Abstract
During the transition from post-exponential to stationary phase, Escherichia coli changes from the motile-planktonic to the adhesive-sedentary “lifestyle.” We demonstrate this transition to be controlled by mutual inhibition of the FlhDC/motility and σS/adhesion control cascades at two distinct hierarchical levels. At the top level, motility gene expression and the general stress response are inversely coordinated by σ70/σFliA/σS competition for core RNA polymerase and the FlhDC-controlled FliZ protein acting as a σS inhibitor. At a lower level, the signaling molecule bis-(3′–5′)-cyclic-diguanosine monophosphate (c-di-GMP) reduces flagellar activity and stimulates transcription of csgD, which encodes an essential activator of adhesive curli fimbriae expression. This c-di-GMP is antagonistically controlled by σS-regulated GGDEF proteins (mainly YegE) and YhjH, an EAL protein and c-di-GMP phosphodiesterase under FlhDC/FliA control. The switch from motility-based foraging to the general stress response and curli expression requires σS-modulated down-regulation of expression of the flagellar regulatory cascade as well as proteolysis of the flagellar master regulator FlhDC. Control of YhjH by FlhDC and of YegE by σS produces a fine-tuned checkpoint system that “unlocks” curli expression only after down-regulation of flagellar gene expression. In summary, these data reveal the logic and sequence of molecular events underlying the motile-to-adhesive “lifestyle” switch in E. coli.
Keywords: Adhesion, c-di-GMP, flagella, GGDEF, EAL, RpoS
Depending on environmental conditions, bacteria can exhibit very different “lifestyles.” They can occur in a motile single-cellular or planktonic state or as sedentary cells that use adhesive fimbriae to cluster together and form biofilms on surfaces (O’Toole et al. 2000; Hall-Stoodley et al. 2004). With Escherichia coli grown in complex medium, these states can be observed successively, with cells transiently becoming highly motile during the post-exponential growth phase (Adler and Templeton 1967; Amsler et al. 1993; Zhao et al. 2007) followed by the induction of adhesive curli fimbriae during entry into stationary phase (Olsén et al. 1989). Motility and adhesion can also be expected to be mutually exclusive—being sticky seems counterproductive for swimming around, whereas adhesion and settling down might require reduced activity of the force-generating flagellar “propellers.” A convenient way to realize such an inverse coordination would be to endow the flagellar/motility control system with at least one component that plays an inhibitory role in the adhesion control system and vice versa.
In E. coli, both motility and curli-mediated adhesion are under the control of regulatory feedforward cascades, each with a master regulator at the top, which acts as a massive environmental signal integrator. For flagellar expression and motility, this master regulator is the FlhDC complex (FlhD4C2, with the flhDC operon being defined as the flagellar class 1 operon) (Liu and Matsumura 1994; Wang et al. 2006). FlhDC activates the expression of class 2 operons, which encode the inner part of the flagellum (i.e., the hook basal body that also acts as a secretion system for the outer components) as well as the flagellar σ subunit of RNA polymerase (RNAP), FliA (σ28 or σF), and its anti-σ factor, FlgM. Upon secretion of FlgM, which occurs as soon as the basal body secretion system is functional, FliA is released to activate class 3 operons that encode the outer subunits of the flagellum, additional proteins required for flagellar function and chemotaxis, as well as a number of proteins of still unknown function (Chilcott and Hughes 2000; Karlinsey et al. 2000; Aldrigde et al. 2006).
Adhesive curli fimbriae, on the other hand, are expressed during entry into stationary phase (in cells that grow below 30°C). Curli fimbriae are involved in both cell–cell aggregation and surface adhesion (Barnhart and Chapman 2006). The curli control cascade is a module within the general stress response, for which the σS (RpoS) σ subunit of RNAP acts as the master regulator (Hengge-Aronis 2000). In parallel, σS-containing RNAP (EσS) activates the expression of MlrA (a MerR-like regulator) and YdaM (a GGDEF protein, see below), which, together with EσS again, are essential to activate transcription of the csgD gene. The CsgD protein (also called AgfD in Salmonella) is an essential activator for the curli structural gene operon (csgBAC), and cooperates with the vegetative RNAP at the csgB promoter (Römling et al. 2000; Brown et al. 2001; Gerstel et al. 2003; Weber et al. 2006).
Another key player in the control of motility and curli expression—i.e., in bacterial “lifestyle” switching—is the signaling molecule bis-(3′–5′)-cyclic-diguanosine monophosphate (c-di-GMP) (for recent reviews, see Römling et al. 2005; Jenal and Malone 2006; Ryan et al. 2006; Tamayo et al. 2007). Overproduction of certain diguanylate cyclases (DGC; characterized by GGDEF domains) interferes with motility and strongly activates the expression of curli fimbriae and the biofilm matrix component cellulose in enteric bacteria, whereas overproduction of certain c-di-GMP-degrading phosphodiesterases (PDE; carrying EAL domains) produces the opposite phenotype. In recent mutational and biochemical analyses, specific DGCs and PDEs have been assigned antagonistic roles in the regulation of curli expression (Kader et al. 2006; Weber et al. 2006; Simm et al. 2007). For E. coli , these are YdaM (the GGDEF protein already mentioned above) and YciR (a GGDEF + EAL protein), which control csgD transcription (Weber et al. 2006). Another EAL protein, YhjH, was shown to play a positive role in motility (Ko and Park 2000; Rychlik et al. 2002; Frye et al. 2006; Ryjenkov et al. 2006). However, the molecular details of c-di-GMP action in the control of curli expression and motility have not been characterized.
