Abstract
Shugoshin-2 (SGOL2) is one of the two mammalian orthologs of the Shugoshin/Mei-S322 family of proteins that regulate sister chromatid cohesion by protecting the integrity of the multiprotein cohesin complexes. This protective system is essential for faithful chromosome segregation during mitosis and meiosis, which is the physical basis of Mendelian inheritance. Regardless of its evolutionary conservation from yeast to mammals, little is known about the in vivo relevance and specific role that SGOL2 plays in mammals. Here we show that disruption of the gene encoding mouse SGOL2 does not cause any alteration in sister chromatid cohesion in embryonic cultured fibroblasts and adult somatic tissues. Moreover, mutant mice develop normally and survive to adulthood without any apparent alteration. However, both male and female Sgol2-deficient mice are infertile. We demonstrate that SGOL2 is necessary for protecting centromeric cohesion during mammalian meiosis I. In vivo, the loss of SGOL2 promotes a premature release of the meiosis-specific REC8 cohesin complexes from anaphase I centromeres. This molecular alteration is manifested cytologically by the complete loss of centromere cohesion at metaphase II leading to single chromatids and physiologically with the formation of aneuploid gametes that give rise to infertility.
Keywords: Cohesion, chromosome segregation, Shugoshin-2, mouse, mitosis, meiosis
Loss of sister chromatid cohesion is probably one of the most dramatic events during eukaryotic cell division and is the pivotal process that enables the equal delivery of the replicated genetic material to daughter cells. Sister chromatid cohesion is mediated during both mammalian mitosis and meiosis by a cohesin complex constituted by two members of the family of structural maintenance of chromosome proteins (SMC1 and SMC3), one kleisin subunit (RAD21 at mitosis and/or REC8 at meiosis) that is the substrate of the cysteine protease separase, and a HEAT repeat domain protein (SA1/SA2 at mitosis or STAG3 at meiosis) (Watanabe 2005; Hirano 2006). These proteins create a gigantic ring structure that is presumed to entrap the sister chromatids (Gruber et al. 2003).
During mitosis, most of the cohesin complexes at the arms are released by the phosphorylation of its SA2 subunit by the PLK1 kinase (Waizenegger et al. 2000; Hauf et al. 2005; Losada and Hirano 2005). The remaining cohesin complexes at centromeres and arms are lost from chromosomes during the metaphase/anaphase transition, once all chromosomes have bioriented and the spindle assembly checkpoint (SAC) is satisfied, since separase is activated by the APC (Anaphase-Promoting Complex) (Huang and Moazed 2006) and then cleaves the cohesin subunit RAD21.
The meiotic cycle is governed by similar principles as the mitotic one, but the existence of two consecutive rounds of segregation after a single round of DNA replication explains why the dissociation of cohesin must be tightly regulated in a spatial and time-dependent manner. During the first meiotic division, recombined homologs segregate to opposite poles since cohesin complexes at the arms are cleaved by separase. The cohesin complexes remaining at centromeres are then proteolyzed by separase during the metaphase II/anaphase II transition to allow the segregation of single chromatids (Buonomo et al. 2000; Petronczki et al. 2003). In this sense, the second meiotic division mimics mitosis in that sister chromatids segregate to opposite poles. It has been proposed that the protection of centromeric cohesion during meiosis I is essential for chromosome mono-orientation at metaphase I and biorientation at metaphase II (Page and Hawley 2003; Hauf and Watanabe 2004; Kudo et al. 2006).
In yeast, flies, and vertebrates, a family of centromeric proteins identified as key regulators of chromosome segregation named Shugoshins (Sgo1 and Sgo2/Mei-S322), are responsible for the effective protection of the centromeric RAD21/REC8 cohesin complexes from removal by PLK1 during mitosis and cleavage by separase during meiosis I by recruiting the phosphatase PP2A. This enzyme in turn neutralizes the phosphorylation ability of PLK1 onto SA2 during mitosis (Kitajima et al. 2004, 2006; McGuinness et al. 2005). The shugoshin family of proteins is composed of a single gene in Saccharomyces cerevisiae and Drosophila melanogaster, and two paralogs in the fission yeast Schizosaccharomyces pombe and mammals (Shugoshin-1, or SGOL1, and Shugoshin-2, or SGOL2) (Kitajima et al. 2004). In S. pombe, Sgo1 is only required for meiosis completion, and its lack generates random segregation at meiosis II, whereas Sgo2-depleted cells show both mitotic and meiotic alterations, like chromosome biorientation defects and alterations in the mono-orientation of sister chromatids, respectively (Rabitsch et al. 2004; Vaur et al. 2005; Vanoosthuyse et al. 2007).
The role of SGOL1 and SGOL2 in mammals is still poorly understood, and their function in chromatid cohesion has been only functionally analyzed by means of RNAi-mediated repression in HeLa cells, giving rise to controversial reports (Kitajima et al. 2006; Huang et al. 2007). In addition, the presence of two members has added a new level of difficulty in the course of their functional characterization. Despite the implication of human SGOL1 in maintaining centromeric cohesion in mitosis, a similar function has also been proposed for SGOL2 in HeLa cells. Moreover, both SGOL1 and SGOL2 cooperate with PP2A at the centromere, where they antagonize the kinase activity of PLK1 onto the cohesin complex (McGuinness et al. 2005; Kitajima et al. 2006; Rivera and Losada 2006). Yet, little is known about the functional contribution of SGOL2 in chromatid cohesion in a somatic cell lineage, even less about its role in embryogenesis, organogenesis, and tissue homeostasis. In this study, we demonstrate that SGOL2 is not essential for mouse embryogenesis and adult somatic development. Accordingly, SGOL2 deficiency does not promote any mitotic defect in primary cultured somatic cells. However, both adult male and female mice, despite not showing any overt phenotype, are infertile. We provide evidence that such infertility is caused by the premature loss of centromeric cohesion at meiosis that leads to the formation of gametes with an aberrant number of chromatids.
Results
Generation of Sgol2 mutants
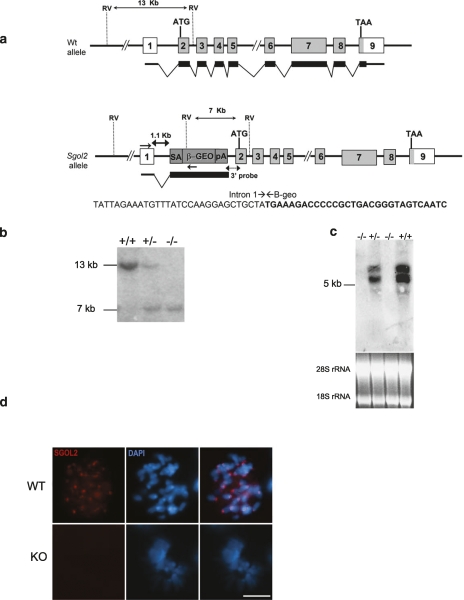
To address the in vivo role of SGOL2, we searched the gene trap database (http://www.knockoutmouse.org) for insertional mutations that could disrupt the transcription of the mouse Sgol2 gene. We selected the line D025B05 (GGTC) in which the cassette rFlpROSA-βgeo (Schnütgen et al. 2005) was inserted into the first intron of the Sgol2 gene. This mutation was further characterized and confirmed by cloning the insertion of the retrovirus into intron 1 (Fig. 1a). We generated founder mice from this embryonic stem (ES) cell line, and following heterozygote intercrossing, Sgol2-deficient (knockout [KO], −/−), heterozygous (+/−), and wild-type (+/+) mice were obtained in the expected Mendelian ratio. Homozygosity for the mutation was demonstrated by Southern blot, and the lack of both transcript and protein was verified by Northern blot, RT–PCR, and immunofluorescence (Fig. 1b–d; Supplemental Fig. S1a,b), indicating that the insertional mutation is a null allele. In addition, since it has been recently reported that SGOL2 is needed for the loading of the mitotic centromere-associated kinesin (MCAK) at centromeres (Huang et al. 2007), we analyzed the localization of MCAK in mouse embryonic fibroblasts (MEFs) of both genotypes. Our results showed that MCAK was present at the inner centromere domain in wild-type dividing MEFs. In contrast, and as expected from a loss-of-function Sgol2 allele, MCAK was not detected at the centromeres of Sgol2−/− mitotic chromosomes (Supplemental Fig. S2a).
Figure 1.
