Abstract
Yeast initiation factor eIF3 (eukaryotic initiation factor 3) has been implicated in multiple steps of translation initiation. Previously, we showed that the N-terminal domain (NTD) of eIF3a interacts with the small ribosomal protein RPS0A located near the mRNA exit channel, where eIF3 is proposed to reside. Here, we demonstrate that a partial deletion of the RPS0A-binding domain of eIF3a impairs translation initiation and reduces binding of eIF3 and associated eIFs to native preinitiation complexes in vivo. Strikingly, it also severely blocks the induction of GCN4 translation that occurs via reinitiation. Detailed examination unveiled a novel reinitiation defect resulting from an inability of 40S ribosomes to resume scanning after terminating at the first upstream ORF (uORF1). Genetic analysis reveals a functional interaction between the eIF3a-NTD and sequences 5′ of uORF1 that is critically required to enhance reinitiation. We further demonstrate that these stimulatory sequences must be positioned precisely relative to the uORF1 stop codon and that reinitiation efficiency after uORF1 declines with its increasing length. Together, our results suggest that eIF3 is retained on ribosomes throughout uORF1 translation and, upon termination, interacts with its 5′ enhancer at the mRNA exit channel to stabilize mRNA association with post-termination 40S subunits and enable resumption of scanning for reinitiation downstream.
Keywords: Translation initiation, reinitiation, eIF3, 40S ribosomal subunit, GCN4, short uORF
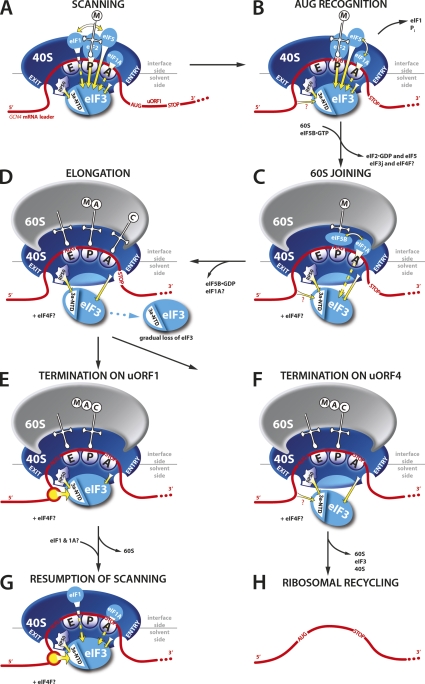
General translation initiation (GTI) in eukaryotes consists of several steps that ultimately lead to formation of the 80S ribosome with the anti-codon of methionyl initiator tRNA (Met-tRNAiMet) base-paired to the AUG start codon in the ribosomal P-site. In the first step, Met-tRNAiMet is bound by eukaryotic initiation factor 2 (eIF2) in its GTP form to produce the ternary complex (TC). The TC is subsequently recruited to the small ribosomal subunit with the help of eIF3, eIF1, eIF5, and eIF1A, forming the 43S preinitiation complex (PIC). The 43S PIC then interacts with the 5′ end of mRNA in a reaction stimulated by eIF4F, eIF4B, PABP, and eIF3. The 48S PIC thus formed scans the mRNA leader until the AUG start codon is recognized (for review, see Marintchev and Wagner 2005). Scanning is promoted by eIF1, eIF1A, and eIF4F in a mammalian reconstituted system (Pestova and Kolupaeva 2002), whereas yeast genetic data indicate that eIF3 and eIF5 also participate in vivo (Nielsen et al. 2004; Yamamoto et al. 2005). The GTP bound to eIF2 is partially hydrolyzed to GDP and Pi, dependent on the GAP eIF5, but the Pi is not released from the scanning complex until initiation codon–anti-codon base-pairing induces dissociation or displacement of eIF1 (Algire et al. 2005; Cheung et al. 2007). Met-tRNAiMet is then released into the P-site, and the 60S subunit can join the 40S-mRNA-Met-tRNAiMet PIC in a reaction stimulated by GTP-bound eIF5B (Pestova et al. 2000). Subunit joining is thought to facilitate ejection of all eIFs but eIF1A (Unbehaun et al. 2004). Finally, GTP-hydrolysis on eIF5B triggers its dissociation, producing an active 80S ribosome poised for elongation (summarized in Fig. 6, below).
Figure 6.
A yeast model for eukaryotic reinitiation following translation of a short uORF. (A,B) eIF3 association with the scanning 48S PIC is stabilized by supporting contacts with eIF1, eIF1A, eIF5, and the TC. (C) Upon subunit joining, eIF3 (and possibly also eIF4F) remains bound to the 80S ribosomes owing to its strategic position on the solvent-exposed side of the 40S ribosome and the contacts that it makes with 40S ribosomal components (e.g., eIF3a-NTD with RPS0A) and presumably also with mRNA. (D) During the first few rounds of elongation, weakly associated eIF3 gradually dissociates from the 80S as a function of length and complexity of the translated region. (E,F) After translation of a short uORF, a certain proportion of 80S ribosomes terminating at its stop codon still contains eIF3, the presence of which is required for resumption of scanning. (E,G) Binding of the eIF3a-NTD directly to the specific eIF3a-RS 5′ of uORF1 greatly stabilizes association of the post-termination 40S subunit with mRNA following dissociation of the 60S subunit in the first stage of the ribosome recycling reaction and thereby promotes efficient resumption of scanning and REI downstream. (F,H) Absence of the stimulatory eIF3a-RS, for example at uORF4, results in completion of ribosomal recycling by the majority of terminating 80S ribosomes.
Various alternative mechanisms to the GTI pathway exist that are mostly utilized by viruses or function in gene-specific translational control. They rely on cis-acting mRNA features and exhibit distinct factor requirements. Reinitiation (REI) is one such mechanism utilized to down- or up-regulate translation of regulatory proteins such as transcription factors and proto-oncogenes in response to various environmental stimuli (Kozak 2005). Ribosomes initiate in the normal way at the first AUG codon; however, at the termination codon, the 40S subunit (40S) remains bound to the mRNA, resumes scanning, and initiates again at a downstream start site. REI depends on de novo recruitment of the TC that is required to recognize the next AUG codon; therefore, it can be delicately regulated by manipulating the eIF2⋅GTP levels (for review, see Hinnebusch 2005).
Except for a few features in mRNA structure, we know very little about the molecular mechanisms underlying REI. It was shown that the ability of the 40S to reinitiate is limited by the size of the upstream ORF (uORF); however, the critical parameter that determines whether the 40S resumes scanning seems to be the time taken to translate the uORF, rather than its length (Kozak 2001; Rajkowitsch et al. 2004). REI is usually inefficient owing to excessive length or other features of the uORF that prevent retention of post-termination 40S ribosomes on mRNA. Hence, short uORFs generally serve to reduce protein production from a major ORF downstream (Kozak 2005). There are, however, a few examples where uORFs bear special characteristics that render them highly permissive for REI (REI-permissive), such as the mRNAs encoding the yeast transcription factors YAP1 (Vilela et al. 1998), GCN4 (Hinnebusch 2005) and its mammalian ortholog ATF4 (Vattem and Wek 2004), and bZIP transcriptional regulator ATF5 (Zhou et al. 2008).
GCN4 is a transcriptional activator of a large number of biosynthetic genes (Hinnebusch 2005). Although GCN4 mRNA is synthesized constitutively, its translation is repressed under nutrient-rich conditions through a REI mechanism involving four upstream open reading frames (uORFs 1–4) that is very sensitive to the TC levels in cells. After translating the first and only REI-permissive uORF1, small ribosomal subunits remain attached to the mRNA, resume scanning, and reinitiate downstream. Under nonstarvation conditions, characterized by high levels of the TC, nearly all of the rescanning 40S ribosomes will rebind the TC before reaching uORFs 2–4, translate one of them, and dissociate from the mRNA. Amino acid starvation leads to phosphorylation of eIF2α by kinase GCN2, converting eIF2.GDP from a substrate to a competitive inhibitor of its GEF, eIF2B, thus reducing the concentration of TC. Low TC levels derepress GCN4 translation by allowing ∼50% of rescanning 40S ribosomes to rebind TC after bypassing uORF4 and reinitiate at GCN4 instead. Failure to induce expression of GCN4 in response to a shortage of amino acids in various mutant cells confers increased sensitivity to inhibitors of amino acid biosynthetic enzymes, and is designated the Gcn− phenotype. Conversely, constitutive expression of GCN4 independent of amino acid levels due to a defect in TC assembly or recruitment overcomes sensitivity to the latter inhibitors in gcn2Δ cells and is called the Gcd− phenotype. A related but not identical mechanism has been shown to govern translation of the mammalian ATF4 and ATF5 transcription factors, indicating that at least basic principles of this regulatory system have been evolutionarily conserved (Vattem and Wek 2004).