The questions addressed by our study are the following: Do the FlhDC/motility and σS/curli control cascades really directly communicate to inversely coordinate their activities? And, if so, what are the factors involved, and are (some of) these factors c-di-GMP control systems; i.e., GGDEF/EAL proteins? With the present study, we identified several such factors acting at different hierarchical levels of the control network established, we provide evidence that throwing the motility-to-adhesion switch requires a precise down-regulation of motility at the transcriptional, proteolytic, and protein activity levels, and we present a framework model for the logic and the sequence of molecular events underlying this phenotypic “lifestyle” change in E. coli during entry into stationary phase.
Results
FliZ, a protein under flagellar control, interferes with σS activity and curli formation
Escherichia coli K-12 strains are quite heterogenous with respect to σS levels and activity (Jishage and Ishihama 1997), curli expression (Bokranz et al. 2005), and motility (Barker et al. 2004). The strain MC4100 used in our previous studies of σS and curli regulation is nonmotile due to a+1 frameshift mutation in the flhDC operon encoding the flagellar master regulator (Barembruch and Hengge 2007). Therefore, we chose another commonly used strain, W3110, which is flhDC+ and exhibits wild-type motility (Supplemental Fig. S1). W3110 reaches very similar σS levels in stationary phase as MC4100 (data not shown) and also expresses the curli genes (assayed using a single-copy csgB∷lacZ reporter fusion) under the same control of σS, MlrA, YdaM, YciR, and CsgD as MC4100 (Supplemental Fig. S1). Interestingly, the control by YdaM seemed even more stringent in W3110, which was a first hint that the presence of the flagellar system may produce an inhibitory influence that has to be overcome by the complex curli control cascade.
In parallel, we also used strain MC4100, in which we complemented the chromosomal flhDC mutation by a low-copy-number plasmid (pFlhDC) that upon IPTG induction expresses FlhDC from the tac promoter (ptac) at similar levels as W3110 (Barembruch and Hengge 2007). While this construct generated motility (Barembruch and Hengge 2007), it surprisingly eliminated curli gene expression as assayed using a csgB∷lacZ reporter fusion (Supplemental Fig. S2A). Moreover, pFlhDC did so too when transformed into W3110 (data not shown). We reasoned that the major difference between strains with a single flhDC+ allele in the chromosome and the pFlhDC constructs is the flhDC expression pattern during transition into stationary phase: Whereas wild-type strains seem to shut down flagellar gene expression during this time (Amsler et al. 1993; Barembruch and Hengge 2007; see below), ptac-driven flhDC continues to be expressed. This suggested that the flagellar FlhDC regulon indeed includes at least one inhibitor of curli expression, and that the strong curli-negative phenotype of the pFlhDC-carrying strain provided a screen for identifying this inhibitor genetically.
As a first step, we introduced a fliA mutation into the pFlhDC-carrying strain and observed a complete suppression of the inhibition of curli gene expression (Supplemental Fig. S2B). We concluded that the inhibitor for curli expression is either (1) FliA itself, which when remaining present in stationary phase at elevated levels may outcompete σS at the RNAP core enzyme, (2) a protein the expression of which is FliA-dependent (i.e., a class 3 flagellar gene product), or (3) a protein expressed from a gene right downstream from fliA onto which the fliA∷cat insertion would exert a polar effect. A candidate for the latter was the fliZ gene, because at least fliZ seems to be part of an operon with fliA, and its molecular function has not been defined. Therefore, we introduced a fliZ mutation into the pFlhDC strain and again observed suppression; i.e., curli genes were expressed (Supplemental Fig. S2C). This result not only eliminated the first two possibilities for the identity of the inhibitor (since in this fliZ mutant, FliA is still expressed under pFlhDC control), but in addition indicated that FliZ itself is the inhibitor, as the fliZ mutation used here is a nonpolar in-frame deletion. Finally, we cloned fliZ alone onto the same low-copy-number ptac vector (pFliZ). When this plasmid was introduced into MC4100, which due to its flhDC mutation does not express any other flagellar gene product besides this plasmid-encoded FliZ, it completely inhibited curli expression during entry into stationary phase even in the absence of inducer (Fig. 1B). We conclude that FliZ is a highly potent inhibitor of curli expression.