Generation and genetic characterization of Sgol2-deficient mice. (a) Schematic representation of the wild-type locus (wt) and insertional mutant at the Sgol2 locus showing the insertion site, the corresponding coding exons (light gray) and noncoding exons (open boxes), and the β-geo cassette (dark gray). Thin (noncoding) and thick (coding sequences) lines under exons represent the expected transcript derived from wild-type and Sgol2 trapped allele. (ATG) Initiation codon; (TAA) stop codon; (SA) splicing acceptor; (β-geo) bacterial β-galactosidase fused to the neomycin resistance gene; (pA) polyadenylation signal. The nucleotide sequence of the insertion site derived from PCR amplification of DNA from the ES cells (D56678) is indicated. Primers are represented by arrows. The junction of the intronic sequence (regular) and from the β-geo cassette (bold) is depicted. (b) Southern blot analysis of genomic DNA from three littermate progeny of Sgol2 heterozygote crosses. Probing of EcoRV-digested DNA revealed 13-kb and 7-kb fragments for wild-type and disrupted alleles, respectively. (+/+), (+/−), and (−/−) designate wild-type, heterozygous, and homozygous knockout animals, respectively. (c) Northern blot analysis of RNA extracts from testis of (+/+), (+/−), and (−/−) animals with a full-length Sgol2 cDNA probe. The corresponding ethidium bromide stained 18S and 28S ribosomal RNA used as a loading control is also shown below. (d) Immunofluorescence of mitotic plates obtained from MEFs from wild-type (WT) and knockout (KO) genotypes using a polyclonal antibody against the C-terminal region of the SGOL2 protein. (d) Bar, 10 μm.
Unexpectedly, and despite of the Sgol2 widespread expression (http://symatlas.gnf.org; Supplemental Fig. S1a,b), the mutant mice developed normally and displayed no overt phenotype. Moreover, observation of cohorts of mice (n = 25) for 12 mo revealed similar adult survival rate for these mice (100%) and their wild-type controls (96%).
Mouse SGOL2 is not essential for mitosis
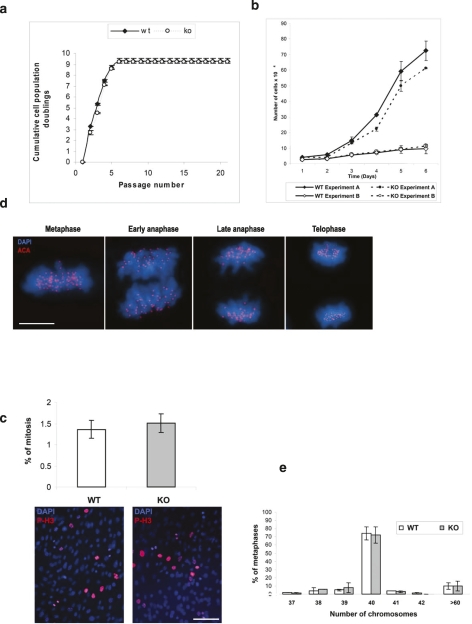
During the prophase pathway, most of the cohesin complexes at chromosome arms are released by PLK1-dependent phosphorylation of their SA2 subunit, whereas centromeric cohesin complexes remain intact until they are proteolyzed at their RAD21 subunit by separase at anaphase (Waizenegger et al. 2000; Hauf et al. 2005). Because it has been reported that human SGOL2 is essential for protecting centromeric cohesin complexes and/or for correcting defective kinetochore–microtubule attachments in mitotic mammalian cells (Kitajima et al. 2006, Huang et al. 2007), we sought to study several mitotic parameters of MEFs lacking SGOL2. Firstly, we studied cell proliferation and transformation, and we found no essential difference in either cellular proliferation rates, mitotic index, or proliferative arrest induced by culture stress in Sgol2−/− MEFs compared with wild-type MEFs (Fig. 2a–c). Partial loss of cohesion in mitosis causes loss of tension across centromeres that leads to anaphase lagging, chromosome mis-segregation, and finally aneuploidy (Nasmyth 2002; Weaver et al. 2003). We extensively analyzed the karyotypes of MEFs and adult somatic cells (cultured thymocytes) lacking SGOL2, and we did not observe significant differences between wild-type and Sgol2−/− in chromatid cohesion defects or any other of those aberrations (Fig. 2e; Supplemental Fig. S1c,d). In addition, and supporting this notion, the karyotype distribution is similar between Sgol2−/− and wild-type MEFs and thymocytes (Fig. 2e; Supplemental Fig. S1c).
Figure 2.
Normal mitosis in somatic cells lacking Sgol2. (a) Serial 3T3 cultivation of primary MEFs of different genotypes. Four independent embryos are shown for each genotype. Cumulative population doublings were measured until immortalization. (b) Proliferation of wild-type (WT) and Sgol2−/− (KO) primary MEFs at two different cell densities (experiments A and B) and counted every following day (see the Materials and Methods). (c) Measurement of the mitotic index (%) by immunostaining with anti-histone H3 phosphorylated at Ser-10 (P-H3-Ser10) in wild-type (WT) and knockout MEFs (KO). Nuclei were counterstained with DAPI. The left panel shows the similar proportion of cells positive for P-H3-Ser10 between Sgol2+/+ and Sgol2−/− MEFs scored in three independent experiments. (d) Immunolabeling of kinetochores (ACA, red) and counterstaning of chromatin (DAPI, blue) of a somatic cell from a Sgol2-deficient mice. (e) Karyotype distribution of wild-type and knockout MEFs. Each data point in a–c, and e represents the mean ± SM. Bars: c, 100 μm; d, 10 μm.
To further analyze the role of SGOL2 in mitosis under stressed conditions, we treated the cell cultures with two antagonistic microtubule poisons such as nocodazole (a microtubule depolimerizing agent) and taxol (a microtubule-stabilizing drug) in order to trigger the activation of the SAC. Similar to their wild-type counterparts, Sgol2-null MEFs have their ability to arrest at metaphase intact (Supplemental Fig. S3d). This result suggests that the fidelity of the SAC is not affected by the absence of SGOL2 under these experimental conditions.
Given the overlapping functions between the SAC and the DNA damage checkpoint and the recent report describing the activation of the SAC in response to DNA damage in an ATM/ATR-dependent manner (Kim and Burke 2008), we determined whether the lack of Sgol2 could be affecting this pathway. For this purpose, we γ-irradiated wild-type and Sgol2−/− MEFs and measured their ability to enter mitosis and to repair their radiation-induced DNA double-strand breaks by looking at the resolution of the γ-H2AX foci after irradiation. We did not observe significant differences at these parameters between cells of both genotypes (Supplemental Fig. S3a–c). Altogether, these findings provide the first in vivo evidence that SGOL2 is dispensable for mitotic chromatid cohesion, cell cycle progression, and proliferation in somatic cells.
Male and female Sgol2-deficient mice are infertile
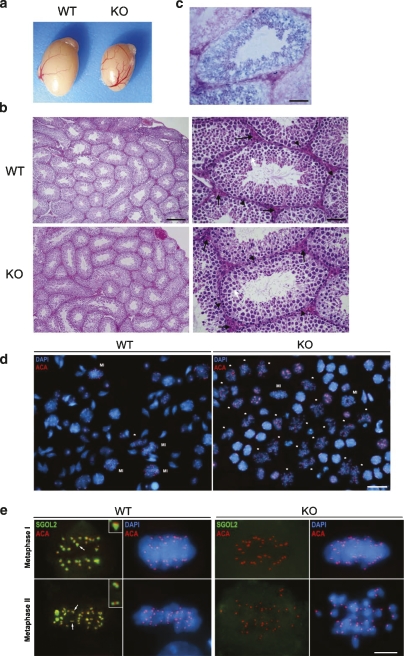
Although adult mice were apparently normal and showed no overt phenotype, they were infertile since they failed to produce offspring when a male or female Sgol2-deficient mouse was mated with a wild type. As controls, these same wild-type males and females were always able to produce offspring when crossed with Sgol2 heterozygotes (data not shown). In order to discard sexual behavior dysfunction as the cause of the observed infertility, we monitored daily for the presence of the vaginal plug in reciprocal crosses between wild-type and knockout mice. In both instances, we observed a similar ratio of vaginal plugs independently of the genotypes. The histopathological analysis of multiple tissues from mutant mice revealed no observable differences with wild-type tissues with the exception of testes, which were ∼35% smaller than their wild-type littermates (Fig. 3a). Although the organization and number of germ cells within the seminiferous tubules appeared normal in the infertile KO mice and no abnormalities were observed in the number and distribution of Sertoli and Leydig cells (Fig. 3b), the number of mature spermatids was reduced, a possible indication of a deficient meiosis. To dissect more precisely the expression pattern of Sgol2 in spermatogenesis, we took advantage of the β-galactosidase–neomycin fusion gene expressed under the control of the Sgol2 promoter. Extensive X-gal staining was observed with the strongest activity corresponding to the spermatocytes (Fig. 3c), supporting the specific role of SGOL2 in spermatogenesis.
Figure 3.