A crucial but vaguely understood feature of GCN4 translational control is the highly disparate capacities of uORF1 and uORF4 to permit efficient resumption of scanning following translation termination. Mutational analyses revealed that AU-rich sequences surrounding the stop codon of uORF1 favor resumption of scanning and REI, whereas GC-rich sequences flanking the uORF4 stop codon likely trigger ribosome release (Grant and Hinnebusch 1994). Sequences 5′ of uORF1 were also shown to be critical for efficient REI (Grant et al. 1995). Virtually nothing is known about what trans-acting factors, if any, function in concert with these mRNA features to promote REI. Mutations in eIF3 subunits b and c, and in eIF1, eIF5, eIF1A, and eIF5B, respectively, were shown to deregulate translational control of GCN4 through their defects in TC recruitment, scanning, AUG selection, or subunit joining (for review, see Hinnebusch 2005); however, no mutations have been isolated that impair retention of post-termination ribosomes at the uORF1 stop codon and the resumption of scanning that is required for REI.
Our previous studies of yeast Saccharomyces cerevisiae eIF3 demonstrated that it plays a stimulatory role in nearly all steps of GTI (for review, see Hinnebusch 2006). It is composed of six subunits (a, b, c, i, g, and j), all of which have corresponding orthologs in mammalian eIF3 (meIF3). In S. cerevisiae, the TC and eIF1, eIF3, and eIF5 can be found in the higher-order ribosome-free assembly called the Multifactor Complex (MFC), and we and others previously demonstrated that there is a substantial cooperation among the eIFs assembled in the MFC in their binding to the 40S as well as their ribosome-associated functions in scanning and AUG recognition (Valášek et al. 2002, 2004; Nielsen et al. 2004, 2006; Yamamoto et al. 2005; Jivotovskaya et al. 2006). An important task is to elucidate the molecular mechanism underlying this extensive cooperation within the MFC.
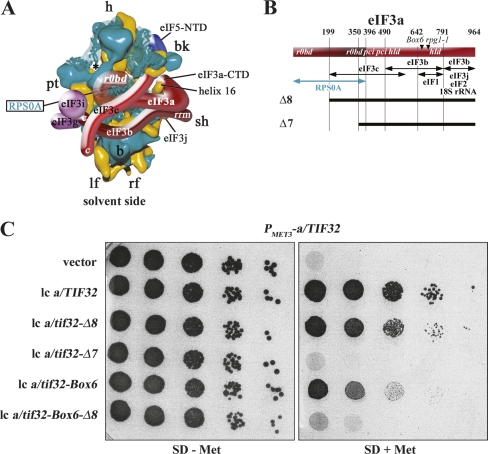
We began addressing this issue by mapping the positions of individual components of the MFC on the 40S (Valášek et al. 2003). We found that deleting the N- and C-terminal domains (NTD and CTD) of eIF3c or the NTD of eIF3a impaired association of otherwise intact eIF3 complexes with the 40S, and, in addition, deleting the eIF3a-CTD reduced 40S association of the MFC when the connection between eIF3 and eIF5/TIF5 in the MFC was impaired by the tif5-7A mutation. In a separate study, the RNA recognition motif (RRM) of the eIF3b-NTD that mediates its interactions with eIF3j and eIF3a was implicated in the ability of eIF3j to stimulate 40S binding by eIF3 (Nielsen et al. 2006). This RRM-eIF3j network is conserved in mammals (ElAntak et al. 2007). Importantly, our findings that the eIF3a-CTD interacts with helices 16–18 of 18S rRNA and that eIF3a-NTD binds to ribosomal proteins RPS0A and RPS10A (Valášek et al. 2003) suggested that yeast eIF3 associates with the solvent-exposed side of the 40S (Fig. 1A), as suggested by others for meIF3 (Srivastava et al. 1992; Siridechadilok et al. 2005).
Figure 1.
Genetic evidence that a partial deletion of the RPS0A-binding domain in the eIF3a-NTD affects cell growth. (A) Hypothetical location of the eIF3 complex on the S. cerevisiae 40S subunit based on Cryo-EM reconstruction adapted from Valášek et al. (2003). The 40S subunit is shown from the solvent side, with RNA segments in yellow and proteins in green. Positions of RPS0A and the RPS0A-binding domain (R0BD) of eIF3a are indicated. The mRNA exit channel is designated by an asterisk. (B) Schematic of eIF3a with arrows delimiting the minimal binding domains for the indicated proteins identified previously. The lines beneath the schematic depict two N-terminally truncated eIF3a-His proteins that were analyzed in this study. The locations of the RPS0A-binding domain (r0bd), the PCI homology domain (pci), the eIF3j/HCR1-like domain (hld), and two mutations are indicated in the colored rectangle. (C) Partial deletion of the R0BD in the eIF3a-NTD (a/tif32-Δ8) produces the Slg− phenotype. Transformants of strain YAH01 bearing the a/TIF32 wild-type allele under control of the MET3 promoter containing empty vector; lc pRSeIF3a-His; lc pRSeIF3a-Δ8-His; lc pRSeIF3a-Δ7-His; lc pRSeIF3a-Box6; and lc pRSeIF3a-Δ8-Box6-His, respectively, were spotted in five serial dilutions on SD medium in the absence (left panel) or presence (right panel) of 2 mM methionine and incubated for 2 d at 30°C.
In this study, we focused on the role of the NTD of eIF3a, known as TIF32 in yeast, in promoting association of eIF3 and other MFC components with the 40S. We found that a partial deletion of the RPS0A-binding site in the a/tif32-NTD (Δ8) is not lethal but reduces the amounts of 40S-bound MFC components in vivo, consistent with the idea that the eIF3a-NTD forms a crucial intermolecular bridge between eIF3 and the 40S. Strikingly, the a/tif32-Δ8 truncation imparted a severe Gcn− phenotype with novel characteristics, providing the first in vivo evidence that eIF3 is required for post-termination retention of 40S ribosomes at the uORF1 stop codon. Genetic epistasis interactions between mutations in the stimulatory sequences upstream of uORF1 and a/tif32-Δ8 strongly indicate that eIF3a interacts with this REI-enhancing element that we named eIF3a-NTD-responsive site (eIF3a-RS). Thus, we propose that establishment of the interaction between eIF3a-NTD and the specific eIF3a-RS 5′ of uORF1 at or near the mRNA exit channel of the post-termination 40S subunit stabilizes its association with mRNA and promotes the resumption of scanning for efficient REI at the downstream ORF.
Results
The eIF3a-NTD constitutes a critical link between eIF3 and the 40S subunit
We recently identified several important domains of eIF3 subunits and eIF5 mediating interaction of the MFC with the 40S that allowed us to predict certain aspects of the organization of the 43S PIC (Fig. 1A; Valášek et al. 2003). Among them, the eIF3a-NTD was found to interact in vivo and in vitro with the CTD of small ribosomal protein RPS0A, which has been located beneath the mRNA exit channel on the solvent side of the 40S (Spahn et al. 2001). Consistently, a partial deletion of the RPS0A-binding domain (R0BD) in a/tif32-Δ8 (Fig. 1B) that did not interfere with the integrity of the MFC (Valášek et al. 2002) completely eliminated binding of the mutant form of the complex (MFC-Δ8) to the 40S in vivo when competing with the wild-type MFC (Valášek et al. 2003). Our previous finding that a/tif32-Δ8 partially complemented the temperature sensitive (Ts−) phenotype of the a/tif32-R731I/rpg1-1 mutant strain (Valášek et al. 2002) prompted us to inquire whether a/tif32-Δ8 can support viability in cells lacking wild-type a/TIF32. If so, we could eliminate the effect of competition and examine the phenotypes of a/tif32-Δ8 in cells lacking wild-type a/TIF32. Thus, we constructed a YAH01 strain in which wild-type a/TIF32 is expressed under control of the MET3 promoter that can be turned off by addition of methionine to the medium. Indeed, plasmid-borne a/tif32-Δ8 supported growth of YAH01 cells in the presence of methionine, albeit producing a slow-growth phenotype (Slg−) with a doubling time (dt) of 2.8 h compared with 1.6 h for the wild-type strain (Fig. 1C, row 3 vs. 2). By contrast, a/tif32-Δ7, with a nearly complete deletion of the R0BD (Fig. 1B), did not complement depletion of wild-type eIF3a (Fig. 1C, row 4), even when expressed from a high-copy plasmid (data not shown).