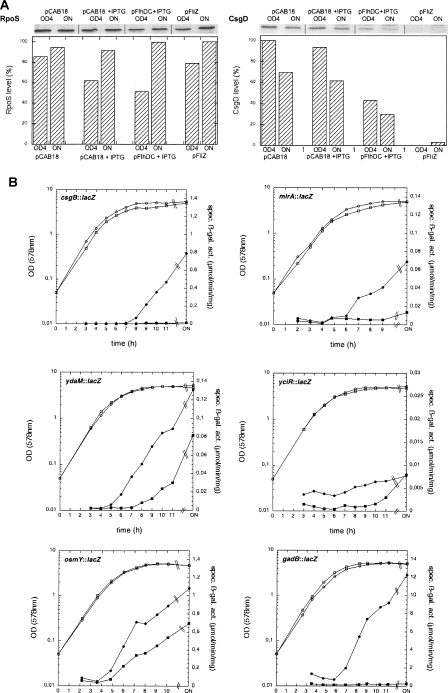
Figure 1.
FliZ is an inhibitor of σS activity. (A) FliZ does not affect σS levels, but strongly down-regulates the σS-controlled curli regulator CsgD. Derivatives of strain MC4100 carrying either pFlhDC, pFliZ, or the vector (pCAB18) alone were grown in the presence or absence of IPTG as indicated. Samples were taken for immunoblot analysis of σS and CsgD at an OD578 of 4.0 and after overnight growth (ON). Densitometric quantification is shown below the blots. (B) FliZ reduces the expression of σS-controlled genes. Expression patterns of the σS-dependent genes csgB, mlrA, ydaM, yciR, osmY, and gadB were recorded using appropriate single-copy reporter fusions in strain MC4100 carrying either pFliZ (squares) or the vector (pCAB18; circles). Cells were grown without IPTG (as even low levels of FliZ are sufficient to interfere with the expression of the genes shown), and OD578 (open symbols) and specific β-galactosidase activities (closed symbols) were determined along the growth curve.
How does FliZ interfere with curli expression, and is this inhibition specific for curli? Along the curli control cascade, there are many potential targets and, as a first step, we tested whether the presence of the inhibitory plasmids pFlhDC and pFliZ affected the cellular levels of the essential curli activators σS and CsgD (Fig. 1A). While σS levels remained unchanged, CsgD levels were reduced by pFlhDC and virtually eliminated by pFliZ (even without inducer). Using single-copy lacZ reporter fusions, we then found that pFliZ reduced the expression of MlrA, YdaM, and YciR (Fig. 1B), which together and highly specifically control csgD transcription (Weber et al. 2006). This explained the effect on CsgD levels (Fig. 1A). In addition, as mlrA, ydaM, and yciR are all under σS control, this finding also raised the possibility that other σS-dependent genes might be affected as well and that FliZ might have a more general role than just controlling curli expression. Indeed, the expression of σS-controlled genes not associated with curli expression was also reduced by pFliZ: While osmY, a directly EσS-controlled gene, was partially affected (Fig. 1B), the expression of gadB—i.e., a gene involved in acid resistance, which is under multiple feedforward control of σS (Weber et al. 2005) in a manner analogous to csgB—was also completely eliminated (Fig. 1B). Thus, while FliZ does not affect σS levels, it reduces σS-dependent gene expression; i.e., σS activity.
A rather general effect on σS-dependent gene expression was further corroborated by genome-wide transcriptional profiling of the pFliZ-carrying strain (Supplemental Table S1): The majority of FliZ-down-regulated genes were identified previously as σS-controlled (Weber et al. 2005; H. Weber and R. Hengge, unpubl., for cells grown at 28°C). Moreover, these FliZ-down-regulated genes showed a strong overlap with genes activated by Crl (Supplemental Table S1)—i.e., a factor that specifically supports σS in its competition with other σ factors for RNAP core (Typas et al. 2007a)—suggesting a role for FliZ in σS control that is functionally antagonistic to that of Crl. Notably, by promoting the formation of EσS, Crl plays a role in the timing of induction of σS-dependent genes (in crl mutants, their activation during entry into stationary phase is delayed) (see Supplemental Fig. S3; Typas et al. 2007a).
What is the role of FliZ in the motility-to-adhesion transition during entry into stationary phase? The essential activating components for transcription of the curli activator CsgD—i.e., EσS, YdaM, and MlrA—are induced in a reproducible temporal order: While EσS-mediated expression of genes with high-affinity promoters such as osmY started as early as an OD578 of 1, ydaM expression started around an OD578 of 2, and mlrA expression finally began only around an OD578 of 3 (Fig. 1B). We observed that the timing of induction of the “late” gene mlrA was most sensitive to FliZ (Fig. 2A): In a fliZ knockout mutant, mlrA expression started more than an hour earlier (at an OD578 even below 2 already) (for comparison, see effects on mlrA expression of mutations that affect EσS levels such as crl in Supplemental Fig. S3). Also, the expression of ydaM reproducibly started somewhat earlier in the fliZ mutant (Fig. 2B). In order to show that fliZ affects σS activity and not some other factor possibly involved in the control of these genes, we also tested a synthetic σS-dependent promoter (synP9, present in a single-copy lacZ fusion), which does not respond to any other regulatory factors besides σS, and is sensitive even to gradual variations in the cellular content of σS-containing RNAP (Typas et al. 2007a). In addition, synP9 is a typical σS-dependent promoter without −35 and with σS-specifically extended −10 regions (Typas et al. 2007b) (the mlrA and ydaM promoter regions are of the same type) (our unpublished data). Stationary phase induction of the synP9::lacZ fusion also started earlier in the fliZ mutant (Fig. 2C).