Testicular characterization showing minor size, normal histology, and metaphase II-like arrest in Sgol2-deficient mice. (a) Testes from a wild-type and a null Sgol2 mouse. (b) Hematoxylin-eosin staining of a section of the testis at 200× (left) showing seminiferous tubules. Higher magnification of seminiferous tubules showing Leydig cells (black arrows), spermatogonia (arrowhead) in a more peripheral position within the tubule, and spermatids (white arrow) in Sgol2−/− and wild-type testes. (c) β-galactosidase staining (blue) counterstained with eosin (red) of a section of a testis from a heterozygote mouse showing blue positive staining in the meiocytes. (d) Low-magnification view of a representative squash preparation of seminiferous tubules showing the accumulation of metaphases II-like in knockout Sgol2 (KO) in comparison with a representative wild-type view (WT, left). The identity of metaphases II (asterisks)/metaphases I (MI) was confirmed by the immunolabeling of kinetochores (ACA, red) and counterstaining with DAPI (blue). (e) Double immunodetection of SGOL2 and kinetochores (ACA) in metaphase I and metaphase II from a wild-type and a Sgol2 KO cell showing an accurate congression during metaphase I but a lack of congression of separated chromatids at the metaphase II-like plate when SGOL2 is lacking. Bars: b (right panels), c, 50 μm; b (left panels), 200 μm; d, 15 μm; e, 5 μm.
To understand the molecular mechanisms underlying the observed infertility in the absence of SGOL2, we examined the accuracy of meiotic divisions in testes using immunofluorescence of whole squashed seminiferous tubules, a technique that enables the analysis of meiosis at any stage in a 3D manner (Prieto et al. 2001). As shown in Figure 3d, the low-magnification view of DAPI and kinetochoric immunofluorescence of squashed spermatocytes from Sgol2−/− testis exhibited a very high proportion of cells arrested at the metaphase II stage (Supplemental Fig. S4), although the remaining cells of the testis looked apparently normal including the mitotic figures of dividing spermatogonial cells. As expected from a null allele, SGOL2, which is localized at the inner centromere domain in wild-type dividing spermatocytes (Gómez et al. 2007), was not detected in Sgol2−/− metaphases I and metaphases II (Fig. 3e). According with this loss of function, the SGOL2-dependent MCAK was also delocalized from the centromeres of metaphase I chromosomes in the knockout meiocytes (Supplemental Fig. S2b). Strikingly, during Sgol2−/− meiosis II, and in contrast to wild type, sister chromatids were not held together through their centromeres and never congressed to a typical metaphase II plate. These secondary spermatocytes are hereafter referred as metaphase II-like spermatocytes (Fig. 3e). Therefore, no canonical anaphases II were observed, due most likely to the lack of biorientation of individualized chromatids (Fig. 4). This observation suggests a premature release of sister chromatid centromere cohesion during meiosis II as the most plausible explanation for the infertile phenotype.
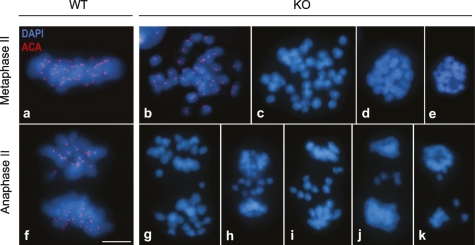
Figure 4.
Defective metaphase II congression and progression to anaphase II. Immunolabeling of kinetochores (ACA, red), and counterstaining with DAPI (blue) in wild-type (WT) and Sgol2−/− (KO) spermatocytes from metaphase II up to anaphase II. (a) Example of a canonical metaphase II in wild-type cells. (b) Metaphase II-like from a Sgol2−/− showing 40 individualized chromatids with a single kinetochore that do not congress at a metaphase II plate like in wild type. (c–e) Examples of degenerating metaphases II without kinetochore labeling. (f) Example of a canonical anaphase II from a wild-type cell. (g–k) Degenerating anaphases II with lagging chromatids from Sgol2−/− spermatocytes. The absence of kinetochoric signals present in c–e, g–j, and k is due to the entrance into a degenerative process with the loss of most centromeric proteins or their anti-genicity. Bar, 5 μm.
The inner domain of metaphase I centromeres is altered in spermatocytes lacking SGOL2
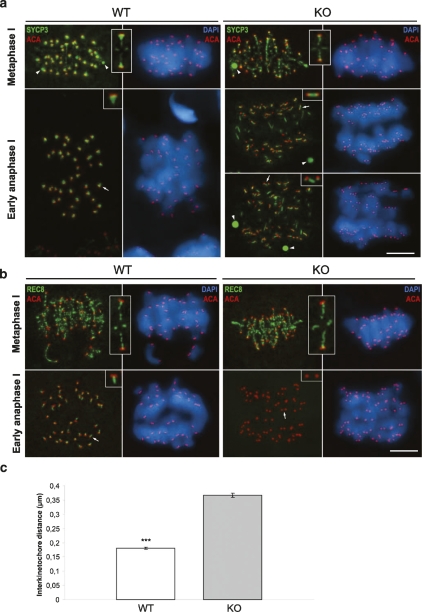
Since the formation of the axial/lateral elements (AEs/LEs) and the synaptonemal complex (SC) are abrogated in several mutant mice with meiotic defects including cohesive ones (Hodges et al. 2005; Xu et al. 2005), we took advantage of the SYCP3 protein marker to follow the development of AEs/LEs and SC during prophase I (Lammers et al. 1994) in the absence of SGOL2. Examination of Sgol2−/− spermatocytes from adult testes by immunofluorescence revealed that the localization of SYCP3 at AEs/LEs was normal during pairing and synapsis of the homologs during zygotene and pachytene (data not shown), and during desynapsis at diplotene and diakinesis (Supplemental Fig. S5). These results indicate that SGOL2 is not necessary for chromosome pairing and recombination during mammalian prophase I. This is the expected situation since in mouse spermatocytes, SGOL2 becomes associated to centromeres during late diplotene (Gómez et al. 2007). However, the distribution of SYCP3 at metaphase I is partially altered in the absence of SGOL2, although bivalents are accurately aligned at the equatorial plate. As shown in Figure 5a, in wild-type metaphase I bivalents, SYCP3 appears preferentially accumulated at the inner centromere domain showing a T-like distribution below the closely associated sister kinetochores, and as faint patches along the surface of contact between sister chromatids (interchromatid domain). SYCP3 maintains a T-like distribution at anaphase I centromeres but redistributes at telophase I to appear as a small bar below or separated from the individualized sister kinetochores (Parra et al. 2004). In contrast, and although the distribution of SYCP3 at the interchromatid domain is unaffected in Sgol2−/− metaphase I bivalents, SYCP3 appears just like a small spot below the closely associated sister kinetochores and not as a T-shape like in wild-type bivalents (inset from KO metaphase I in Fig. 5a). Thus, SGOL2 is not required to maintain the close association between sister kinetochores that allows their monopolar orientation during prometaphase I. Interestingly, in mutant early anaphase I chromosomes, SYCP3 redistributes at the inner centromere domain to appear as small bars connecting clearly separated sister kinetochores (insets from KO early anaphases I in Fig. 5a) resembling the wild-type telophase I distribution, to finally mostly disappear during telophase I.
Figure 5.
Altered localization of SYCP3 and REC8 in the absence of SGOL2 in meiosis I. (a) Double immunolabelings of SYCP3 (green) and kinetochores (ACA, red), and counterstaining with DAPI (blue) in wild-type (WT) and Sgol2−/− spermatocytes (KO) at metaphase I and early anaphase I. Arrowheads indicate the presence of SYCP3 cytoplasmic agglomerates in both wild-type and KO spermatocytes. Insets at the metaphase I panel show magnification of the T-shaped structure delineated by SYCP3 at wild-type bivalents and the small spot in the KO ones. Insets at the early anaphase I panel show SYCP3 at the inner centromere region in wild-type and the bar structure in the KO meiocytes. (b) Double immunolabeling of REC8 (green) with kinetochores (ACA, red) and DAPI (blue) in wild-type and KO spermatocytes at metaphase I and early anaphase I. (Insets) REC8 distribution at the interchromatid domain and at centromeres is similar in wild-type and Sgol2−/− metaphase I bivalents. At early anaphase I, REC8 distribution in wild type persists as a bar (arrow, inset) and is absent in Sgol2−/− spermatocytes, where sister kinetochores are clearly separated (arrow, inset). All images are projections of different focal planes throughout the cell volume. (ACA) Anti-centromeric autoantibody. (c) Centromeres of Sgol2−/− and wild-type anaphase I were immunodetected with ACA, and interkinetochore distances were measured. Error bars represent the mean ± SM (statistical significance is indicated by asterisks, P < 0.001, Student’s t-test). Bar, 5 μm.
