisions (period lengths were not measured across divisions in Fig. 2B,D,F). For example, in a cell expressing the reporter protein from a 19-kb gene (Fig. 2E, blue trajectory), a cell division occurs at ∼13 h (Fig. 2E, red arrow). After cell division, the reporter’s expression eventually continues on the downward course it was headed on before mitosis. An examination of the two daughters produced from this cell division (Fig. 2G) reveals that the sibling cells behave similarly for a window of time (∼8 h), but then the trajectories diverge.
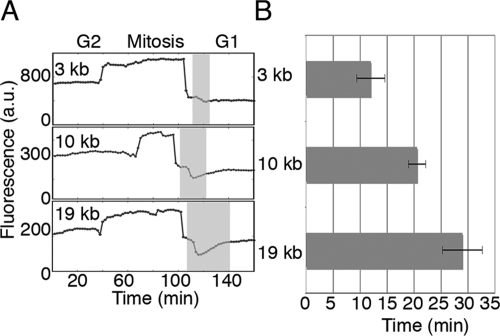
To better understand the impact of mitosis, we examined expression in the absence of autoinhibition by adding doxycycline. By observing gene expression at the higher time resolution of 2 min, we were able to identify dips below a steady level of gene expression during the early G1 phase of the cell cycle that increase in duration with intron length (Fig. 3A). To quantify this difference, we used the nuclear marker to identify when two nascent nuclei are first resolvable. We then measured the time from nuclear emergence (beginning of gray stripes) to the time when the increase in fluorescence slows dramatically (the inflection point, end of gray stripes). From this analysis, we found that the average impact of the cell-cycle constraint increases with intron length: by 16 min as gene length increases by 16 kb (Fig. 3A,B).
Figure 3.
The impact of the mitotic constraint increases with intron length. (A) Cells without autoinhibition were observed at the higher time resolution of 2 min as they went through mitosis. Here, the presented fluorescence values are total fluorescence because segmented object (the nucleus) changes size rapidly during and shortly after mitosis. In early G1, after two nuclei are resolvable, there is a dip in expression (gray stripes, A) that persists for longer lengths of time as the size of the gene increases, quantified for multiple cell division in B (n = 10,10, and 8 for 3-kb, 10-kb, and 19-kb genes). Error bars mark the standard deviation.
The observed times between pulses of gene expression were longer than might be expected. The delay introduced by transcription and translation in this system are expected to be on the order of an hour and modeling where these parameters dominate the delay would predict a period of ∼2 h (Lewis 2003; Monk 2003). However, if these models take into account significant contributions from the slower degradation of the protein and mRNA in eukaryotes, the period can be much longer (Fig. 4B). This is confirmed as we measured the protein half-life to be 45 min, and mRNA half-life is expected to be on the order of 45–60 min (Zubiaga et al. 1995).
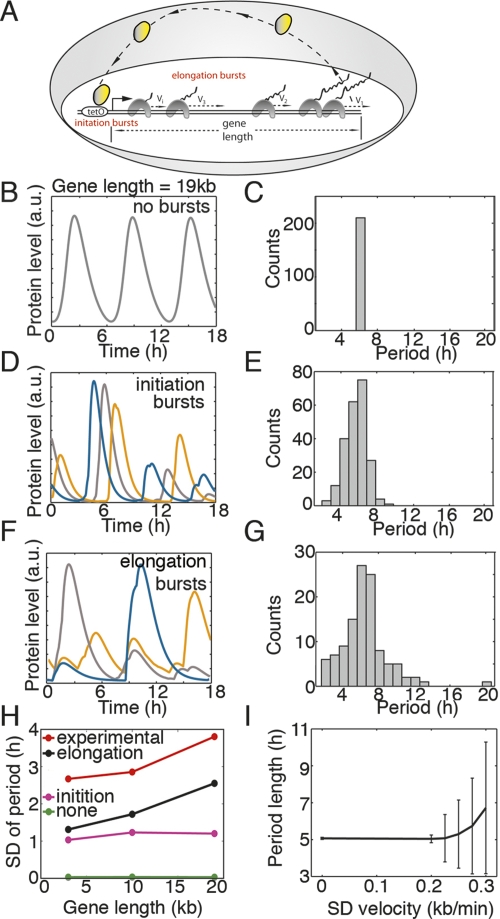
Figure 4.