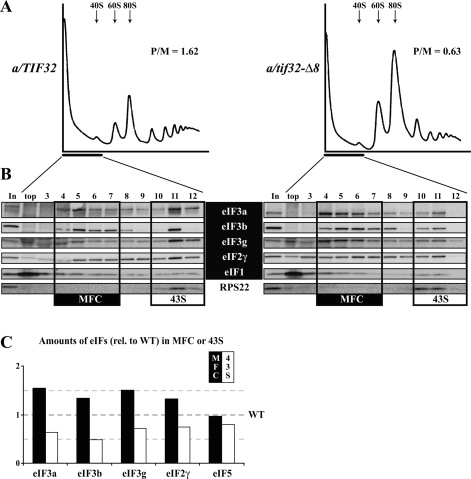
We next wished to confirm that Slg− of the a/tif32-Δ8 cells is associated with a defect in GTI. To that end, we analyzed the polysome content in a/tif32-Δ8 cells pretreated with formaldehyde to cross-link ribosomes on mRNA, by resolving whole-cell extracts (WCEs) by velocity sedimentation through 5%–45% sucrose gradients (Valášek et al. 2007). The a/tif32-Δ8 extracts showed polysomal run-off and an increased amount of 80S monosomes, leading to a ∼2.5-fold decrease in the polysome to monosome (P/M) ratio, which indicates a reduced rate of GTI (Fig. 2A).
Figure 2.
The eIF3a-NTD constitutes a critical link between eIF3 and the 40S ribosome. (A) Isogenic strains YBS47 (GCN2 a/tif32Δ pRSeIF3a-His) and YBS53 (GCN2 a/tif32Δ pRSeIF3a-Δ8-His) were grown in YPD medium at 30°C to an OD600 of ∼1.5 and cross-linked with 1% HCHO prior to harvesting. WCEs were prepared and subsequently separated on a 5%–45% sucrose gradient by centrifugation at 39,000 rpm for 2.5 h. The gradients were collected and scanned at 254 nm to visualize the ribosomal species. Positions of 40S, 60S and 80S species are indicated by arrows and P/M ratios are given above the profiles. (B) The same as in A, except that WCEs were separated on a 7.5%–30% sucrose gradient by centrifugation at 41,000 rpm for 5 h. Proteins from the collected fractions were subjected to Western analysis using antibodies against the proteins listed between the blots. An aliquot of each WCE was analyzed in parallel (In, input), and the first two fractions were combined (top). Rectangles indicate fractions where the Multifactor complex (MFC) constituents (highlighted in black) or the 43S PICs (43S), respectively, sediment. (C) Amounts of the each individual factor in all fractions from three independent experiments were quantified by fluorescence imaging, combined, and the percentage representation of the signal corresponding to the MFC (4–7) or 43S (10–12) fractions was calculated. Values obtained for the a/TIF32 wild-type strain were set to 100%, and relative distribution of individual factors in the MFC and 43S fractions in the a/tif32-Δ8 mutant cells was plotted.
To provide evidence that the eIF3a-NTD represents an important link between the MFC and the 40S, we measured binding of eIF3 subunits and other MFC components to 40S subunits in WCEs of mutant a/tif32-Δ8 cells treated with 1% formaldehyde. This treatment cross-links eIFs to 40S ribosomes in vivo, minimizing dissociation of PICs during sedimentation and thus provides the best available approximation of the native 43S/48S PICs composition in vivo (Valášek et al. 2007). As shown in Figure 2, B and C, we observed ∼40% decreases in the amounts of selected eIF3 subunits sedimenting in the 40S-containing fractions (10–12) with commensurate increases in their levels in fractions 4–7 containing free MFC in the a/tif32-Δ8 cells versus the wild-type control. Similar, but less pronounced, reductions in 40S binding (∼25%) were also observed for eIF5 and the TC component eIF2γ. These results demonstrate that partial deletion of the R0BD of eIF3a reduces stable association of eIF3 and, to a lesser extent, eIF2 and eIF5 with the 40S in vivo.
Further support for the role of the eIF3a-NTD in anchoring eIF3 to the 40S arose from combining a/tif32-Δ8 with a 10-alanine substitution of amino acids 692–701 in the eIF3j/HCR1-like domain (HLD) of eIF3a designated a/tif32-Box6. The a/tif32-Box6 mutation also imparts a Slg− phenotype (dt ∼3.6 h) and reduces association of eIF3 and other MFC constituents with the 40S, suggesting that the HLD also promotes 40S binding of the MFC (L. Burela, L. Valášek, and A.G. Hinnebusch, unpubl.). As shown in Figure 1C, rows 5 and 6, combining the Δ8 and Box6 mutations in the same a/TIF32 allele results in synthetic lethality, consistent with the idea that these mutations disrupt independent contacts of eIF3 with the 40S.
Evidence that a/tif32-Δ8 severely interferes with reinitiation by preventing retention of post-termination ribosomes on mRNA at uORF1
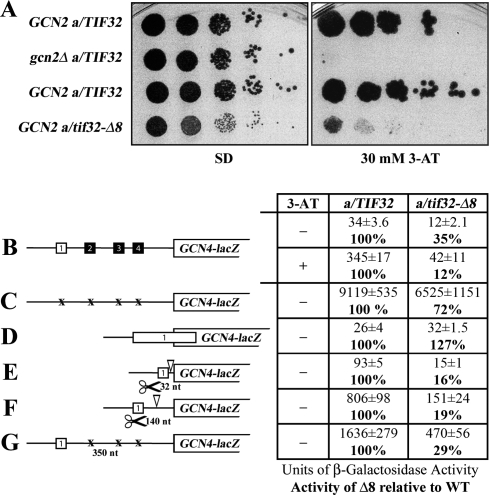
We next wished to confirm our observation that a/tif32-Δ8 reduces association of the TC with the 40S by examining translational regulation of GCN4 expression, which is very sensitive to decreases in the rate of TC loading on 40S subunits in living cells. Based on our biochemical data, we expected that the a/tif32-Δ8 mutation would impart a Gcd− phenotype and allow gcn2Δ cells to grow on 3-aminotriazole (3-AT), an inhibitor of histidine biosynthetic genes. However, this was not observed (data not shown), suggesting that the derepression of GCN4 translation expected from a defect in TC recruitment is being suppressed by another stronger defect in the REI process in this mutant.
In fact, we found that a/tif32-Δ8 GCN2+ cells exhibit a severe Gcn− phenotype, displaying sensitivity to 3-AT nearly as strong as that of the a/TIF32+ gcn2Δ strain where eIF2α cannot be phosphorylated (Fig. 3A). Consistently, derepression of the GCN4-lacZ reporter containing all four uORFs in response to 3-AT was reduced by a factor of about nine compared with the wild-type a/TIF32 strain (Fig. 3B, “+”), fully accounting for the severe 3-ATS phenotype of the a/tif32-Δ8 mutant. This derepression defect resulted partly from a decrease in the induction ratio by a factor of about three and partly from a decrease in the basal level of GCN4-lacZ expression by a factor of about three (Fig. 3B, “−“). The fact that expression from the uORF-less GCN4-lacZ construct was reduced only by a factor of ∼1.3 in a/tif32-Δ8 cells (Fig. 3C) suggests that neither decreased GCN4-lacZ mRNA production nor a general decrease in translation efficiency owing to the 40S binding defect of a/tif32-Δ8 can account for its severe derepression defect. In addition, phosphorylation of eIF2α by GCN2 upon starvation was found to be unaffected in a/tif32-Δ8 cells (data not shown), ruling out this possible mechanism for the Gcn− phenotype. Thus, these results strongly indicate that the eIF3a-NTD plays a critical role in the REI mechanism on GCN4 mRNA, however, by a mechanism distinct from a simple reduction in TC recruitment to the rescanning ribosomes.
Figure 3.