All these results demonstrate that in the absence of SGOL2, the organization of the inner centromere domain, as detected by the SYCP3 labeling, is compromised, but bivalents are able to align correctly at the metaphase I plate. Moreover, the homologs segregate accurately at anaphase I, since we never detected separated chromatids or lagging chromosomes during this stage. In this sense, sister kinetochores lose their close association (hereafter referred as sister kinetochore cohesion) during the metaphase/anaphase I transition, although centromere cohesion is apparently maintained during anaphase I.
SGOL2 protects REC8 centromeric cohesin complexes from degradation during anaphase I
Since we had observed that the lack of SGOL2 promoted an alteration at the inner centromere domain leading to a premature release of sister kinetochore cohesion during the metaphase/anaphase I transition, we next analyzed the distribution of the cohesin subunit REC8. This cohesin subunit of the canonical meiotic complex is cleaved along the chromosome arms by separase through the metaphase/anaphase I transition, but is protected at centromeres, where REC8 is cleaved during the metaphase/anaphase II transition (Watanabe and Nurse 1999; Lee et al. 2003; Kudo et al. 2006). The fact that this protection is carried out by shugoshin-1 orthologs in budding and fission yeast (Katis et al. 2004; Kitajima et al. 2004), Drosophila (Kerrebrock et al. 1995), and maize (Hamant et al. 2005) prompted us to examine the location of REC8 in spermatocytes lacking SGOL2. In both wild-type and Sgol2−/− metaphase I bivalents, REC8 is located at the interchromatid domain and at the inner centromere domain at the vertical region of the T-shaped SGOL2 and SYCP3 signals (cf. Figs. 5b and 3e; Gómez et al. 2007). As metaphase I proceeds and transits to anaphase I, REC8 staining is restricted to a small bar perpendicular to the closely associated sister kinetochores in wild-type chromosomes (Fig. 5b). However, when SGOL2 is not present, REC8 is not detected at centromeres in early anaphase I chromosomes, and sister kinetochores appear clearly individualized (inset from KO anaphase I in Fig. 5b). A similar distribution and behavior were found for the cohesin subunit SMC3 in Sgol2−/− metaphase I and anaphase I spermatocytes (Supplemental Fig. S6). In order to quantify this alteration, we measured the distance between sister kinetochores (wild type vs. KO) and found that it is approximately two times larger in Sgol2−/− anaphase I centromeres than in control ones (Fig. 5c). We therefore conclude from these results that SGOL2 is necessary for protecting REC8 and SMC3-containing cohesin complexes at centromeres of mammalian anaphase I chromosomes. Thus, in the absence of SGOL2, the precocious release of centromeric cohesin complexes during early anaphase I leads to a loss of sister kinetochore cohesion.
Sgol2 deficiency provokes SAC activation during the second meiotic division.
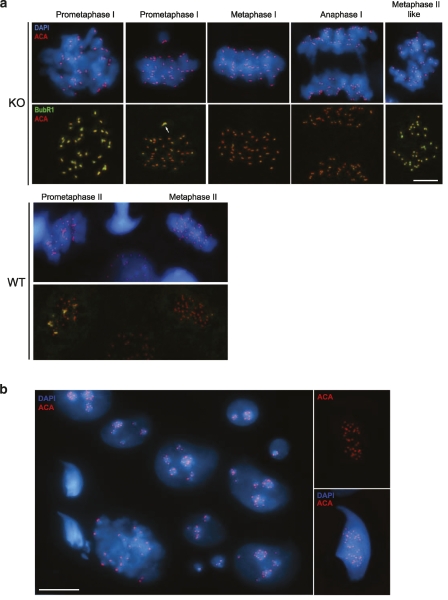
Due to the lack of sister centromere cohesion in metaphase II-like chromosomes when SGOL2 is absent, one would predict that the meiotic SAC should be sensing the inaccurate kinetochore–microtubule attachments, thus avoiding meiotic progression (Fang et al. 1998; Hassold and Hunt 2002; Kouznetsova et al. 2007). To test this, we compared the intensity of the immunofluorescent signals of two proteins involved in this checkpoint (CENP-E and BubR1) from prometaphase I up to metaphase II. As shown in Figure 6a, BubR1 signals are bright at the kinetochores of unaligned bivalents at prometaphase I, but very faint when bivalents have accurately aligned at the metaphase I plate. Likewise, a faint BubR1 labeling is detected at kinetochores of wild-type metaphase II-aligned chromosomes (Fig. 6a). However, in Sgol2-deficient metaphase II-like spermatocytes, we observed a strong labeling of BubR1 and CENP-E in almost all the kinetochores of the separated and unaligned chromatids (Fig. 6a; Supplemental Fig. S7). These results support the hypothesis that the SAC is activated in metaphase II-like arrested spermatocytes, because of the centromere cohesion defect and the incapacity of single chromatids to biorient correctly.
Figure 6.
Abnormal meiosis II and formation of aneuploid spermatids in the presence of an activated SAC in Sgol2−/−spermatocytes. Double immunolabeling of BUBR1 (green), and kinetochores (ACA, red) and counterstaining with DAPI (blue) in wild-type (WT) and Sgol2−/− (KO) spermatocytes. (a) Proper BUBR1 labeling in meiosis I in the absence of SGOL2 showing an unaligned bivalent with intense BUBR1 labeling at their kinetochores during prometaphase I, and a very faint labeling in metaphase I and anaphase I. In metaphase II-like, most of the kinetochores of the separated chromatids show an intense BUBR1 labeling in contrast to the wild type. In wild-type meiosis II spermatocytes, unaligned chromosomes show an intense BUBR1 labeling at their kinetochores in prometaphase II, but the labeling almost disappears in metaphase II. (b) Representative field of round and elongated abnormal spermatids with one up to 36 kinetochore (ACA, red) signals and counterstained with DAPI (blue). Bars: a,b (left panel), 5 μm; b (right panel), 10 μm.
Spermiogenesis is partially altered in the absence of SGOL2
We next examined whether this activation of the SAC at metaphase II-like could lead to aberrant spermatids through an adaptation process (either by slippage through an activated checkpoint or by an eventual inactivation) similar to that observed in mitosis after a prolonged SAC activation (Weaver and Cleveland 2005). Our results show that in the absence of SGOL2, there is an accumulation of metaphase II-like spermatocytes (16% of the meiotic divisions in the wild-type vs. 53.8% in the knockout) that never progress through a standard anaphase II/telophase II stage (Fig. 4; Supplemental Fig. S4). The increased rate of TUNEL-positive spermatocytes detected in the seminiferous tubules from SGOL2-deficient mice in comparison with those in wild-type individuals is also remarkable. This result suggests that a fraction of the Sgol2−/− meiocytes enter into apoptosis (Supplemental Fig. S8). Moreover, our scoring of the percentage of the meiotic divisions in wild-type and mutant individuals on squashed seminiferous tubules shows that 19.3% of metaphase II-like spermatocytes (Fig. 4c–e) and 97.5% of anaphase II-like spermatocytes (Fig. 4g–k) do not present any kind of kinetochore labeling by immunofluorescence and show hypercondensed and lagging chromatids (Supplemental Fig. S4). In order to follow the spermatid maturation, we undertook the quantification of the number of kinetochores per spermatid. We found the presence of round spermatids with a number of kinetochores ranging from one up to 40 (Fig. 6b; Supplemental Fig. S9). In addition, when we looked at elongated spermatids, the percentage of cells with >20 kinetochores drastically increased (45% in round spermatids vs. 79% in elongated spermatids). These observations raise the question of whether mature sperm can still be produced from the differentiation of the abnormal aneuploid spermatids. The results from the histological and sperm counts performed in Sgol2−/− epididymis show that although the knockout display close to a 40% reduction in the number of sperm, they still were able to produce sperm morphologically similar to their wild-type counterparts (Supplemental Fig. S10). Altogether, these results indicate that the absence of SGOL2 results in an aberrant meiotic exit and partially altered spermiogenesis that leads to a reduction in the sperm production.
Female mutant mice for Sgol2 are infertile and also show a cohesion defect in metaphase II
It is well accepted from genetic analysis of human fetuses with chromosomal abnormalities and mouse genetic engineered models that mammalian female meiosis is more prone to error than male meiosis (Hassold and Hunt 2002). To better understand this gender difference, we undertook a meiotic chromosome analysis in female Sgol2−/− mice. By taking advantage of the meiotic maturation process in mammalian oogenesis, we directly analyzed metaphase II-arrested oocytes obtained from superovulated Sgol2−/− and wild-type females. Despite the fact that female Sgol2−/− mice are infertile, the histological analysis of adult ovaries showed that the number and structure of follicles and corpus lutei were similar to those found in wild-type ovaries (Supplemental Fig. S11a). This result indicates that mutant females ovulate physiologically. In addition, we collected a similar number of oocytes from mutant and wild-type females when they were superovulated (data not shown). C-banded metaphase II chromosomes obtained from mutant oocytes showed 40 individualized chromatids, whereas wild-type presented 20 chromosomes, arguing to a similar defect in centromeric cohesion in both male and female meiosis as the cause of infertility (Supplemental Fig. S11b). This conclusion is consistent with a very recent report (which appeared while this manuscript was being edited) using a knockdown strategy of Sgol2 in cultured mouse oocytes (Lee et al. 2008). Taken together, these findings provide in vivo evidence that mammalian SGOL2 is also protecting the removal of REC8 from the meiotic centromeres at oogenesis, since its deficiency also provokes premature chromatid disjunction at metaphase II.