Simulations of delayed autoinhibition suggest that bursting accumulates during transcription elongation. (A) Schematic of delayed autoinhibition that highlights bursting originating from either transcription initiation or transcription elongation involving polymerases that transcribe with a broad distribution of velocities (vi). (B) Trajectories from simulations of delayed autoinhibition without bursting exhibit little variation in period (shown in C). (D) Trajectories from simulation of delayed autoinhibition that incorporate transcription initiation bursts exhibit a broad distribution of period lengths (shown in E). (F) When simulations incorporate transcription bursts due to congestion and traffic jams during elongation, broad distributions of the pulse period are observed (G,H). (H) As gene length increases in simulations, only those that incorporate elongation bursts exhibit a similar length sensitive increase in the standard deviation of period lengths observed experimentally. (I) In simulations that involve slow leading RNA polymerases, the standard deviation of the pulse periods increases rapidly when the standard deviation of polymerase velocities exceeds 0.2 kb/min.
RNA and protein production occur in bursts (Fig. 4A; Golding et al. 2005; Cai et al. 2006; Raj et al. 2006; Yu et al. 2006). In the absence of bursting, pulses from simulated delayed autoinhibition occur with precise regularity and increase with time delay (impact of time delay shown previously) (Fig. 4B,C; Lewis 2003; Monk 2003). The precision was robust when time intervals between transcription initiation events fall within a Gaussian distribution, even for standard deviations greater than the mean (simulations not shown) (Monk 2003). Additionally, we performed simulations of our delayed autoinhibition system to ask whether the broad distributions of pulse lengths are consistent with bursting during gene expression (described in the Supplemental Material). As extensions to the prior modeling of delayed autoinhibition in biology (Lewis 2003; Monk 2003), we introduced bursting to simulations using two approaches. In the first approach, we applied the observed transcriptional bursting characteristics measured in Escherichia coli (Golding et al. 2005) and assumed that this bursting originates during transcription initiation events (Fig. 4A). Simulations show trajectories that behave similarly to our experimental observations in that the lengths of time between pulses of gene expression are irregular and broadly distributed (Fig. 4D,E, different colors are different runs of the simulation). Unlike our experimental observations, the standard deviation of burst length remains relatively constant as gene length (time delay) increases in the model (Fig. 4H, purple line).
To explore an alternative contribution to transcriptional bursting, we tested the hypothesis that bursting is a consequence of RNA polymerase congestion or traffic jams (Fig. 4A; MacDonald et al. 1968; Epshtein and Nudler 2003; Swinburne and Silver 2008). To produce congestion events in silico, we simulated transcription elongation through different lengths of genes where heterogeneities in transcription velocities cause polymerases to accumulate behind a slow leading polymerase (Adelman et al. 2002; Tolic-Norrelykke et al. 2004). Trajectories from simulations using this approach are irregular like the pulses we observed experimentally (Fig. 4F, different colors are different runs of the simulation). The distribution of pulse lengths from many simulations is broad, and the standard deviation in the period of gene expression increases with gene length (Fig. 4G,H, black line). This is consistent with the observed increase in the standard deviation of period lengths in our experimental samples (3 kb: 146 min, 10 kb: 165 min, 19 kb: 223 min) (comparison in Fig. 4H). In conclusion, the experimentally observed broad distributions of gene expression pulses generated by delayed autoinhibition are consistent with bursting that is enhanced during transcription elongation and thus elevated by greater gene length.
Our engineered reporter gene provides a novel perspective on gene networks because it exhibits expression dynamics for a negative feedback loop that are responsive to gene length. With the traffic rules used in our simulations, the impact of the elongation bursting on delayed autoinhibition is very sensitive to the distribution of polymerase velocities. The standard deviation of periods increases rapidly as the standard deviation of polymerase velocities increases above 0.2 kb/min (Fig. 4I). As introns do not appear to introduce fixed time delays, the study of burst propagation during transcription elongation is relevant for its potential impact on the precision of timing and dynamics of networks that rely on transcriptional components. Additionally, it is well documented that pausing occurs during transcription. Future work will need to resolve the relationship between pausing and bursting.
Our results show that intron length can indeed affect the dynamics of transcriptionally controlled feedback loops; such effects may be important in many contexts, such as somitogenesis during development and responses to immunological signals such as NF-κb (Hoffmann et al. 2002; Lewis 2003; Monk 2003). The system presented here offers considerable potential for further study of the kinetic effects of other under-characterized aspects of expression timing unique to eukaryotic genomes such as histone modifications, the relationship between gene length and splicing rates, and alternative splicing.
Materials and methods
Generation of cell lines
The gene, as outlined in Figure 1A, was assembled by standard molecular cloning strategies. A detailed presentation of the cloning and sequence of the gene is present in the Supplemental Material. The gene was made in the backbone of pcDNA5/FRT (Invitrogen). The expression plasmid was then cotransfected with a flipase-expressing plasmid into Flp-In mouse 3T3 fibroblasts (Invitrogen), from which stable clones were then selected with hygromycin (200 μg/mL). The clones also stably express an integrated histone-mCherry. This nuclear signal was used in the analysis of the movies for image segmentation.