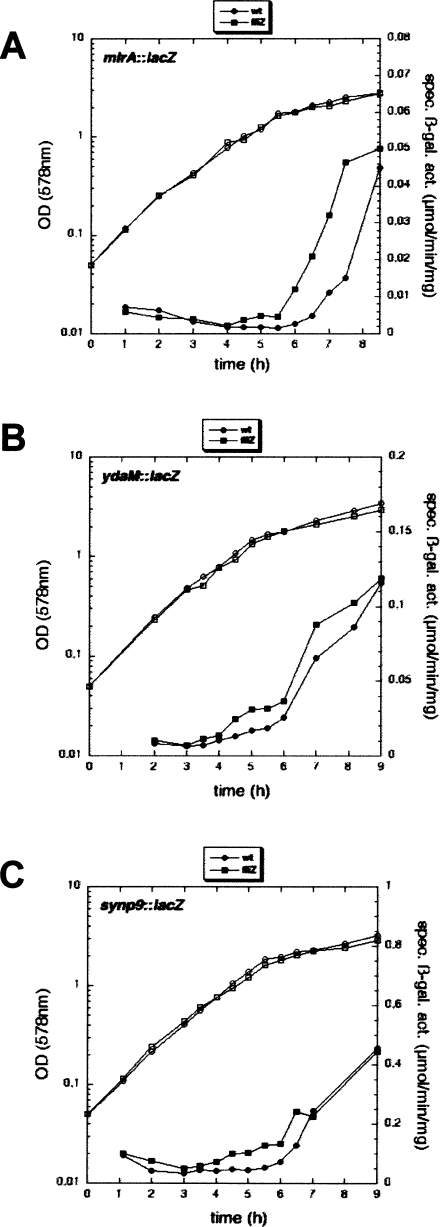
Figure 2.
Mutations in fliZ affect the timing of induction of σS-dependent genes. Two directly σS-controlled genes encoding MlrA (A) and YdaM (B) (i.e., the factors essential for the expression of the curli activator CsgD), as well as a construct with a synthetic promoter that responds exclusively to σS, synp9 (C), were assayed. Derivatives of strain W3110 carrying single-copy lacZ reporter fusions to mlrA, ydaM, and synp9 and the corresponding fliZ knockout mutants were grown in LB (for symbols, see the figure). OD578 (open symbols) and specific β-galactosidase activities (closed symbols) were determined.
Taken together, these data indicate that during the post-exponential phase, FliZ transiently interferes with σS activity; i.e., either with EσS formation or with EσS activity directly at σS-controlled promoters. Thereby, FliZ can act as a timing device for curli expression, since among the essential activators of csgD transcription, MlrA exhibits the latest expression and is most delayed by FliZ.
Down-regulation of motility by a σS-controlled c-di-GMP module comprising the GGDEF proteins YegE/YedQ/YeaJ and the EAL protein YhjH
With FliZ, the flagellar system contains a factor that during the post-exponential growth phase interferes with the general stress response and therefore also with adhesive curli fimbriae expression. In addition, there is another “promotility” factor—i.e., the EAL protein YhjH—that is expressed from a completely FliA-dependent class 3 gene (Fig. 4D, below; Frye et al. 2006). Overexpression of YhjH could rescue motility of an otherwise nonmotile hns mutant (Ko and Park 2000). The E. coli yhjH mutant exhibited strongly reduced motility on soft agar plates (Fig. 3A). Purified YhjH was active as a c-di-GMP-cleaving phosphodiesterase (Fig. 3B), and the glutamic acid in its ELL sequence (corresponding to the generic EAL motif in this protein family) was essential for its enzymatic activity in vitro (Fig. 3B) as well as for its biological activity in vivo (i.e., for complementation of a yhjH mutation; data not shown). Both in Salmonella and E. coli, the effects of the yhjH mutation on motility depend on the presence of the YcgR protein (Fig. 3A; Ryjenkov et al. 2006; Girgis et al. 2007), which is also under flagellar control (Frye et al. 2006) and belongs to the family of PilZ proteins that function as c-di-GMP-binding effector proteins (Ryjenkov et al. 2006; Benach et al. 2007; Christen et al. 2007). Reduced motility in liquid cultures could actually be observed directly under the microscope. While all strains tested finally became nonmotile in stationary phase (probably due to reduced energy supply), the yhjH mutant began to reduce speed in the swimming phase at an OD578 of 3.5–3.6 already, followed by the parental strain (W3110) about an hour later (at an OD578 of 3.7–3.8), whereas the ycgR mutant began to reduce swimming speed only at an OD578 beyond 4.0; i.e. >2 h later than the parental strain (data not shown).