Evidence that a/tif32-Δ8 severely interferes with the reinitiation process by preventing post-termination retention of the 40S ribosome on mRNA. (A) a/tif32-Δ8 imparts a severe Gcn− phenotype implicating eIF3 in regulation of translational control of GCN4 expression that occurs via REI. Isogenic strains H2880 (GCN2 a/TIF32; row 1), and H2881 (gcn2Δ a/TIF32; row 2), YBS47 (GCN2 a/tif32Δ pRSeIF3a-His; row 3), YBS53 (GCN2 a/tif32Δ pRSeIF3a-Δ8-His; row 4), respectively, were spotted in five serial dilutions on SD (left panel) or SD containing 30 mM 3-AT (right panel) and then incubated for 3 and 7 d at 30°C, respectively. (B) a/tif32-Δ8 reduces basal expression of GCN4-lacZ and prevents its full derepression upon starvation. Isogenic strains YBS47 (a/TIF32) and YBS53 (a/tif32-Δ8) were transformed with p180, grown in minimal media for 6 and 8 h, respectively, and the β-galactosidase activities were measured in the WCEs and expressed in units of nanomoles of o-nitrophenyl-b-D-galactopyranoside hydrolyzed per minute per milligram of protein. To induce GCN4-lacZ expression, a/TIF32 and a/tif32-Δ8 transformants grown at the minimal media for 2 h were treated with 10 mM 3-AT for 6 or 16 h, respectively. The mean values and standard deviations obtained from at least six independent measurements with three independent transformants, and activity in a/tif32-Δ8 relative to wild type, respectively, are given in the table to the left of schematics. (C,D) The failure of a/tif32-Δ8 to derepress GCN4-lacZ is not caused by leaky scanning. YBS47 and YBS53 were transformed with p227 (C) and pM226 (D), respectively, and analyzed as in B, except that they were not treated with 3-AT. (E–G) The failure of a/tif32-Δ8 to derepress GCN4-lacZ is not caused by slow or defective scanning. YBS47 and YBS53 were transformed with pG67 (E), pM199 (F), or p209 (G), respectively, and analyzed as in C and D.
The reinitiation process can be divided into two phases: the initial REI-specific phase and the following GTI-like phase. The REI-specific steps occur following translation of uORF1 and include (1) 80S ribosome recycling after polypeptide termination, (2) post-termination retention of the 40S subunit, and (3) recruitment of factors required for resumption of scanning. The GTI-like phase is represented by (1) scanning, (2) TC recruitment, (3) GTP-hydrolysis by TC, (4) AUG recognition, and (5) subunit joining. To determine what step is perturbed by a/tif32-Δ8 and leads to impairment of GCN4 derepression, we analyzed a series of GCN4-lacZ reporters varying in their GCN4 mRNA leader sequences. It should be noted here that the GCN4-lacZ mRNA levels for all constructs used throughout this study were found to be indistinguishable between the wild-type and a/tif32-Δ8 cells (Supplemental Fig. 1). Defects in recognition of the AUG start codon or subunit joining could result in the bypass of uORF1 by scanning PICs (leaky scanning) with initiation at REI-nonpermissive uORFs 2–4 instead. To examine this possibility, the β-galactosidase activity was measured in WCEs from wild-type and mutant cells bearing a reporter plasmid in which uORF1 is elongated and overlaps the beginning of GCN4. The a/tif32-Δ8 mutation produced only a small increase in expression from this construct (Fig. 3D), which cannot account for the strong derepression of wild-type GCN4-lacZ for the following reason: By comparing the results in panels C and D, it can be deduced that only a negligible amount of 40S ribosomes (<1%) that scan from the 5′ end leaky scan the uORF1 AUG, in both mutant and wild-type cells.
Another way to explain the derepression defect in a/tif32-Δ8 cells is to propose that rescanning ribosomes progress more slowly from uORF1 to uORF4 in the mutant cells. This slower rate of scanning would compensate for the reduction in TC recruitment that we observed by providing the scanning 40Ss sufficient time to rebind TC before reaching uORF4. To test that, we analyzed constructs carrying only uORF1 at three different positions relative to GCN4-lacZ (Fig. 3E–G). As observed previously for prt1-1 (eIF3b) (Nielsen et al. 2004), slow scanning would be expected to increase GCN4-lacZ expression from these constructs, particularly those in which uORF1 is closer than normal to the GCN4 AUG start codon (Fig. 3E,F). Importantly, a/tif32-Δ8 strongly decreases, not increases, expression from all three constructs (Fig. 3E–G), arguing against the slow scanning mechanism. It is noteworthy that a/tif32-Δ8 reduces expression from all three constructs by nearly the same factor (71%–84%), despite the fact that the uORF1–GCN4 interval varies dramatically from 32 nucleotides (nt) to 350 nt. This phenotype is also inconsistent with less processive scanning, leading to dissociation of 40S reinitiation complexes (RICs) as they travel downstream from uORF1. Reduced processivity would elicit a much greater reduction in REI efficiency for the construct with the 350-nt spacer (Fig. 3G) versus that with the 32-nt spacer (Fig. 3E), which is not the case.
Hence, we propose that a/tif32-Δ8 impairs the ability of the 40S to either remain attached to GCN4 mRNA following peptide termination at uORF1 or to acquire factors, such as eIF1 and eIF1A, which promote scanning. Either defect would reduce the number of 40Ss that resume scanning and reinitiate at GCN4. This model readily explains why the REI efficiency is reduced by nearly the same factor for the constructs in Figure 3, E–G, in a/tif32-Δ8 mutant cells. The implication of this model is that the specialized ability of uORF1 to promote efficient REI depends on the eIF3a-NTD and, hence, on the presence of eIF3 on the terminating ribosome at the uORF1 stop codon. The a/tif32-Δ8 mutation thus establishes a novel class of Gcn− mutants affecting translational control of GCN4 by impairing the initial specific steps of REI.
A functional interaction between the eIF3a-NTD and sequences 5′ of uORF1 promotes resumption of scanning by post-termination 40S subunits
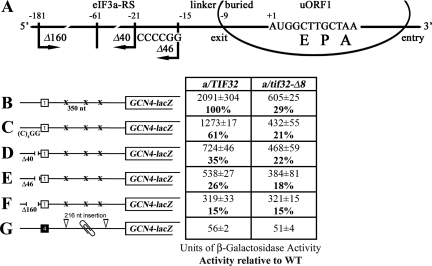
It was shown previously that the high propensity of uORF1 for REI requires sequences upstream of its start codon as well as its last codon and 6 nt following the stop codon (Grant and Hinnebusch 1994; Grant et al. 1995); however, the molecular mechanism of their function(s) is unknown. Having established that the eIF3a-NTD promotes association of eIF3 with the 40S and is critical for REI, we wished to examine whether the eIF3a-NTD acts together with specialized sequences that impart the high REI potential of uORF1. Based on its interaction with RPS0A, the eIF3a-NTD is expected to bind in the vicinity of the mRNA exit channel on the solvent side of the 40S (Spahn et al. 2001; Valášek et al. 2003). Whereas the 3′ 12-nt sequence feature surrounding the stop codon of uORF1 should be buried in the mRNA-binding cleft of the terminating 80S ribosome, sequences beginning at nucleotide −10 upstream of the uORF1 AUG would most likely have emerged from the mRNA exit channel and be accessible to the eIF3a-NTD (Fig. 4A; Wang and Sachs 1997). Hence, it is conceivable that interaction of the eIF3a-NTD with sequences upstream of −9 would stabilize binding of the 40S to the mRNA following termination at uORF1 and thereby promote its ability to resume scanning.
Figure 4.
The eIF3a-NTD functionally interacts with the 5′ sequences of uORF1 to ensure efficient resumption of scanning. (A) Schematic showing predicted position of the 40S ribosome terminating at the stop codon of uORF1 from the GCN4 mRNA leader (data adapted from Wang and Sachs 1997). E, P, and A sites of the 40S ribosomes are aligned with the last two coding triplets and the TAA stop codon. Location of the eIF3a-NTD-responsive site (eIF3a-RS), linker, and buried parts of the sequences upstream of uORF1 are indicated at the top; 3′ boundary of the Δ40 deletion (identical to Δ160), Δ46 deletion, and the (C)4GG multiple substitution are shown below the line depicting mRNA. (B–F) Same as in Figure 3B, except that YBS47 and YBS53 were transformed with p209, pBS64, pBS62, pVM11, and pBS63, respectively, and analyzed without 3-AT treatment. Activities relative to wild type are given in bold in the table to the left of schematics. (G) a/tif32-Δ8 does not affect GCN4-lacZ expression after translating the REI-nonpermissive uORF4. YBS47 and YBS53 were transformed with pA80z and analyzed as in Figure 3B.