Discussion
We generated knockout mice for Sgol2 in order to analyze the in vivo role that this protein plays during both mitosis and meiosis. We found that these mice develop normally and reach adulthood but are infertile. Our data on the proliferation and transformation rates, mitotic index, and conservation of normal karyotypes on MEFs and thymocytes suggest that SGOL2 is dispensable for mitosis. On the other hand, in these mutant individuals, pairing, synapsis, and recombination during prophase I are normal, as well as bivalent biorientation and alignment at the metaphase I plate. However, in the absence of SGOL2, there is a loss of centromeric cohesin complexes at the onset of anaphase I that promotes the separation between sister kinetochores, although the segregation of chromosomes to opposite poles is normal. During meiosis II, chromatids lose their centromeric cohesion, appear separated, and do not biorient at the equatorial plate. Some of the metaphase II-like and anaphase II-like spermatocytes degenerate by apoptosis, but other spermatocytes escape the SAC and give rise to aneuploid spermatids with one up to 40 chromatids. Altogether, our data suggest that SGOL2 protects centromeric cohesin complexes until the metaphase II/anaphase II transition and is thus essential for an accurate gametogenesis.
Mitosis is not altered in SGOL2-deficient mice
The analysis of the mutant mice demonstrated surprisingly that despite the wide expression of SGOL2 in somatic tissues, homozygous mutant mice were viable and did not show any overt phenotype. Accordingly, all the mitotic parameters analyzed in cultured MEFs including centromeric cohesion and chromosome stability were unaltered. In addition, we found that the absence of SGOL2 was dispensable for the development of the tissular architecture of both the ovary and the testis including the highly proliferative spermatogonial compartment that sustains the spermatogenesis (de Rooij and de Boer 2003) as well as the proliferation capacity of cultured thymocytes. These results are in marked contrast with the assumption that human SGOL2 participates together with SGOL1 in protecting the centromeric cohesion in human HeLa cells, since when Sgol2 was knocked down, a high incidence of prematurely chromatid separation was observed (Kitajima et al. 2006). Apart from the few putative species-specific differences between humans (HeLa) and mouse (MEFs), the fact that HeLa cells are not primary cell cultures but transformed cells with mutations in several important oncogenic and tumor-suppressor pathways such as p53, a tumor suppressor that interacts with the SAC pathway (Oikawa et al. 2005), might underlie some of the observed discrepancies. In this same regard, Mei-S322, the only shugoshin ortholog in Drosophila, is not essential for somatic cell division, embryonic development, and adult homeostasis (Kerrebrock et al. 1995).
Moving backward in terms of complexity, the fission yeast with two paralogs (Sgo1 involved solely in meiosis and Sgo2 involved in mitosis and meiosis) and the budding yeast with a single member (Sgo1) constitute very well-known systems to study this hypothesis. Despite controversial interpretations regarding the function of shugoshins in protecting centromeric cohesion (Goulding and Earnshaw 2005; Wang and Dai 2005) fueled by the strong meiotic phenotype observed in their mutants, the deficiency of either Sgo1 or Sgo2 did not elicit an obvious cohesion defect in mitosis (Katis et al. 2004; Kitajima et al. 2004; Marston et al. 2004). Moreover, these mutants shared properties at the mitotic level with other SAC mutants such as MAD2, like the presence of chromosome instability and aneuploidy as well as of an increased sensitivity to depolymerizing drugs (Indjeian et al. 2005). It thus seems from the more recent findings in yeast that shugoshins act by sensing tension across centromeres by interacting with members of the chromosomal passenger complex, enabling chromosome biorientation (Kawashima et al. 2007; Vanoosthuyse et al. 2007). We cannot rule out that in the absence of Sgol2, its paralog Sgol1 could be playing a major role in protecting centromeric cohesion in mammalian mitosis constitutively or because of a compensatory mechanism. This possibility cannot be empirically tested in the absence of mice deficient for SGOL1. However, it has been reported recently that the Cre-dependent loss of the SAC protein BUB1 in MEFs resulted in the functional delocalization of SGOL1 from mitotic centromeres, while their chromatid cohesion was only modestly altered (Perera et al. 2007). In addition, the SGOL2 centromere loading is also dependent on BUB1 (Huang et al. 2007), a fact that would indicate that the simultaneous loss of function of both mouse shugoshins (in addition to BUB1) in this primary cell culture model does not abruptly compromise mitotic cohesion. Further development of a loss-of-function mouse models of SGOL1 will be required to assess this hypothesis.
SGOL2 is necessary for protecting centromeric cohesion during the metaphase I/anaphase I transition
Our analysis of Sgol2 mutant mice has allowed us to demonstrate unambiguously that mammalian SGOL2 is responsible for protecting the dissociation of centromeric cohesin complexes during meiosis I. This mechanism of protection is most likely due to the colocalization of SGOL2 and cohesin complexes at the inner domain of metaphase I centromeres (Gómez et al. 2007). In the absence of SGOL2, we found that REC8 and SMC3 are lost from centromeres at the metaphase I/anaphase I transition concomitant to the separation between sister kinetochores. However, and strikingly, sister chromatids remain associated at their centromeres, and anaphase I segregation proceeds normally. A similar chromosome behavior was previously found in mutants for Mei-S322 in Drosophila meiosis I (Kerrebrock et al. 1995). These observations are apparently contradictory since we could expect that the loss of centromeric cohesin complexes would lead to the complete separation of chromatids during anaphase I and then to an aberrant segregation. In our opinion, in Sgol2 mutants, chromosomes can segregate normally during anaphase I with separated kinetochores given that they were previously attached to microtubules emanating from the same pole at metaphase I, when they were closely associated. There are several possibilities that may explain why the loss of centromere cohesion is only detected at metaphase II-like spermatocytes and oocytes when cohesin complexes with REC8 and SMC3 are released from centromeres during the metaphase I/anaphase I transition. One possibility is that other RAD21-based cohesin complexes also located at the inner domain of metaphase I centromeres (Parra et al. 2004; Gómez et al. 2007) are not affected in the absence of SGOL2, and could maintain centromere cohesion during anaphase I while allowing the loss of association between sister kinetochores. Since it has been reported that RAD21 redistributes and is lost from centromeres during telophase I (Parra et al. 2004), only in this moment could sister centromeres separate. Another not exclusive possibility consists in the persistence of a topoisomerase II-dependent cohesion mechanism during anaphase I when REC8-containing cohesin complexes have been lost. Future studies are required to test these hypotheses.
It is tempting to speculate that the additional processes required for the stepwise loss of cohesion in yeast should also be necessary in mammals, such as homologous recombination, the REC8 phosphorylation status by the interplay between phosphatases (PP2A) and kinases (Polo kinases) at meiosis I, but not meiosis II, and the separase-dependent cleavage of the phosphorylated form of REC8. Supporting this notion, it has been elegantly demonstrated in a conditional mouse model of separase, that cohesin dissociation from chromosome arms at anaphase I is dependent on the proteolytic activity of separase onto REC8 (Kudo et al. 2006).
REC8 is considered as a specific cohesin subunit involved in the formation of the SC in yeast, flies, and mammals (Watanabe and Nurse 1999; Lee et al. 2003). However, it remains controversial that the mammalian function of REC8 is constrained to the germline since mice lacking REC8 show perinatal mortality and adult infertility, whereas another reported loss-of-function mutation only displays infertility (Bannister et al. 2004; Xu et al. 2005). Likely, previous unpublished results reported in Lee et al. (2008) comments about the lethality of deficient Sgol2. In relation with the additional functions of cohesins, very recent evidence supports a noncanonical function in the integration of DNA sequence with the epigenetic state of the cell through the DNA-binding protein CTCF (Parelho et al. 2008).
Phylogenetic relationships of the shugoshin family of proteins
Genetic analysis in flies and later on in fission and budding yeast has shed essential knowledge on the chromosome cohesion field with the functional identification of the members of the shugoshin family of proteins (Kerrebrock et al. 1995; Kitajima et al. 2004). However, the confusing nomenclature used leads to the establishment of some putative erroneous phylogenetic relationships between their members attending to their numeration. This is specially significant in the case of the mammalian shugoshins (Sgol1 and Sgol2) with respect to their yeast counterparts, which has hampered a true functional relationship among them. In addition, the very low sequence identity between the yeast and the mammalian counterparts has not allowed elucidation of their orthology relationship. Thus, and from the functional data accumulated in this study, essentially the major involvement of mouse SGOL2 in meiosis and its role in protecting centromeric cohesion in spermatocytes, we suggest that mammalian SGOL2 is the functional ortholog of S. pombe Sgo1. We can speculate that the remaining mammalian member, mouse SGOL1, will share functional abilities with its fission yeast counterpart Sgo2.