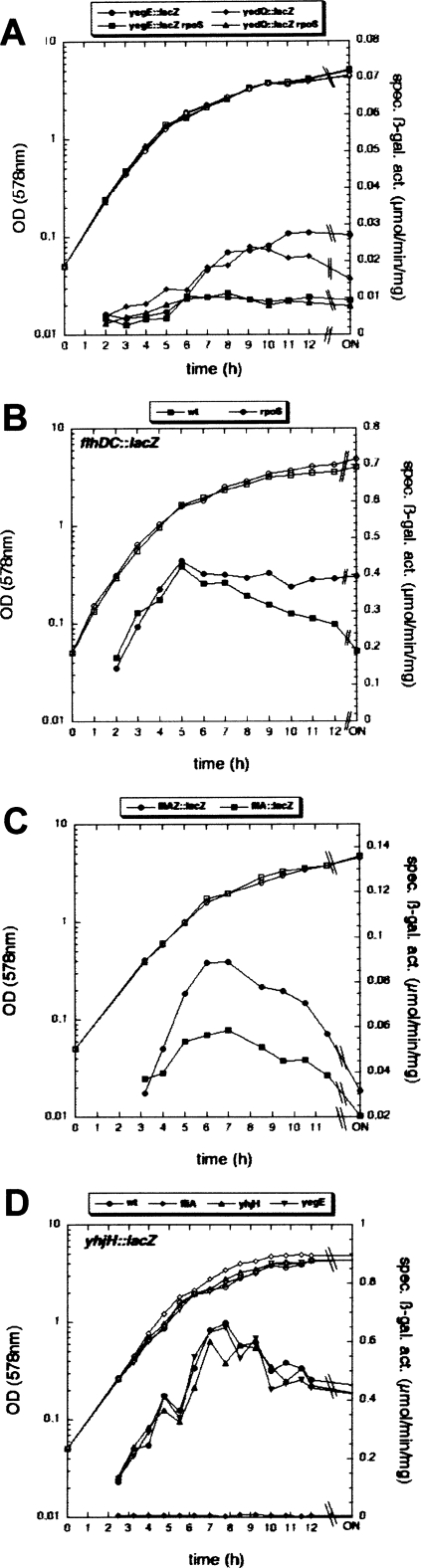
Figure 4.
Expression patterns of genes relevant for the motility-to-adhesion switch. Derivatives of strain W3110 carrying single-copy lacZ reporter fusions in yegE and yedQ (A), flhDC (B; fusion inserted early in flhC), fliA and fliAZ (C; the latter fusion inserted early in fliZ), and yhjH (D) as well as knockout mutations in the genes indicated in the boxes above the single graphs, were used for the determination of OD578 (open symbols) and specific β-galactosidase activities (closed symbols) along the growth curve.
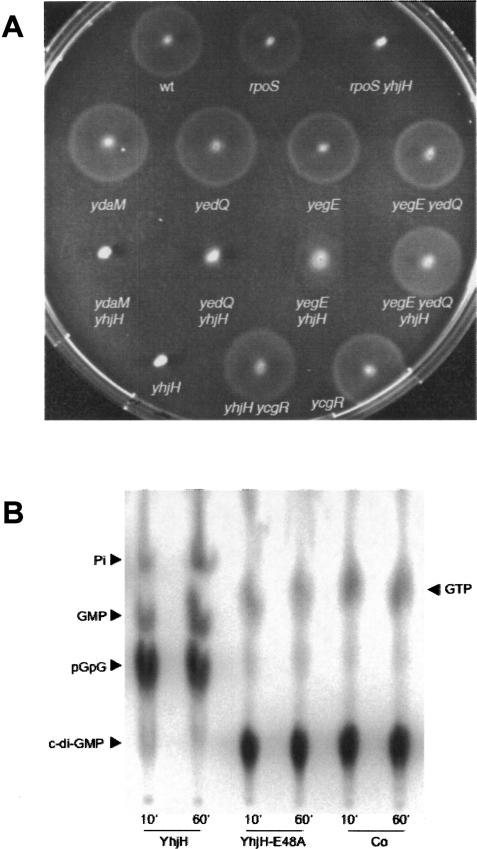
Figure 3.
A complex c-di-GMP control module interferes with motility. (A) Knockout mutations in the genes for the GGDEF proteins YegE and YedQ (but not YdaM) and the c-di-GMP effector YcgR suppress the nonmotile phenotype of strains defective for the EAL domain protein YhjH. Single, double, and triple mutants defective in the genes indicated were tested for motility on swim plates incubated at 28°C. (B) YhjH is a c-di-GMP phosphodiesterase (PDE). Purified YhjH [whose EAL(48–50) domain actually features an ELL sequence] and YhjHE48A were tested for PDE activity in vitro using radiolabeled c-di-GMP. Cleavage products are indicated.