Time-lapse microscopy
Roughly 2 × 104 cells were plated in the wells of 12-well glass coverslip bottom polylysine-coated plates (MatTek) and grown with DMEM, 10%FBS, and 2.5 μg/mL doxycycline for 4 d prior to imaging. Before imaging, cells were washed 3× with 37°C PBS and then grown in DMEM without phenol red and supplemented with 10% FBS that had been screened for the absence of tetracycline (Clontech, 631101). Long-term imaging was performed in a heated incubation chamber (37°C, 5% CO2). Images were acquired on a Nikon TE2000-E equipped with a 20×, NA 0.75 phase objective (Nikon), Hamamatsu Orca AG cooled CCD camera, Prior Scientific Proscan motorized stage, Metamorph software, YFP (41,028, Chroma), and RFP filter sets (G-2E/C, Nikon). Images were acquired every 6 min for most experiments and every 2 min for the observation of the cell cycle constraint.
Image analysis
Custom Matlab (The Mathworks) programs were adapted from previous approaches to track and segment nuclei using the histone-mCherry reporter (detailed description in Supplemental Material). When small bumps occurred around a minimum, the lowest minimum was chosen to delineate periods. It was not determined which small bumps are biological (possibly due to bursting) and which are due to technical artifacts (bulb flicker, cell movements, etc.).
Measuring pulse lengths
Pulse lengths are times between expression minima. Periods were not measured through mitotic events because of the impact of the mitotic constraint.
Modeling
A description of the modeling is presented in the Supplemental Material. Simulations were run using Matlab (The Mathworks, code available upon request).
Acknowledgments
We are thankful for the advice and insights provided by Johan Paulsson, Michael Greenberg, and Jeff Parvin. We thank Rebecca Ward for comments on the manuscript and Jennifer Waters, Lara Petrak, and Cassandra Rogers for assistance with image acquisition and maintenance of the Harvard Systems Biology Microscope Facility. We also thank A. Francis Stewart and Konstantinos Anastassiadis for providing reagents. This work was supported by grants from the National Institutes of Health. D.G.M. acknowledges financial support of a post-doctoral fellowship from the Minesterio de Educación y Ciencia of Spain.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1696108.
References
- Adelman K., La Porta A., Santangelo T.J., Lis J.T., Roberts J.W., Wang M.D. Single molecule analysis of RNA polymerase elongation reveals uniform kinetic behavior. Proc. Natl. Acad. Sci. 2002;99:13538–13543. doi: 10.1073/pnas.212358999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anastassiadis K., Kim J., Daigle N., Sprengel R., Scholer H.R., Stewart A.F. A predictable ligand regulated expression strategy for stably integrated transgenes in mammalian cells in culture. Gene. 2002;298:159–172. doi: 10.1016/s0378-1119(02)00979-4. [DOI] [PubMed] [Google Scholar]
- Becskei A., Serrano L. Engineering stability in gene networks by autoregulation. Nature. 2000;405:590–593. doi: 10.1038/35014651. [DOI] [PubMed] [Google Scholar]
- Cai L., Friedman N., Xie X.S. Stochastic protein expression in individual cells at the single molecule level. Nature. 2006;440:358–362. doi: 10.1038/nature04599. [DOI] [PubMed] [Google Scholar]
- Dublanche Y., Michalodimitrakis K., Kummerer N., Foglierini M., Serrano L. Noise in transcription negative feedback loops: Simulation and experimental analysis. Mol. Syst. Biol. 2006;2:41. doi: 10.1038/msb4100081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epshtein V., Nudler E. Cooperation between RNA polymerase molecules in transcription elongation. Science. 2003;300:801–805. doi: 10.1126/science.1083219. [DOI] [PubMed] [Google Scholar]
- Goldbeter A. Computational approaches to cellular rhythms. Nature. 2002;420:238–245. doi: 10.1038/nature01259. [DOI] [PubMed] [Google Scholar]
- Golding I., Paulsson J., Zawilski S.M., Cox E.C. Real-time kinetics of gene activity in individual bacteria. Cell. 2005;123:1025–1036. doi: 10.1016/j.cell.2005.09.031. [DOI] [PubMed] [Google Scholar]
- Goodwin B.C. Oscillatory behavior in enzymatic control processes. Adv. Enzyme Regul. 1965;3:425–438. doi: 10.1016/0065-2571(65)90067-1. [DOI] [PubMed] [Google Scholar]