Alterations in the SGOL2 pathway can lead to meiosis II segregation defects and altered gametogenesis
Most of the human chromosome abnormalities found in fetuses result from nondisjunction during female meiosis I. In addition, there is a group of less frequent, but also important, trisomies (chromosome 18, and to a lesser extent chromosomes 13, 14, and 15, and 20% of the trisomies at chromosome 21) that arise because of meiosis II errors (Hassold et al. 2007). It has been previously postulated that partial alterations in the SGO/REC8/Separase cohesion pathway could lead to metaphase II nondisjunction in human gametogenesis and thus to aneuploidy (Orr-Weaver 1996; Warren and Gorringe 2006). However, the in vivo analysis of mutations affecting this meiotic pathway has impacted our understanding of aneuploidy in a limited way due to the lack of viability of the model or because most mutations lead to prophase I arrest and subsequent apoptosis (de Rooij and de Boer 2003). The SGOL2-deficient mouse model analyzed in this work constitutes an exception in which the meiotic alterations (loss of centromeric cohesion) do not apparently compromise prophase I, and the gametogenesis is completed in both sexes. Thus, this mutant provides new experimental evidence into the association between loss of chromosome cohesion, which would ultimately result in an increased susceptibility of premature chromosome segregation, and aneuploidy.
In this context, it has been reported in human and mouse that a gradual loss of chromosome cohesion in females is associated with chronological age coupled with a reduced expression of the SAC genes MAD2 and BUB1 (Cukurcam et al. 2004; Steuerwald 2005). This age-dependent missegregation has also been observed in a more rapid time scale in the SMC1β mutant mouse (Hodges et al. 2005). We postulate that subtle alterations in the cohesin pathway due to either single nucleotide polymorphisms (SNPs) and/or age-dependent accumulated errors could account for a fraction of cases of mechanistically orphan aneuploidies. In this model, the Sgol2-deficient mice would display a full penetrance phenotype with complete infertility.
In summary, our results establish for the first time in a mammalian organism that SGOL2 deficiency does not alter mitotic progression. However, SGOL2 is essential for meiosis, and its deficiency provokes premature removal of REC8-containing cohesin complexes during anaphase I that leads to a loss of sister chromatid centromere cohesion during meiosis II, resulting in mice infertility.
Materials and methods
Mapping the integration site and genotyping
Insertion of the β-geo cassette was verified by XL–PCR (with primers exon 1 sense, 5′-ACCCTTCTCGGTAGCCACTCCTG-3′; and a universal LacZ antisense, 5′-GTGCTGCAAGGCGAT TAAGTTGG-3′) using as template genomic DNA obtained from the embryonic cell line D025B05 and an Expand long template PCR polymerase kit (Roche). The PCR product was cloned in the EcoRV site of pBS (Stratagene), and the integration site was sequenced using universal primers.
Genotyping of the colony was performed by Southern blot analysis using as probe a PCR fragment amplified from genomic DNA using primers exon 2 sense, 5′-CTGTTACCTCTGG AATTCAGAG-3′; and intron 2 antisense, 5′-CACTTGGC TCTCCTTGGCATACC-3′. The 870-bp fragment enclosed exon 2 and part of the adjacent intron 2. The probe was labeled with α-dCTP32 (Perkin Elmer) by random priming. The probe recognized a 13-kb EcoRV restriction band in wild-type DNA and a 7-kb restriction band in the Sgol2 mutant allele due to an EcoRV site located in the β-geo cassette.
The ES cell clone was used to generate chimeric mice by standard microinjection procedures at the University of Salamanca Transgenic facility. Two highly chimeric males were bred to C57BL/6 females transmitting the Sgol2 allele by germline. The genetic background under which the mutation was analyzed is a mixed BL6/129. The handling, maintenance and care of the animals, as well as all procedures performed in this study, were in accordance with institutional guidelines (CSIC and USAL).
Fertility assessment
We investigated the reproductive capacities of Sgol2−/− and wild-type mice by mating one male with two females for 2 wk. Female mice were examined for vaginal plugs each morning, and litter size was recorded on delivery after three successive matings.
Cell culture and proliferation assays
MEFs were derived from embryonic day 13.5 (E13.5) embryos following standard procedures. Cells were cultured at atmospheric oxygen pressure in Dulbecco’s modified Eagle’s medium (Gibco) supplemented with 10% fetal bovine serum (Sigma), 1% nonessential amino acids, and 2 mM Gln. Serial 3T3 cultivation was carried out following standard procedures. Briefly, 106 cells were plated on 10-cm diameter dishes, and 3 d later, the total number of cells in the dish was counted and 106 cells were replated again. This procedure was repeated for 25 passages. The increase in population doubling level (PDL) was calculated according to the formula PDL = log(nf/n0)/log 2, where n0 is the initial number of cells and nf is the final number of cells.
For cell proliferation assays, 2.5 × 104 or 4 × 104 cells/well were seeded in 12-well plates with 10% fetal bovine serum. The number of cells was counted daily.
Immunofluorescence microscopy
For immunofluorescence, mitotic cells were fixed in cold 4% paraformaldehyde in PBS, rinsed in PBS, permeabilized with 0.2% Triton X-100, and incubated with the corresponding primary antibodies diluted in 7% FCS in PBS. The Rhodamine and FITC-conjugated secondary antibodies were from Jackson. Nuclei were counterstained with DAPI diluted in Vectashield (Vector Laboratories).
Testes from adult male Sgol2+/+, Sgol2+/−, and Sgol2−/− mice were used. Testes were removed and detunicated, and seminiferous tubules were fixed for squashing and subsequent immunofluorescence as previously described (Page et al. 1998). The slides were rinsed three times for 5 min in PBS and incubated for 45 min at room temperature or 12 h at 4°C with primary antibodies diluted in PBS. The slides were then incubated for 30 min at room temperature with either a fluorescein isothiocyanate (FITC)-conjugated donkey anti-rabbit IgG (Jackson) secondary antibody (1:150) in PBS, a FITC-conjugated donkey anti-sheep IgG (Jackson) secondary antibody (1:40) in PBS, a FITC-conjugated donkey anti-mouse IgG (Jackson) secondary antibody (1:150) in PBS, or a Texas Red-conjugated donkey anti-human IgG (Jackson) secondary antibody (1:150) in PBS. The slides were counterstained with DAPI and mounted with Vectashield (Vector Laboratories). Kinetochores were revealed with a purified human anti-centromere autoantibody (ACA) serum (Antibodies Incorporated, 15-235) at a 1:50 dilution. SGOL2 was detected with the rabbit polyclonal serum K1059 against the C terminus of mouse SGOL2 (Gómez et al. 2007) at a 1:20 dilution. The cohesin subunit REC8 was detected with a rabbit polyclonal serum against the C terminus of mouse REC8 kindly provided by J. Lee at a 1:10 dilution. To detect SYCP3, we used a mouse monoclonal antibody (Abcam, ab-12452) at a 1:100 dilution. An affinity-purified sheep polyclonal antibody against human BubR1 (SBR1.1) kindly provided by Stephen S. Taylor was used at a 1:50 dilution. To detect CENP-E, we used a polyclonal rabbit serum (pAb1.6) that recognizes the neck region (amino acids 256–817) of human CENP-E kindly provided by T. Yen at a 1:100 dilution. The antibody against anti-histone H3 phosphorylated at Ser-10 (P-H3-Ser10) was from Upstate Biotechnologies and was used at 1:400.
Immunofluorescence image stacks were collected on an Olympus BX61 microscope equipped with epifluorescence optics, a motorized z-drive, and an Olympus DP70 digital camera controlled by analySIS software (Soft Imaging System). Stacks were analyzed and processed using the public domain ImageJ software (National Institutes of Health; http://rsb.info.nih.gov/ij) and VirtualDub (VirtualDub.org; http://www.virtualdub.com). Final images were processed with Adobe Photoshop 7.0 software.
Histology and β-gal histochemistry
For histological analysis, samples were fixed with cold 4% paraformaldehyde in PBS, processed into serial paraffin sections, and stained with H&E. Histochemical analysis of β-galactosidase reporter activity in frozen sections was performed following standard procedures.