The di-guanylate cyclase(s)—i.e., the GGDEF protein(s)—that provide the c-di-GMP necessary to reduce motility have not been characterized, but a previously observed partial suppression of the motility impairment of a yhjH mutant by a yegE mutation suggested that the GGDEF+EAL protein YegE may be involved (Girgis et al. 2007). Here, we tested whether knockout mutations in any of the 19 GGDEF domain-encoding genes could suppress the nonmotile phenotype of the yhjH mutant. When cells were grown at 28°C, two mutations, yegE and to a minor extent yedQ, were found to be partially effective (Fig. 3A). The yegE yedQ double knockout completely restored motility of the yhjH mutant, indicating that the two proteins function in a redundant and additive manner, with YegE being the major player. At 37°C (where curli fimbriae are not expressed), another GGDEF gene, yeaJ, exhibited most pronounced suppression when knocked out alone, and in order to obtain wild-type motility, yeaJ, yegE, and yedQ had to be mutated all together (Supplemental Fig. S4). These data indicate that, depending on growth temperature, YegE (28°C) or YeaJ (37°C), and to a minor extent, YedQ, are the GGDEF proteins that provide the c-di-GMP that via YcgR reduces motility.
Notably, the GGDEF protein YdaM, which is the diguanylate cyclase absolutely required for curli expression (see Fig. 6C, below; Weber et al. 2006), does not contribute to motility control, as its knockout mutation did not produce even traces of suppression of the yhjH nonmotility phenotype (Fig. 3A; it should be noted that the ydaM gene is induced already at an OD578 of 0.3 in the tryptone medium used for motility plates; data not shown). In addition, eliminating the EAL protein and phosphodiesterase YciR, which acts as an antagonist to YdaM in curli control (Weber et al. 2006), had no effect at all on motility (Supplemental Fig. S5); i.e., its presence in the yhiH mutant cannot compensate for the absence of YhjH. Thus, the YdaM/YciR c-di-GMP control module does not “cross-talk” into motility regulation.
Figure 6.
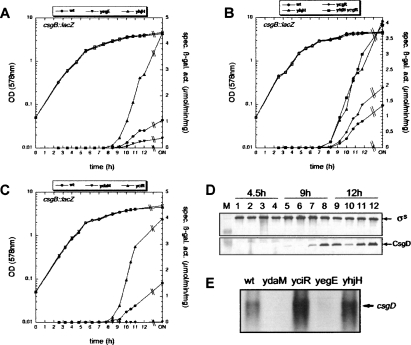
The GGDEF/EAL proteins YegE and YhjH affect curli expression by controlling csgD transcription, but in this process do not act via the c-di-GMP-binding protein YcgR. (A–C) csgB∷lacZ expression was assayed in derivatives of strain W3110 carrying mutations in yegE, yhjH, ycgR, ydaM, or yciR as indicated (for symbols, see figure). OD578 (open symbols) and specific β-galactosidase activities (closed symbols) were determined along the growth curve. (D) σS and CsgD levels were determined by immunblotting of strain W3110 (lanes 1,5,9) and its derivatives with mutations in yegE (lanes 2,6,10), yegE and yhjH (lanes 3,7,11), and yhjH (lanes 4,8,12). Samples were taken 4.5, 9, and 12 h after inocculation (corresponding to an OD578 of 1.0, 3.0, and 4.0, respectively). (E) csgD mRNA levels were determined by Northern analysis in W3110 and its mutant derivatives as indicated, with samples taken at an OD578 of 4.0.
We then tested whether the GGDEF proteins YegE and YedQ that down-regulate motility might somehow be linked to the σS/curli control cascade. Using single-copy lacZ reporter fusions, we observed that both yegE and yedQ exhibited low basal expression during log phase that increases during transition into stationary phase, and that this induction was dependent on σS (Fig. 4A). Thus, by stimulating the expression of the GGDEF proteins YegE and YedQ, σS seems to positively contribute to the cellular c-di-GMP balance.
Switching from motility to adhesion requires down-regulation of expression of the flagellar system
Our finding that continued ectopic expression of the flagellar master regulator FlhDC under ptac control during entry into stationary phase strongly inhibited curli expression (Supplemental Fig. S2) indicated that in a wild-type strain, down-regulation of flagellar gene expression is a prerequisite for the activation of the curli genes. In order to monitor such a physiological stop of expression for all relevant regulatory proteins at the different hierarchy levels, we used the single-copy reporter fusions flhDC∷lacZ, fliA∷lacZ, fliAZ∷lacZ, and yhjH∷lacZ (the latter also represents class 3 expression in general). Their expression patterns (Fig. 4B–D) demonstrated that all these genes (and therefore most likely the entire flagellar system) were activated during the early post-exponential phase. Then, however, expression of these genes stopped at an OD578 between 2 and 2.5, with the class 3 gene yhjH reproducibly shutting down later than the class 1 and 2 operons. As flhDC down-regulation was less pronounced in a rpoS mutant (Fig. 4B), σS, which is induced during this phase of the growth cycle, directly or indirectly contributed to this stop of expression of flagellar genes.