It was demonstrated that deletion of 40 nt from nucleotide −21 upstream of the 5′ sequences of uORF1 (Δ40) dramatically reduced (by ∼65%) the induction of GCN4-lacZ expression in wild-type cells (Fig. 4D), whereas replacing the −15 to −1 region 5′ of ORF1 (“linker” and “buried” sequences in Fig. 4A) with the corresponding region upstream of uORF4 had no effect (Miller and Hinnebusch 1989; Grant et al. 1995). Nearly complete elimination of the 5′ sequences in Δ160 then further diminished GCN4-lacZ expression, showing only 15% of the wild-type activity derived most probably from the intact 3′ feature (Fig. 4F; Grant et al. 1995). We found, in addition, that replacement of the −21 AAAATT −16 stretch with CCCCGG [(C)4GG] also significantly reduced β-galactosidase activity by ∼40% in wild-type cells (Fig. 4C). Combining the latter mutation with Δ40 in the Δ46 construct carrying deletion of 46 nt from nucleotide −61 to −15 resulted in an additive effect (∼75% reduction; Fig. 4E). Thus, we reasoned that if the region upstream of nucleotide −15 contains the eIF3a-NTD-binding site, then all these mutations should have a smaller to no effect on REI in a/tif32-Δ8 versus wild-type cells because the interaction would be already disrupted by a/tif32-Δ8. Accordingly, (C)4GG, Δ40, and Δ46 mutations conferred significantly smaller reductions of GCN4-lacZ expression in a/tif32-Δ8 cells, whereas the most destructive Δ160 mutation completely eliminated the negative impact of a/tif32-Δ8 on REI efficiency, producing identical residual activities in both wild-type and a/tif32-Δ8 cells (Fig. 4C–F). These findings of genetic epistasis strongly suggest that the 5′ sequences from nucleotides −181 to −16 constitute an eIF3a-NTD-responsive site (eIF3a-RS), perhaps a direct binding domain, which is required for efficient REI.
To further underscore this point, we analyzed a construct containing solitary uORF4 with its 5′ and 3′ flanking sequences that was placed precisely at the normal position of uORF1 (Fig. 4G). Since it was previously shown that sequences functionally equivalent to the eIF3a-RS of uORF1 are lacking upstream of uORF4, and thus this short uORF allows only a negligible level of REI (Grant and Hinnebusch 1994), we predicted that a/tif32-Δ8 should not affect GCN4-lacZ expression from the latter construct when compared with wild type. Indeed, Figure 4G clearly demonstrates that the deletion of eIF3a-NTD has no impact on REI in the absence of the eIF3a-RS of uORF1.
Proper placement of the eIF3a-RS of uORF1 relative to the 40S mRNA exit channel is critical for efficient reinitiation
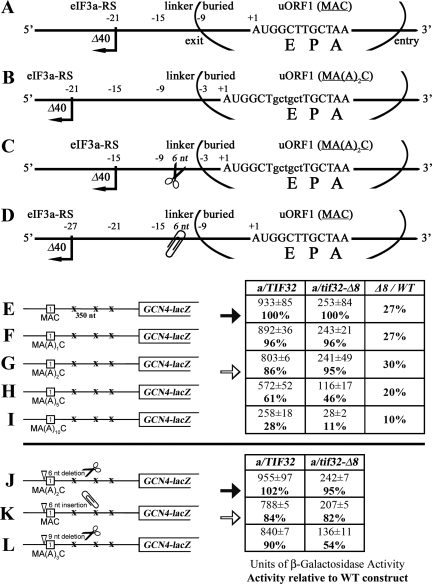
Reinitiation was found to decrease as the uORF was lengthened up to 35 codons, or the rate of translation elongation was slowed down (Kozak 2001; Rajkowitsch et al. 2004). To explain this phenomenon, it was proposed that REI depends on retention of certain initiation factors that gradually dissociate from 80S ribosomes during the course of elongation, thereby reducing the efficiency of REI (Kozak 2005). The results presented so far allow us to propose that eIF3 is a factor that stays bound temporarily to the elongating ribosome, owing to its strategic position on the solvent-exposed “back” side of the 40S subunit. Furthermore, our finding of the specific eIF3a-RS 5′ of uORF1 that promotes REI suggested to us that lengthening uORF1 could decrease REI efficiency by a combination of two effects: (1) displacing this potential eIF3a-NTD-binding site from the mRNA exit channel when the ribosome reaches the stop codon, and (2) by progressive reduction of the eIF3 occupancy on elongating ribosomes.
Since the effect of progressive lengthening of uORF1 has not been investigated before, we analyzed the effect of adding an increasing number of alanine codons to uORF1 on GCN4-lacZ expression in wild-type a/TIF32 cells. As expected, extending the MAC coding sequence of uORF1 with additional Ala codons led to a gradual decrease in β-galactosidase activity (Fig. 5E–I, left column). Addition of two Ala codons [Fig. 5B,G, MA(A)2C] reduced activity by 14%, whereas addition of 10 Ala codons resulted in a ∼72% reduction (Fig. 5I; MA(A)10C). Assuming that spacing between the eIF3a-RS of uORF1 and its stop codon affects REI efficiency and that the two-Ala extension will reduce eIF3 occupancy on the elongating ribosomes only by a small margin, we should be able to restore the REI activity of the MA(A)2C construct at least partially by shortening the linker sequence by 6 nt (Fig. 5C). Indeed, the 6-nt linker deletion in MA(A)2C produced 102% of wild-type activity (Fig. 5, J vs. E, closed arrows). Consistently, moving the eIF3a-RS further upstream by extending the linker by 6 nt in the MAC construct (Fig. 5D) decreased GCN4-lacZ expression to a level comparable with the MA(A2)C construct (Fig. 5, K vs. G, open arrows). In contrast, correcting the spacing in MA(A)3C by the deletion of 9 nt no longer restored the wild-type activity (Fig. 5L), most probably due to the more pronounced reduction in eIF3 occupancy on the elongating 80S as judged from a more dramatic impact on REI activity in a/tif32-Δ8 cells (see also below). The fact that similar results were obtained by testing some of these uORF1 extensions and deletions in a construct containing uORF1 and uORF4 that suffice for wild-type regulation of GCN4 expression (Supplemental Fig. 2) further support our aforementioned suggestion of the compound effect of lengthening of uORF1 on efficiency of REI.
Figure 5.
Proper placement of the eIF3a-NTD-responsive site (eIF3a-RS) in 5′ of ORF1 relative to the mRNA exit channel is a critical requirement for efficient REI. (A–D) Schematics as in Figure 4A except that the change in a position of the 40S ribosome on the mRNA of uORF1 extended by two codons (B), extended by two codons and at the same time shortened by 6 nt in the linker region (C), and shortened by 6 nt in the linker only (D) are shown. (E–I) Progressive extension of the uORF1 reading frame results in a gradual decrease in the GCN4-lacZ expression in the wild-type but not in a/tif32-Δ8 strains. Same as in Figure 4, B–F, except that YBS47 and YBS53 transformed with pM128 or pBS71 through pBS74, respectively, were examined. In addition, activities in a/tif32-Δ8 relative to wild type, respectively, are given in the right column of the table. (J–L) Spacing between the eIF3a-RS and the uORF1 stop codon is a critical determinant of REI. Same as above, except that the strains were transformed with pBS75, pBS77, and pBS94, respectively. Closed and open arrows highlight β-galactosidase activities obtained with constructs with the eIF3a-RS in its native position or shifted by 6 nt upstream, respectively.
Addition of two Ala codons to uORF1 did not reduce GCN4-lacZ expression as much in a/tif32-Δ8 cells as in the wild type; however, the longer extensions of five or 10 Ala codons produced a larger decrease in β-galactosidase activity in the mutant versus wild-type strains (Fig. 5E–I; middle and far right columns). This ostensibly paradoxical result can be understood by recalling that a/tif32-Δ8 is expected to reduce REI by two distinct mechanisms: (1) impairing functional interaction of eIF3 with the eIF3a-RS 5′ of uORF1 and (2) decreasing retention of eIF3 by elongating ribosomes translating uORF1 by reducing the binding of eIF3 to 40S subunits. We observed that addition of five Ala codons was required to detect a marked decrease in GCN4-lacZ activity from a construct containing REI-nonpermissive uORF4 only (B. Szamecz and L. Valášek, unpubl.). Hence, we can stipulate that the retention of eIF3 is not significantly reduced by increasing the length of uORF1 by only one or two Ala codons. This would mean that the attenuation in GCN4-lacZ expression from the MA(A)1C and MA(A)2C constructs in wild-type cells results primarily from displacement of the 5′ sequence feature away from the mRNA exit site of a post-termination ribosome. We showed above that the a/tif32-Δ8 mutation diminishes the effect of disrupting the 5′ eIF3a-RS by the (C)4GG, Δ40, and Δ46 mutations; hence, a/tif32-Δ8 should likewise blunt the effects of the MA(A)1C and MA(A)2C mutations on REI, as we observed in Figure 5G. We further stipulate that the larger increases in uORF1 length of five or 10 codons lead to a significant reduction in eIF3 occupancy by post-termination 40S subunits. Because a/tif32-Δ8 weakens binding of eIF3 to the 40S subunit, it can be expected that this mutation would exacerbate the effect of lengthening uORF1, as observed in Figure 5, H and I, for the MA(A)5C and MA(A)10C mutations. Taken together, these findings strongly support the idea that, at least for REI-permissive uORF1, it is not only the length of the coding region but also a proper placement of the 5′ eIF3a-RS relative to the mRNA exit channel on the post-termination 40S ribosome that represent two critical parameters for efficient REI.