Superovulation of female mice
At least three individuals of each genotype were induced successfully to superovulate using an standard procedure. Pregnant mare’s serum gonadotropin (PMSG; Folligon; Intervet) and human chorionic gonadotropin (HCG; Chorulon; Intervet) were used to induce efficient superovulation. Briefly, 4-wk-old-C57BL/6J females were injected intraperitonally with 7 U.I. PMSG resuspended in 0.1 mL of saline solution at 2 p.m. into each animal, and the same dosage of HCG was administered 46 h later. Unfertilized oocytes enclosed in cumulus were collected 21 h post-HCG administration, digested with hialuronidase under microscope, and immediately used for chromosome preparation. Meiotic chromosome figures were obtained following the classical air-dried technique and C-banded.
Karyotyping of MEFs
Metaphases were obtained from MEFs at passage 4–5 by standard procedures. Active growing cultures were arrested using colcemide (1 μg/mL) for 4 h, trypsinized, treated with hypotonic solution (0.75 mM KCl), and fixed with Methanol/Acetic. At least 50 metaphases were counted from four independent embryos of each genotype.
Acknowledgments
We acknowledge Dr. C. López-Otín and Dr. A. Folgueras for their advice and Dr. A.K.Tarkowski for helpful comments. We express our sincere thanks to Drs. J. Lee, S.S. Taylor, T. Yen, L. Wordeman, M. Serrano, and M. Balbín for providing antibodies and reagents. This work was supported by SAF-2005-01993, J CyLe (SA), CSIC 2006 2 01 051, and Fundación Mutua Madrileña. C.G.C. and Y.S. are supported by FIS and FPI fellowships, respectively. E.L. and M.S.M. are recipients of a Ramón y Cajal Research contract. M.S.M. is supported by FIS and Junta de Castilla y León. J.A.S. and R.G. are supported by grants BFU2005-05668-C03-01/BCM from MEC and CCG06-UAM/SAL-0260 from UAM/Comunidad de Madrid, and an FPI predoctoral fellowship, respectively. A.C. and J.L.B. are supported by BFU2006-04406/BMC from MEC and P-BIO-0189-2006 from CAM.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.475308.
References
- Bannister L.A., Reinholdt L.G., Munroe R.J., Schimenti J.C. Positional cloning and characterization of mouse mei8, a disrupted allelle of the meiotic cohesin Rec8. Genesis. 2004;40:184–194. doi: 10.1002/gene.20085. [DOI] [PubMed] [Google Scholar]
- Buonomo S.B., Clyne R.K., Fuchs J., Loidl J., Uhlmann F., Nasmyth K. Disjunction of homologous chromosomes in meiosis I depends on proteolytic cleavage of the meiotic cohesin Rec8 by separin. Cell. 2000;103:387–398. doi: 10.1016/s0092-8674(00)00131-8. [DOI] [PubMed] [Google Scholar]
- Cukurcam S., Sun F., Betzendahl I., Adler I.D., Eichenlaub-Ritter U. Trichlorfon predisposes to aneuploidy and interferes with spindle formation in vitro maturing mouse oocytes. Mutat. Res. 2004;564:165–178. doi: 10.1016/j.mrgentox.2004.08.008. [DOI] [PubMed] [Google Scholar]
- de Rooij D.G., de Boer P. Specific arrests of spermatogenesis in genetically modified and mutant mice. Cytogenet. Genome Res. 2003;103:267–277. doi: 10.1159/000076812. [DOI] [PubMed] [Google Scholar]
- Fang G., Yu H., Kirschner M.W. The checkpoint protein MAD2 and the mitotic regulator CDC20 form a ternary complex with the anaphase-promoting complex to control anaphase initiation. Genes & Dev. 1998;12:1871–1883. doi: 10.1101/gad.12.12.1871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gómez R., Valdeolmillos A., Parra M.T., Viera A., Carreiro C., Roncal F., Rufas J.S., Barbero J.L., Suja J.A. Mammalian SGO2 appears at the inner centromere domain and redistributes depending on tension across centromeres during meiosis II and mitosis. EMBO Rep. 2007;8:173–180. doi: 10.1038/sj.embor.7400877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulding S.E., Earnshaw W.C. Shugoshin: A centromeric guardian senses tension. Bioessays. 2005;27:588–591. doi: 10.1002/bies.20240. [DOI] [PubMed] [Google Scholar]
- Gruber S., Haering C.H., Nasmyth K. Chromosomal cohesin forms a ring. Cell. 2003;112:765–767. doi: 10.1016/s0092-8674(03)00162-4. [DOI] [PubMed] [Google Scholar]
- Hamant O., Golubovskaya I., Meeley R., Fiume E., Timofejeva L., Schleiffer A., Nasmyth K., Cande W.Z. A REC8-dependent plant Shugoshin is required for maintenance of centromeric cohesion during meiosis and has no mitotic functions. Curr. Biol. 2005;15:948–954. doi: 10.1016/j.cub.2005.04.049. [DOI] [PubMed] [Google Scholar]
- Hassold T., Hunt P. Rescuing distal crossovers. Sex matters in meiosis. Science. 2002;296:2181–2183. doi: 10.1126/science.1071907. [DOI] [PubMed] [Google Scholar]
- Hassold T., Hall H., Hunt P. The origin of human aneuploidy: Where we have been, where we are going. Hum. Mol. Genet. 2007;16:203–208. doi: 10.1093/hmg/ddm243. [DOI] [PubMed] [Google Scholar]
- Hauf S., Watanabe Y. Kinetochore orientation in mitosis and meiosis. Cell. 2004;119:317–327. doi: 10.1016/j.cell.2004.10.014. [DOI] [PubMed] [Google Scholar]
- Hauf S., Roitinger E., Koch B., Dittrich C.M., Mechtler K., Peters J.M. Dissociation of cohesin from chromosome arms and loss of arm cohesion during early mitosis depends on phosphorylation of SA2. PLoS Biol. 2005;3:419–432. doi: 10.1371/journal.pbio.0030069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano T. At the heart of the chromosome: SMC proteins in action. Nat. Rev. Mol. Cell Biol. 2006;7:311–322. doi: 10.1038/nrm1909. [DOI] [PubMed] [Google Scholar]
- Hodges C.A., Revenkova E., Jessberger R., Hassold T.J., Hunt P.A. SMC1β-deficient female mice provide evidence that cohesins are a missing link in age-related non-disjunction. Nat. Genet. 2005;37:1351–1355. doi: 10.1038/ng1672. [DOI] [PubMed] [Google Scholar]
- Huang J., Moazed D. Sister chromatid cohesion in silent chromatin: Each sister to her own ring. Genes & Dev. 2006;20:132–137. doi: 10.1101/gad.1398106. [DOI] [PubMed] [Google Scholar]
- Huang H., Feng J., Famulski J., Rattner J.B., Liu S.T., Kao G.D., Muschel R., Chan G.K., Yen T.J. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J. Cell Biol. 2007;177:413–424. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indjeian V.B., Stern B.M., Murray A.W. The centromeric protein Sgo1 is required to sense lack of tension on mitotic chromosomes. Science. 2005;307:130–133. doi: 10.1126/science.1101366. [DOI] [PubMed] [Google Scholar]
- Katis V.L., Galova M., Rabitsch K.P., Gregan J., Nasmyth K. Maintenance of cohesin at centromeres after meiosis I in budding yeast requires a kinetochore-associated protein related to MEI-S332. Curr. Biol. 2004;14:560–572. doi: 10.1016/j.cub.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Kawashima S.A., Tsukahara T., Langegger M., Hauf S., Kitajima T.S., Watanabe Y. Shugoshin enables tension-generating attachment of kinetochores by loading Aurora to centromeres. Genes & Dev. 2007;21:420–435. doi: 10.1101/gad.1497307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerrebrock A.W., Moore D.P., Wu J.S., Orr-Weaver T.L. Mei-S332, a Drosophila protein required for sister-chromatid cohesion, can localize to meiotic centromere regions. Cell. 1995;83:247–256. doi: 10.1016/0092-8674(95)90166-3. [DOI] [PubMed] [Google Scholar]
- Kim E.M., Burke D.J. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. PLoS Genet. 2008;4:e1000015. doi: 10.1371/journal. pgen.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitajima T.S., Kawashima S.A., Watanabe Y. The conserved kinetochore protein shugoshin protects centromeric cohesion during meiosis. Nature. 2004;427:510–517. doi: 10.1038/nature02312. [DOI] [PubMed] [Google Scholar]
- Kitajima T.S., Sakuno T., Ishiguro K., Iemura S., Natsume T., Kawashima S.A., Watanabe Y. Shugoshin collaborates with protein phosphatase 2A to protect cohesin. Nature. 2006;441:46–52. doi: 10.1038/nature04663. [DOI] [PubMed] [Google Scholar]