We then wondered whether this stop of expression of the flagellar system might also be linked to the activity of the YegE+YedQ/YhjH/YcgR system, which reduces motility as described above. However, neither the yegE nor the yhjH knockout mutations had any effect on the expression of the yhjH∷lacZ fusion (which is used here as a representative for the overall class 3 output of the flagellar control system; Fig. 4D). Additional class 2 and 3 reporter fusions tested (flgA∷lacZ and flgM∷lacZ, respectively) were also not affected (data not shown).
We conclude that the reduction of motility during entry into stationary phase is based on two separate components: (1) a shut-down of flagellar gene expression during entry into stationary phase, which is required for curli expression to occur, and which is triggered by signals that act independently from c-di-GMP; and (2) a reduction of flagellar activity; i.e., altered motor function and reduced swimming speed, which is modulated by the c-di-GMP control module consisting of YegE+YedQ/YhjH/YcgR.
Switching from motility to adhesion requires ClpXP-mediated proteolysis
Although the shut-down of de novo expression of flagellar proteins is necessary for switching from motility to curli-mediated adhesion during entry into stationary phase, it may actually not be sufficient, since the already present “promotility” regulatory proteins may not be sufficiently diluted by future cell divisions any more (after shut-down of flagellar gene expression at an OD578 of 2.5, the remaining culture cell mass increases until stationary phase just twofold). Thus, these regulators should be inactivated or even proteolytically degraded. For the master regulator of flagellar gene expression—i.e. the FlhDC complex—rapid proteolysis has indeed been shown already (Tomoyasu et al. 2003), although the physiological function of FlhDC degradation has remained unknown.
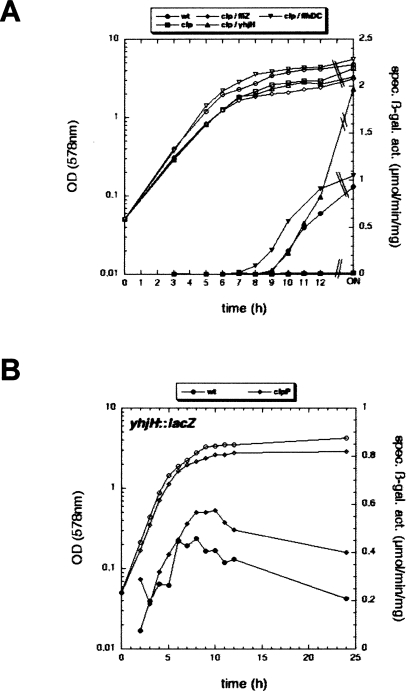
Here, we observed that a clpP mutant, which lacks the proteolytic subunit of the ClpXP and ClpAP proteases, was completely defective for curli expression (Fig. 5A)—even though σS—i.e., the master regulator in the curli control cascade—is itself a ClpXP substrate and therefore accumulates earlier and in higher amounts in the clpP mutant (Hengge-Aronis 2002). The inability of the clpP mutant to express curli was completely suppressed by the flhDC knockout mutation (Fig. 5A), indicating that the interference of the flagellar system with curli expression was responsible for this phenotype. The relevant component, however, was not FliZ, as a fliZ mutation did not suppress the curli-negative phenotype of the clpP mutant (Fig. 5A). In contrast, knocking out fliA (no matter whether polar on fliZ or not; data not shown) and yhjH (Fig. 5A) resulted in complete suppression. This suggested that, with respect to curli expression, the problem in the clpP mutant is a failure to down-regulate YhjH appropriately. Moreover, it also indicated that c-di-GMP controlled by YhjH not only reduces motility but also has a positive influence on curli expression (see further below).
Figure 5.
The Clp protease plays a key role in down-regulating YhjH, which is a prerequisite for inducing curli expression during entry into stationary phase. (A) A clpP derivative of W3110 (carrying csgB∷lacZ as a reporter) does not induce curli expression, and this defect is suppressed by secondary mutations in flhDC and yhjH, but not in fliZ (for symbols, see figure). (B) Under the same conditions, the expression of a single-copy yhjH∷lacZ fusion was tested in clpP+ and clpP mutant backgrounds. OD578 (open symbols) and specific β-galactosidase activities (closed symbols) were determined along the growth curve of LB-grown cells.
A failure of the clpP mutant to appropriately down-regulate YhjH could mean that either YhjH is itself a Clp protease substrate, or that prolonged presence of the flagellar master activator FlhDC, a known ClpXP substrate, may result in a failure to shut down the expression of the yhjH gene in due time. The second possibility seems correct, as YhjH protein levels were only slightly reduced after the stop of expression around an OD578 of 2.5 (consistent with dilution by still ongoing cell division) (Supplemental Fig. S6A), YhjH did not rapidly disappear upon spectinomycin-induced cessation of total protein synthesis (Supplemental Fig. S6B), and the clpP mutation did not aggravate the partial down-regulation of curli expression by low-level ectopic YhjH expression (i.e., uncoupled from its natural control by FlhDC) (Supplemental Fig. S6C). On the other hand, the expression of yhjH in the clpP mutant was prolonged by ∼2 h during entry into stationary phase (Fig. 5B). This indicated that a precise shut-down of YhjH expression around an OD578 of 2.5 in a wild-type strain requires FlhDC proteolysis and is essential for the ability of the concomitantly induced GGDEF proteins YegE/YedQ to overcome the PDE activity of YhjH and thereby shift the c-di-GMP balance as required for the induction of curli.