Discussion
The major part of this study was driven by our efforts to address the longstanding question of what endows uORF1 from the GCN4 mRNA leader with its unique ability to allow highly efficient REI. We began by demonstrating that the N-terminal domain of eIF3a forms an important intermolecular bridge between eIF3 and the 40S ribosomal subunit, most probably by its previously reported interaction with RPS0A (Valášek et al. 2003). The a/tif32-Δ8 mutation also reduced 40S association of TC and eIF5, consistent with the role of eIF3 in stimulating 40S binding of other MFC components. We observed a mild defect in 40S biogenesis in a/tif32-Δ8 cells, evident from the increased ratio of free 60S to 40S subunits (Fig. 2A; B. Szamecz and L. Valášek, unpubl.), which might contribute to the Slg− phenotype. However, the fact that combining a/tif32-Δ8 with a/tif32-Box6, which is known to affect 40S binding of eIF3 but not ribosome biogenesis (L. Burela, L. Valášek, and A.G. Hinnebusch, unpubl.), resulted in synthetic lethality supports our conclusion that the eIF3a-NTD is required primarily for an important contact between eIF3 and the 40S. Previously, mutations in eIF3b and eIF3c were shown to affect binding of eIF3 to the 40S (Valášek et al. 2001, 2004; Nielsen et al. 2006). Unlike a/tif32-Δ8, however, these mutations destabilized the MFC and thus might impair 40S binding indirectly. Hence, our results provide the first in vivo evidence implicating a particular domain of a core subunit of eIF3 in its 40S-binding activity.
Although a/tif32-Δ8 decreases the efficiency of TC recruitment to 40S subunits in vivo, it does not constitutively derepress GCN4 translation manifested by the Gcd− phenotype. Rather, a/tif32-Δ8 strongly impairs derepression of GCN4 translation in starved cells. Our genetic analysis ruled out several mechanisms to explain this Gcn− phenotype that were established previously for mutations in other eIFs, including leaky scanning of uORF1, a slow rate of scanning, and less processive scanning (Fig. 3). The simplest explanation of our results is that a/tif32-Δ8 reduces the ability of 40S subunits to resume scanning after terminating at uORF1, neutralizing the specialized features that optimize uORF1 for this key first step in REI and that are lacking at uORF4. These are the first results directly implicating eIF3 in this critical aspect of the canonical REI mechanism involving short uORFs.
There is previous evidence that REI diminishes with increasing uORF length or in response to other perturbations that increase the time required to translate the uORF (Kozak 2001; Rajkowitsch et al. 2004). Accordingly, Kozak (2001) proposed that contacts between one or more eIFs and the 40S established in the PIC are not disrupted at subunit joining and, instead, decay stochastically during elongation. This would allow persistence of an eIF in association with post-termination 40S subunits following translation of a short uORF and thereby facilitate the resumption of scanning and REI. We found that elongating uORF1 by one or two Ala codons has only a small effect, whereas addition of five or 10 Ala codons produces a marked reduction in REI efficiency. Importantly, a/tif32-Δ8 exacerbates the reduction in REI produced by lengthening uORF1 by five or 10 codons. Considering that a/tif32-Δ8 impairs 40S binding of eIF3, these last data provide strong genetic evidence that eIF3 is a factor that remains bound to the 40S subunits of elongating 80S ribosomes as they translate uORF1, and that this interaction is crucial for REI by post-termination ribosomes.
Based on structural (Srivastava et al. 1992; Siridechadilok et al. 2005) and biochemical data (Valášek et al. 2003), it appears that both mammalian and yeast eIF3 bind primarily to the solvent-exposed side of the 40S (Figs. 1A, 6A). Hence, it is plausible to suggest that eIF3 remains associated with the 40S, at least temporarily, after subunit joining (Fig. 6B–D). The interaction should be relatively weak, however, owing to the absence of supporting contacts that eIF3 makes with several eIFs that are ejected at subunit joining (summarized in Hinnebusch 2006). Presumably, eIF3 relies on direct interactions with 40S components—e.g., the eIF3a-NTD binding to RPS0A—and possibly also on contacts with mRNA residues near the entry and exit sites on the solvent side of the 40S subunit. Indeed, meIF3 and mRNA were shown to mutually stabilize their 40S interactions in vitro (Kolupaeva et al. 2005). Moreover, UV-cross-linking of 48S complexes revealed that human eIF3a specifically interacted with mRNA positions −14 and −17 and eIF3d cross-linked to positions −8 through −17, suggesting that eIF3 forms an extension of the mRNA-binding channel (Pisarev et al. 2008).
It is noteworthy, however, that a/tif32-Δ8 does not exacerbate the small reductions in REI produced by lengthening uORF1 by only one or two codons. Likewise, a/tif32-Δ8 does not diminish the minimal level of REI that wild-type uORF4 (also three codons in length) allows despite lacking stimulatory sequences of uORF1 (Fig. 4G). These results make it unlikely that a/tif32-Δ8 reduces REI by decreasing eIF3 association with elongating ribosomes during translation of wild-type, three-codon uORF1. Presumably, the effect of a/tif32-Δ8 in weakening eIF3 binding to the 40S is not great enough to significantly reduce eIF3 occupancy on elongating ribosomes until they have translated more than five codons, as with the elongated versions of uORF1. Hence, we conclude that a/tif32-Δ8 interferes with a step in REI after termination at the stop codon of wild-type uORF1.
It was shown previously that sequences located upstream of uORF1, and also the last codon and sequences immediately 3′ to the uORF1 stop codon, stimulate REI. The absence of such sequences at uORF4 (Supplemental Fig. 3) contributes to the much lower REI frequency of uORF4 versus uORF1. The fact that a/tif32-Δ8 has no effect on the low level of REI after uORF4 translation indicates that a/tif32-Δ8 neutralizes one or more of the features of uORF1 that confer a high REI potential and are absent at uORF4. Importantly, we detected a genetic interaction between a/tif32-Δ8 and mutations in sequences 5′ of uORF1, wherein the deleterious effect of a/tif32-Δ8 on REI is blunted or even eliminated by mutations upstream of uORF1 (Fig. 4). These epistatic interactions strongly suggest that the eIF3a-NTD promotes REI by mediating the stimulatory function of the 5′ sequences of uORF1, which we thus refer to as the eIF3a-NTD-responsive site (eIF3a-RS).
We also presented genetic data supporting the idea that the eIF3a-RS of uORF1 must be located at a specific distance upstream of the uORF1 stop codon (Fig. 5). This finding strongly suggests that the efficiency of REI conferred by the eIF3a-RS strictly depends on its proper positioning relative to the 40S mRNA exit channel. Our previous observation that the eIF3a-NTD interacts directly with RPS0A, and the fact that RPS0A occurs near the 40S mRNA exit channel (Spahn et al. 2001), lead us to propose that the eIF3a-NTD binds directly to the stimulatory eIF3a-RS, present at uORF1 but lacking at uORF4, on the solvent side of the 40S when the ribosome is positioned at the uORF1 stop codon (Fig. 6, E vs. F). This interaction would help stabilize association of the 40S subunit with mRNA following dissociation of the 60S subunit in the first stage of the ribosome recycling reaction (Pisarev et al. 2007) and thereby promote efficient resumption of scanning and REI downstream from uORF1 (Fig. 6, G vs. H).
Why is this mechanism so sensitive to the proper placement of the eIF3a-RS? Perhaps the eIF3a-NTD binds to a special mRNA structure whose proper assembly and orientation is critical because the eIF3a-NTD attachment with the 40S is not flexible enough to accommodate even minor alterations. Thus far, we were unable to detect direct binding between purified eIF3a-NTD and the sequences 5′ of uORF1 using conventional UV-cross-linking assays. However, it seems reasonable to suppose that this interaction would be stabilized by simultaneous binding of eIF3 to the back side of the 40S and of mRNA to the mRNA-binding cleft of the 40S (Fig. 6E), making it impossible to detect with eIF3a and mRNA alone. Moreover, the proposed interaction should not be so strong as to impede the rapid resumption of scanning by the post-termination 40S⋅eIF3 complex.