- Hoffmann A., Levchenko A., Scott M.L., Baltimore D. The IκB-NF-κB signaling module: Temporal control and selective gene activation. Science. 2002;298:1241–1245. doi: 10.1126/science.1071914. [DOI] [PubMed] [Google Scholar]
- Kondo S. Circadian variation of bronchial caliber and antigen-induced late asthmatic response. Chest. 1993;104:801–805. doi: 10.1378/chest.104.3.801. [DOI] [PubMed] [Google Scholar]
- Lahav G., Rosenfeld N., Sigal A., Geva-Zatorsky N., Levine A.J., Elowitz M.B., Alon U. Dynamics of the p53-Mdm2 feedback loop in individual cells. Nat. Genet. 2004;36:147–150. doi: 10.1038/ng1293. [DOI] [PubMed] [Google Scholar]
- Lewis J. Autoinhibition with transcriptional delay: A simple mechanism for the zebrafish somitogenesis oscillator. Curr. Biol. 2003;13:1398–1408. doi: 10.1016/s0960-9822(03)00534-7. [DOI] [PubMed] [Google Scholar]
- Li X., Zhao X., Fang Y., Jiang X., Duong T., Fan C., Huang C.C., Kain S.R. Generation of destabilized green fluorescent protein as a transcription reporter. J. Biol. Chem. 1998;273:34970–34975. doi: 10.1074/jbc.273.52.34970. [DOI] [PubMed] [Google Scholar]
- MacDonald C.T., Gibbs J.H., Pipkin A.C. Kinetics of biopolymerization on nucleic acid templates. Biopolymers. 1968;6:1–5. doi: 10.1002/bip.1968.360060102. [DOI] [PubMed] [Google Scholar]
- Manak J.R., Dike S., Sementchenko V., Kapranov P., Biemar F., Long J., Cheng J., Bell I., Ghosh S., Piccolboni A., et al. Biological function of unannotated transcription during the early development of Drosophila melanogaster. Nat. Genet. 2006;38:1151–1158. doi: 10.1038/ng1875. [DOI] [PubMed] [Google Scholar]
- Monk N.A. Oscillatory expression of Hes1, p53, and NF-κB driven by transcriptional time delays. Curr. Biol. 2003;13:1409–1413. doi: 10.1016/s0960-9822(03)00494-9. [DOI] [PubMed] [Google Scholar]
- Nagai T., Ibata K., Park E.S., Kubota M., Mikoshiba K., Miyawaki A. A variant of yellow fluorescent protein with fast and efficient maturation for cell-biological applications. Nat. Biotechnol. 2002;20:87–90. doi: 10.1038/nbt0102-87. [DOI] [PubMed] [Google Scholar]
- Raj A., Peskin C.S., Tranchina D., Vargas D.Y., Tyagi S. Stochastic mRNA synthesis in mammalian cells. PLoS Biol. 2006;4:e309. doi: 10.1371/journal.pbio.0040309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenfeld N., Elowitz M.B., Alon U. Negative autoregulation speeds the response times of transcription networks. J. Mol. Biol. 2002;323:785–793. doi: 10.1016/s0022-2836(02)00994-4. [DOI] [PubMed] [Google Scholar]
- Rothe M., Pehl M., Taubert H., Jackle H. Loss of gene function through rapid mitotic cycles in the Drosophila embryo. Nature. 1992;359:156–159. doi: 10.1038/359156a0. [DOI] [PubMed] [Google Scholar]
- Shermoen A.W., O’Farrell P.H. Progression of the cell cycle through mitosis leads to abortion of nascent transcripts. Cell. 1991;67:303–310. doi: 10.1016/0092-8674(91)90182-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swinburne I.A., Silver P.A. Intron delays and transcriptional timing during development. Dev. Cell. 2008;14:324–330. doi: 10.1016/j.devcel.2008.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tennyson C.N., Klamut H.J., Worton R.G. The human dystrophin gene requires 16 hours to be transcribed and is cotranscriptionally spliced. Nat. Genet. 1995;9:184–190. doi: 10.1038/ng0295-184. [DOI] [PubMed] [Google Scholar]
- Tolic-Norrelykke S.F., Engh A.M., Landick R., Gelles J. Diversity in the rates of transcript elongation by single RNA polymerase molecules. J. Biol. Chem. 2004;279:3292–3299. doi: 10.1074/jbc.M310290200. [DOI] [PubMed] [Google Scholar]
- Venter J.C., Adams M.D., Myers E.W., Li P.W., Mural R.J., Sutton G.G., Smith H.O., Yandell M., Evans C.A., Holt R.A., et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- Yu J., Xiao J., Ren X., Lao K., Xie X.S. Probing gene expression in live cells, one protein molecule at a time. Science. 2006;311:1600–1603. doi: 10.1126/science.1119623. [DOI] [PubMed] [Google Scholar]
- Zubiaga A.M., Belasco J.G., Greenberg M.E. The nonamer UUAUUUAUU is the key AU-rich sequence motif that mediates mRNA degradation. Mol. Cell. Biol. 1995;15:2219–2230. doi: 10.1128/mcb.15.4.2219. [DOI] [PMC free article] [PubMed] [Google Scholar]