- Kouznetsova A., Lister L., Nordenskjöld M., Herbert M., Höög C. Bi-orientation of achiasmatic chromosomes in meiosis I oocytes contributes to aneuploidy in mice. Nat. Genet. 2007;39:966–968. doi: 10.1038/ng2065. [DOI] [PubMed] [Google Scholar]
- Kudo N.R., Wassmann K., Anger M., Schuh M., Wirth K.G., Xu H., Helmhart W., Kudo H., McKay M., Maro B., et al. Resolution of chiasmata in oocytes requires separase-mediated proteolysis. Cell. 2006;126:135–146. doi: 10.1016/j.cell.2006.05.033. [DOI] [PubMed] [Google Scholar]
- Lammers J.H., Offenberg H.H., van Aalderen M., Vink A.C., Dietrich A.J., Heyting C. The gene encoding a major component of the lateral elements of synaptonemal complexes of the rat is related to X-linked lymphocyte-regulated genes. Mol. Cell. Biol. 1994;14:1137–1146. doi: 10.1128/mcb.14.2.1137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Iwai T., Yokota T., Yamashita M. Temporally and spatially selective loss of Rec8 protein from meiotic chromosomes during mammalian meiosis. J. Cell Sci. 2003;116:2781–2790. doi: 10.1242/jcs.00495. [DOI] [PubMed] [Google Scholar]
- Lee J., Kitajima T.S., Tanno Y., Yoshida K., Morita T., Miyano T., Miyake M., Watanabe Y. Unified mode of centromeric protection by shugoshin in mammalian oocytes and somatic cells. Nat. Cell Biol. 2008;10:42–52. doi: 10.1038/ncb1667. [DOI] [PubMed] [Google Scholar]
- Losada A., Hirano T. Dynamic molecular linkers of the genome: The first decade of SMC proteins. Genes & Dev. 2005;19:1269–1287. doi: 10.1101/gad.1320505. [DOI] [PubMed] [Google Scholar]
- Marston A.L., Tham W.H., Shah H., Amon A. A genome-wide screen identifies genes required for centromeric cohesion. Science. 2004;303:1367–1370. doi: 10.1126/science.1094220. [DOI] [PubMed] [Google Scholar]
- McGuinness B.E., Hirota T., Kudo N.R., Peters J.M., Nasmyth K. Shugoshin prevents dissociation of cohesin from centromeres during mitosis in vertebrate cells. PLoS Biol. 2005;3:433–449. doi: 10.1371/journal.pbio.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. Segregating sister genomes: The molecular biology of chromosome separation. Science. 2002;297:559–565. doi: 10.1126/science.1074757. [DOI] [PubMed] [Google Scholar]
- Oikawa T., Okuda M., Ma Z., Goorha R., Tsujimoto H., Inokuma H., Fukasawa K. Transcriptional control of BubR1 by p53 and suppression of centrosome amplification by BubR1. Mol. Cell. Biol. 2005;25:4046–4061. doi: 10.1128/MCB.25.10.4046-4061.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orr-Weaver T. Meiotic non-disjunction does the two-step. Nat. Genet. 1996;14:374–376. doi: 10.1038/ng1296-374. [DOI] [PubMed] [Google Scholar]
- Page S.L., Hawley R.S. Chromosome choreography: The meiotic ballet. Science. 2003;301:785–789. doi: 10.1126/science.1086605. [DOI] [PubMed] [Google Scholar]
- Page J., Suja J.A., Santos J.L., Rufas J.S. Squash procedure for protein immunolocalization in meiotic cells. Chromosome Res. 1998;6:639–642. doi: 10.1023/a:1009209628300. [DOI] [PubMed] [Google Scholar]
- Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H.C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., et al. Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell. 2008;132:422–433. doi: 10.1016/j.cell.2008.01.011. [DOI] [PubMed] [Google Scholar]
- Parra M.T., Viera A., Gómez R., Page J., Benavente R., Santos J.L., Rufas J.S., Suja J.A. Involvement of the cohesin Rad21 and SCP3 in monopolar attachment of sister kinetochores during mouse meiosis I. J. Cell Sci. 2004;117:1221–1234. doi: 10.1242/jcs.00947. [DOI] [PubMed] [Google Scholar]
- Perera D., Tilston V., Hopwood J.A., Barchi M., Boot-Handford R.P., Taylor S.S. Bub1 maintains centromeric cohesion by activation of the spindle checkpoint. Dev. Cell. 2007;13:566–579. doi: 10.1016/j.devcel.2007.08.008. [DOI] [PubMed] [Google Scholar]
- Petronczki M., Siomos M.F., Nasmyth K. Un ménage à quatre: The molecular biology of chromosome segregation in meiosis. Cell. 2003;112:423–440. doi: 10.1016/s0092-8674(03)00083-7. [DOI] [PubMed] [Google Scholar]
- Prieto I., Suja J.A., Pezzi N., Kremer L., Martínez-A C., Rufas J.S., Barbero J.L. Mammalian STAG3 is a cohesin specific to sister chromatid arms during meiosis I. Nat. Cell Biol. 2001;3:761–766. doi: 10.1038/35087082. [DOI] [PubMed] [Google Scholar]
- Rabitsch K.P., Gregan J., Schleiffer A., Javerzat J.P., Eisenhaber F., Nasmyth K. Two fission yeast homologs of Drosophila Mei-S332 are required for chromosome segregation during meiosis I and II. Curr. Biol. 2004;14:287–301. doi: 10.1016/j.cub.2004.01.051. [DOI] [PubMed] [Google Scholar]
- Rivera T., Losada A. Shugoshin and PP2A, shared duties at the centromere. Bioessays. 2006;8:775–779. doi: 10.1002/bies.20448. [DOI] [PubMed] [Google Scholar]
- Schnütgen F., Schnütgen F., De-Zolt S., Van Sloun P., Hollatz M., Floss T., Hansen J., Altschmied J., Seisenberger C., Ghyselinck N.B., et al. Genome wide production of multipurpose alleles for the functional analysis of the mouse genome. Proc. Natl. Acad. Sci. 2005;102:7221–7226. doi: 10.1073/pnas.0502273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steuerwald N. Meiotic spindle checkpoints for assessment of aneuploid oocytes. Cytogenet. Genome Res. 2005;111:256–259. doi: 10.1159/000086897. [DOI] [PubMed] [Google Scholar]
- Vanoosthuyse V., Prykhozhij S., Hardwick K.G. Shugoshin 2 regulates localization of the chromosomal passenger proteins in fission yeast mitosis. Mol. Biol. Cell. 2007;18:1657–1669. doi: 10.1091/mbc.E06-10-0890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaur S., Cubizolles F., Plane G., Genier S., Rabitsch P.K., Gregan J., Nasmyth K., Vanoosthuyse V., Hardwick K.G., Javerzat J.P. Control of Shugoshin function during fission-yeast meiosis. Curr. Biol. 2005;15:2263–2270. doi: 10.1016/j.cub.2005.11.034. [DOI] [PubMed] [Google Scholar]
- Waizenegger I.C., Hauf S., Meinke A., Peters J.M. Two distinct pathways remove mammalian cohesin from chromosome arms in prophase and from centromeres in anaphase. Cell. 2000;103:399–410. doi: 10.1016/s0092-8674(00)00132-x. [DOI] [PubMed] [Google Scholar]
- Wang X., Dai W. Shugoshin, a guardian for sister chromatid segregation. Exp. Cell Res. 2005;310:1–9. doi: 10.1016/j.yexcr.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Warren W.D., Gorringe K.L. Molecular model for sporadic human aneuploidy. Trends Genet. 2006;22:218–224. doi: 10.1016/j.tig.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Watanabe Y. Sister chromatid cohesion along arms and at centromeres. Trends Genet. 2005;21:405–412. doi: 10.1016/j.tig.2005.05.009. [DOI] [PubMed] [Google Scholar]
- Watanabe Y., Nurse P. Cohesin Rec8 is required for reductional chromosome segregation at meiosis. Nature. 1999;400:461–464. doi: 10.1038/22774. [DOI] [PubMed] [Google Scholar]
- Weaver B.A., Cleveland D.W. Decoding the links between mitosis, cancer, and chemotherapy: The mitotic checkpoint, adaptation and cell death. Cancer Cell. 2005;8:7–12. doi: 10.1016/j.ccr.2005.06.011. [DOI] [PubMed] [Google Scholar]
- Weaver B.A., Bonday Z.Q., Putkey F.R., Kops G.J., Silk A.D., Cleveland D.W. Centromere-associated protein-E is essential for the mammalian mitotic checkpoint to prevent aneuploidy due to single chromosome loss. J. Cell Biol. 2003;162:551–563. doi: 10.1083/jcb.200303167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu H., Beasley M.D., Warren W.D., van der Horst G.T., McKay M.J. Absence of mouse REC8 cohesin promotes synapsis of sister chromatids in meiosis. Dev. Cell. 2005;8:949–961. doi: 10.1016/j.devcel.2005.03.018. [DOI] [PubMed] [Google Scholar]