Alternatively, it is conceivable that eIF3 bound to the post-termination 40S subunit recruits another factor that interacts with the sequences 5′ of uORF1 and possibly even with the eIF3a-NTD itself. Weakening this interaction by a/tif32-Δ8 that also impairs 40S binding of eIF3 would reduce the recruitment of this hypothetical factor and still diminish the effect on REI of mutating these sequences, as we observed. Interestingly, it was reported that eIF4G/eIF4A must be involved in translating a short uORF to observe efficient REI at a downstream ORF in rabbit reticulocyte lysates (Pöyry et al. 2004). Since eIF3 mediates interaction of eIF4G with the 40S in mammals (Korneeva et al. 2000), eIF3 attachment to the back side of the 40S could allow eIF4G to remain bound temporarily to the ribosome during elongation of a short uORF. As eIF4G has mRNA-binding activity (Lomakin et al. 2000), it could help stabilize the binding of post-termination 40S subunits to the mRNA, as proposed for eIF3; however, eIF4G could also facilitate scanning to the downstream start codon based on its role in ribosomal scanning (Pestova and Kolupaeva 2002). It is true that a direct eIF3–eIF4G interaction has not been observed in yeast; however, this does not exclude a possibility that it occurs only in the ribosomal context, and the hypothetical involvement of eIF4G in translational control of GCN4 expression has never been systematically investigated.
Our finding that eIF3 is critical for resumption of scanning by post-termination ribosomes and our identification of the eIF3a-RS upstream of the uORF1 stop codon that promotes REI resonate with recent findings on the mechanism of REI after translation of a long ORF. Only recently, Jackson and colleagues (Pöyry et al. 2007) identified eIF3 as a key factor required for REI after translation of a long ORF2 that overlaps the beginning of a downstream ORF3 on polycistronic subgenomic mRNA of feline calicivirus. They found that REI is enhanced by an 87-nt element at the 3′ end of ORF2 that functions at least partly as a binding site for eIF3 (Pöyry et al. 2007). Considering that eIF3 has been implicated in releasing eukaryotic ribosomes from the mRNA after termination of translation at stop codons (Pisarev et al. 2007), it was suggested that the resulting post-termination eIF3/40S complexes at the ORF2 stop codon do not dissociate from the mRNA, but instead get transferred to the 87-nt element by virtue of the eIF3-mRNA contact. This interaction would stabilize mRNA-40S binding and also position the AUG codon of ORF3 in the ribosomal P-site for efficient REI (Pöyry et al. 2007). This mechanism differs considerably from the canonical REI after a short uORF studied here in that (1) ORF2 is too long to retain eIF3 during translation; (2) eIF3 must be acquired de novo; and (3) there appears to be no scanning after termination of ORF2 before REI at ORF3. Nevertheless, it resembles the model we propose for GCN4 uORF1 by involving a cis-acting enhancer of REI located upstream of the stop codon of the first ORF, which interacts with eIF3 to prevent dissociation of post-termination 40S subunits and stimulate REI.
The fact that translation in general is a rather conserved process in both lower and higher eukaryotes may suggest that our yeast model of REI illustrated in Figure 6 also applies to mammals. However, there are certain aspects by which yeast and mammals differ that ought to be mentioned here: (1) meIF3 contains seven subunits in addition to the yeast six that are conserved (Hinnebusch 2006). Moreover, meIF3a has an extended C terminus compared with its yeast counterpart, though, interestingly, the N termini show the highest degree of similarity. (2) Yeast ribosomes are less sensitive to the start codon context and more sensitive to inhibition by secondary structures (Kozak 2005). (3) REI efficiency in mammalian systems seems to fall off less drastically as the uORF is lengthened in the three to 13 codon range than in yeast (Fig. 5; Kozak 2001). This last fact might indicate that the interaction between eIF3 and the 40S subunit in mammals is stronger and persists longer after initiation at a short uORF, presumably owing to additional contacts that the seven extra mammalian subunits make with the 40S. (4) To our knowledge, there is virtually no information regarding the importance of 5′ and 3′ sequences flanking short uORFs for REI in mammals. Our attempt to compare sequences 5′ of uORF1 of yeast GCN4 and mammalian ATF4 and ATF5 mRNAs, respectively, also did not yield evidence suggesting that the nature of the uORF1-specific post-termination 40S retention on the latter mammalian mRNAs is mechanistically similar to that described for GCN4. Thus, whereas it seems fair to propose that at least the basic principles drawn in Figure 6 are shared between the yeast and mammals, the REI mechanism in both systems will likely differ in numerous details.
Our conclusion that eIF3 allows post-termination 40S subunits to remain attached to the mRNA and resume scanning after translation of uORF1 seems at odds with the function of eIF3 in dissociating post-termination ribosomes identified by Pestova and colleagues in a reconstituted system (Pisarev et al. 2007). Moreover, if eIF3 is recruited to all post-termination 80S subunits to participate in ribosome recycling, then why isn’t REI efficient after translation of any ORF, regardless of length? Assuming that eIF3 functions in ribosome recycling in vivo, it could be proposed that eIF3 that is acquired de novo from the cytoplasmic pool by post-termination 80S ribosomes at long ORFs differs functionally from eIF3 that is transferred from initiation complexes to elongating ribosomes during translation of a short uORF. In particular, the j-subunit of eIF3 and mRNA are mutually antagonistic for binding to 40S subunits (Unbehaun et al. 2004; Fraser et al. 2007), which might indicate that eIF3j dissociates from eIF3 prior to subunit joining (Fig. 6B). Moreover, eIF3j was shown to promote release of post-termination eIF3⋅40S complexes from mRNA in the last step of ribosome recycling (Pisarev et al. 2007). Hence, if the eIF3 transferred from the initiation complex to translating ribosomes lacks eIF3j, it would be defective for the last step of ribosome recycling. The persistence of this eIF3j-depleted eIF3 on 80S ribosomes terminating at uORF1 could still promote the eIF3-dependent release of the 60S subunit from the post-termination 80S ribosome (Pisarev et al. 2007), but not dissociation of the 40S from mRNA. Instead, it would stabilize the 40S⋅mRNA complex (by virtue of eIF3’s interaction with eIF3a-RS of uORF1) and promote scanning and reassembly of the 48S PIC for REI downstream.
Materials and methods
Yeast strains and plasmids
Lists of strains and plasmids used in this study and details of their construction can be found in the Supplemental Material.
Biochemical methods
Polysome profile analysis, 1% HCHO-cross-linking, WCE preparation, and fractionation of extracts for analysis of preinitiation complexes were carried out as described by Valášek et al. (2007). β-galactosidase assays were conducted as described previously (Grant and Hinnebusch 1994).
Acknowledgments
We are thankful to Libor Krásný for critical reading of the manuscript, to the members of the Valášek and Krásný laboratories for helpful suggestions, and to Olga Krýdová, Anna Delijannisová, and Ilona Krupičková for technical and administrative assistance. This research was supported for the most part by The Wellcome Trusts Grant 076456/Z/05/Z, and partly also by the Howard Hughes Medical Institute, by NIH Research Grant R01 TW007271 funded by the Fogarty International Center, by the Fellowship of Jan E. Purkyne from the Academy of Sciences of the Czech Republic, by the Institute Research Concept AV0Z50200510, and by the Intramural Research Program of the National Institutes of Health.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.480508.
References
- Algire M.A., Maag D., Lorsch J.R. Pi release from eIF2, not GTP hydrolysis, is the step controlled by start-site selection during eukaryotic translation initiation. Mol. Cell. 2005;20:251–262. doi: 10.1016/j.molcel.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Cheung Y.N., Maag D., Mitchell S.F., Fekete C.A., Algire M.A., Takacs J.E., Shirokikh N., Pestova T., Lorsch J.R., Hinnebusch A.G. Dissociation of eIF1 from the 40S ribosomal subunit is a key step in start codon selection in vivo. Genes & Dev. 2007;21:1217–1230. doi: 10.1101/gad.1528307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ElAntak L., Tzakos A.G., Locker N., Lukavsky P.J. Structure of eIF3b RNA recognition motif and its interaction with eIF3j: Structural insights into the recruitment of eIF3b to the 40 S ribosomal subunit. J. Biol. Chem. 2007;282:8165–8174. doi: 10.1074/jbc.M610860200. [DOI] [PubMed] [Google Scholar]
- Fraser C.S., Berry K.E., Hershey J.W., Doudna J.A. 3j is located in the decoding center of the human 40S ribosomal subunit. Mol. Cell. 2007;26:811–819. doi: 10.1016/j.molcel.2007.05.019. [DOI] [PubMed] [Google Scholar]
- Grant C.M., Hinnebusch A.G. Effect of sequence context at stop codons on efficiency of reinitiation in GCN4 translational control. Mol. Cell. Biol. 1994;14:606–618. doi: 10.1128/mcb.14.1.606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant C.M., Miller P.F., Hinnebusch A.G. Sequences 5′ of the first upstream open reading frame in GCN4 mRNA are required for efficient translational reinitiation. Nucleic Acids Res. 1995;23:3980–3988. doi: 10.1093/nar/23.19.3980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A.G. eIF3: A versatile scaffold for translation initiation complexes. Trends Biochem. Sci. 2006;31:553–562. doi: 10.1016/j.tibs.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Jivotovskaya A., Valášek L., Hinnebusch A.G., Nielsen K.H. Eukaryotic translation initiation factor 3 (eIF3) and eIF2 can promote mRNA binding to 40S subunits independently of eIF4G in yeast. Mol. Cell. Biol. 2006;26:1355–1372. doi: 10.1128/MCB.26.4.1355-1372.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva V.G., Unbehaun A., Lomakin I.B., Hellen C.U., Pestova T.V. Binding of eukaryotic initiation factor 3 to ribosomal 40S subunits and its role in ribosomal dissociation and anti-association. RNA. 2005;11:470–486. doi: 10.1261/rna.7215305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneeva N.L., Lamphear B.J., Hennigan F.L., Rhoads R.E. Mutually cooperative binding of eukaryotic translation initiation factor (eIF) 3 and eIF4A to human eIF4G-1. J. Biol. Chem. 2000;275:41369–41376. doi: 10.1074/jbc.M007525200. [DOI] [PubMed] [Google Scholar]
- Kozak M. Constraints on reinitiation of translation in mammals. Nucleic Acids Res. 2001;29:5226–5232. doi: 10.1093/nar/29.24.5226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. doi: 10.1016/j.gene.2005.06.037. [DOI] [PubMed] [Google Scholar]
- Lomakin I.B., Hellen C.U., Pestova T.V. Physical association of eukaryotic initiation factor 4G (eIF4G) with eIF4A strongly enhances binding of eIF4G to the internal ribosomal entry site of encephalomyocarditis virus and is required for internal initiation of translation. Mol. Cell. Biol. 2000;20:6019–6029. doi: 10.1128/mcb.20.16.6019-6029.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marintchev A., Wagner G. Translation initiation: Structures, mechanisms and evolution. Q. Rev. Biophys. 2005;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- Miller P.F., Hinnebusch A.G. Sequences that surround the stop codons of upstream open reading frames in GCN4 mRNA determine their distinct functions in translational control. Genes & Dev. 1989;3:1217–1225. doi: 10.1101/gad.3.8.1217. [DOI] [PubMed] [Google Scholar]
- Nielsen K.H., Szamecz B., Valášek L.J., Shin B.S., Hinnebusch A.G. Functions of eIF3 downstream of 48S assembly impact AUG recognition and GCN4 translational control. EMBO J. 2004;23:1166–1177. doi: 10.1038/sj.emboj.7600116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen K.H., Valášek L., Sykes C., Jivotovskaya A., Hinnebusch A.G. Interaction of the RNP1 motif in PRT1 with HCR1 promotes 40S binding of eukaryotic initiation factor 3 in yeast. Mol. Cell. Biol. 2006;26:2984–2998. doi: 10.1128/MCB.26.8.2984-2998.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Kolupaeva V.G. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes & Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova T.V., Lomakin I.B., Lee J.H., Choi S.K., Dever T.E., Hellen C.U.T. The joining of ribosomal subunits in eukaryotes requires eIF5B. Nature. 2000;403:332–335. doi: 10.1038/35002118. [DOI] [PubMed] [Google Scholar]
- Pisarev A.V., Hellen C.U.T., Pestova T.V. Recycling of eukaryotic posttermination ribosomal complexes. Cell. 2007;131:286–299. doi: 10.1016/j.cell.2007.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisarev A.V., Kolupaeva V.G., Yusupov M.M., Hellen C.U.T., Pestova T.V. Ribosomal position and contacts of mRNA in eukaryotic translation initiation complexes. EMBO J. 2008;27:1609–1621. doi: 10.1038/emboj.2008.90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry T.A., Kaminski A., Jackson R.J. What determines whether mammalian ribosomes resume scanning after translation of a short upstream open reading frame? Genes & Dev. 2004;18:62–75. doi: 10.1101/gad.276504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pöyry T.A., Kaminski A., Connell E.J., Fraser C.S., Jackson R.J. The mechanism of an exceptional case of reinitiation after translation of a long ORF reveals why such events do not generally occur in mammalian mRNA translation. Genes & Dev. 2007;21:3149–3162. doi: 10.1101/gad.439507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajkowitsch L., Vilela C., Berthelot K., Ramirez C.V., McCarthy J.E. Reinitiation and recycling are distinct processes occurring downstream of translation termination in yeast. J. Mol. Biol. 2004;335:71–85. doi: 10.1016/j.jmb.2003.10.049. [DOI] [PubMed] [Google Scholar]
- Siridechadilok B., Fraser C.S., Hall R.J., Doudna J.A., Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- Spahn C.M., Beckmann R., Eswar N., Penczek P.A., Sali A., Blobel G., Frank J. Structure of the 80S ribosome from Saccharomyces cerevisiae—tRNA–ribosome and subunit–subunit interactions. Cell. 2001;107:373–386. doi: 10.1016/s0092-8674(01)00539-6. [DOI] [PubMed] [Google Scholar]
- Srivastava S., Verschoor A., Frank J. Eukaryotic initiation factor 3 does not prevent association through physical blockage of the ribosomal subunit–subunit interface. J. Mol. Biol. 1992;220:301–304. doi: 10.1016/0022-2836(92)90946-h. [DOI] [PubMed] [Google Scholar]
- Unbehaun A., Borukhov S.I., Hellen C.U., Pestova T.V. Release of initiation factors from 48S complexes during ribosomal subunit joining and the link between establishment of codon–anticodon base-pairing and hydrolysis of eIF2-bound GTP. Genes & Dev. 2004;18:3078–3093. doi: 10.1101/gad.1255704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L., Phan L., Schoenfeld L.W., Valášková V., Hinnebusch A.G. Related eIF3 subunits TIF32 and HCR1 interact with an RNA recoginition motif in PRT1 required for eIF3 integrity and ribosome binding. EMBO J. 2001;20:891–904. doi: 10.1093/emboj/20.4.891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L., Nielsen K.H., Hinnebusch A.G. Direct eIF2–eIF3 contact in the multifactor complex is important for translation initiation in vivo. EMBO J. 2002;21:5886–5898. doi: 10.1093/emboj/cdf563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L., Mathew A., Shin B.S., Nielsen K.H., Szamecz B., Hinnebusch A.G. The yeast eIF3 subunits TIF32/a and NIP1/c and eIF5 make critical connections with the 40S ribosome in vivo. Genes & Dev. 2003;17:786–799. doi: 10.1101/gad.1065403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L., Nielsen K.H., Zhang F., Fekete C.A., Hinnebusch A.G. Interactions of eukaryotic translation initiation factor 3 (eIF3) subunit NIP1/c with eIF1 and eIF5 promote preinitiation complex assembly and regulate start codon selection. Mol. Cell. Biol. 2004;24:9437–9455. doi: 10.1128/MCB.24.21.9437-9455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valášek L., Szamecz B., Hinnebusch A.G., Nielsen K.H. In vivo stabilization of preinitiation complexes by formaldehyde cross-linking. Methods Enzymol. 2007;429:163–183. doi: 10.1016/S0076-6879(07)29008-1. [DOI] [PubMed] [Google Scholar]
- Vattem K.M., Wek R.C. Reinitiation involving upstream ORFs regulates ATF4 mRNA translation in mammalian cells. Proc. Natl. Acad. Sci. 2004;101:11269–11274. doi: 10.1073/pnas.0400541101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilela C., Linz B., Rodrigues-Pousada C., McCarthy J.E. The yeast transcription factor genes YAP1 and YAP2 are subject to differential control at the levels of both translation and mRNA stability. Nucleic Acids Res. 1998;26:1150–1159. doi: 10.1093/nar/26.5.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Sachs M. Ribosome stalling is responsible for arginine-specific translational attenuation in Neurospora crassa. Mol. Cell. Biol. 1997;17:4904–4913. doi: 10.1128/mcb.17.9.4904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto Y., Singh C.R., Marintchev A., Hall N.S., Hannig E.M., Wagner G., Asano K. The eukaryotic initiation factor (eIF) 5 HEAT domain mediates multifactor assembly and scanning with distinct interfaces to eIF1, eIF2, eIF3, and eIF4G. Proc. Natl. Acad. Sci. 2005;102:16164–16169. doi: 10.1073/pnas.0507960102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou D., Pallam L.R., Jiang L., Narasimhan J., Staschke K.A., Wek R.C. Phosphorylation of eIF2 directs ATF5 translational control in response to diverse stress conditions. J. Biol. Chem. 2008;283:7064–7073. doi: 10.1074/jbc.M708530200. [DOI] [PubMed] [Google Scholar]