The GGDEF/EAL proteins YegE and YhjH connect inhibition of motility to activation of curli expression
The involvement of YhjH in curli repression in the clpP mutant suggested that the entire YegE+YedQ/YhjH/YcgR c-di-GMP control module may not only regulate motility, but also curli expression. Indeed, while the yegE mutation reduced curli expression by ∼60%, the yhjH mutation increased curli expression (Fig. 6A). The yedQ mutation had no effect (also when introduced in addition to the yegE mutation; data not shown). Strikingly, however, YcgR is not the c-di-GMP-binding effector responsible for YegE-mediated activation of curli expression, since the ycgR mutation even slightly increased curli expression (Fig. 6B). Moreover, increased curli expression in the yhjH mutant was not suppressed by the ycgR mutation (Fig. 6B), in contrast to the nonmotile phenotype of the yhjH mutant (Fig. 3A). We conclude that YegE/YhjH-controlled c-di-GMP activates curli expression, but that it does so not via the effector protein YcgR. Thus, YegE/YhjH-controlled c-di-GMP affects different processes via different effector components.
Finally, we wanted to know at which level of the σS/curli regulatory cascade YegE/YhjH-controlled c-di-GMP comes into play. The yegE and yhjH mutations did not affect σS levels, but did alter CsgD levels (Fig. 6D). In addition, the yegE and yhjH mutations did not change the expression of the regulators of csgD transcription, i.e., YdaM, YciR, and MlrA (assayed with their respective lacZ fusions) (Supplemental Fig. S7); i.e., YegE and YhjH do not have a general influence on σS activity and therefore the general stress response. However, YegE/YhjH did affect csgD mRNA levels (Fig. 6E). As ectopic expression of CsgD from exactly the same csgD mRNA transcribed from the pBAD promoter was not altered by the yegE and yhjH mutations (Supplemental Fig. S8), we conclude that the YegE/YhjH system controls csgD transcription rather than csgD mRNA degradation, and by affecting CsgD levels, regulates curli expression.
Discussion
Switching from motility to adhesion is based on mutual inhibition of the flagellar and curli control cascades
The natural change from the motile-planktonic to the adhesive-sedentary state during transition from the post-exponential to the stationary phase of growth of E. coli provides a convenient model system for studying a bacterial “lifestyle” switch. It should be noted, however, that “adhesion” or “sedentary state” are not semantically exchangeable with the term “biofilm.” Rather, in a developing biofilm, motile and sedentary subpopulations can coexist (Klausen et al. 2006), and it may be that the molecular switch mechanisms described here, which produce a temporal succession of the motile and adhesive states in a liquid culture, are also involved in the differentiation into spatially separated motile and sessile subpopulations in a growing biofilm.
We show here that the two complex transcription factor cascades in E. coli that control expression and function of the motility system, and the expression of the adhesive curli fimbriae, respectively, operate in a mutually exclusive way by negatively “cross-talking” at different hierarchical levels (summarized in Fig. 7). The FlhDC/motility control system contains at least three components that down-regulate the σS/curli control system (Fig. 7, red arrows): (1) FliZ interferes with σS activity, (2) FliA(σF) competes with σS for RNAP core enzyme, and (3) the EAL protein and phosphodiesterase YhjH eliminates c-di-GMP that otherwise (via YcgR) inhibits motility and (via a second still unknown effector component) activates csgD transcription and therefore curli expression. On the other hand, σS, which is strongly up-regulated during entry into stationary phase, contributes to down-regulation of motility (Fig. 7, green arrows) (1) by increasingly competing with σ70 and FliA for RNAP core enzyme, and (2) by inducing the GGDEF proteins YegE and YedQ that by producing c-di-GMP counteract the activity of YhjH.
Figure 7.
A model summarizing the communication network between the FlhDC/motility and σS/curli regulatory cascades and the role of different c-di-GMP control modules. At the top of the network, motility and the entire general stress response are inversely coordinated by competition of σ70, σFliA, and σS for RNAP core, with FliZ being a “promotility” component that interferes with σS activity. At lower levels of the network hierarchy, motility and adhesion (via curli fimbriae) are inversely coordinated by two separately acting c-di-GMP control modules involving GGDEF/EAL proteins: YegE(+YedQ)/YhjH down-regulates flagellar activity and positively modulates csgD transcription, whereas YdaM/YciR acts specifically in and is essential for csgD transcription only (for more details, see Discussion). Also included is a third GGDEF/EAL module (i.e., YaiC/YoaD) that is expressed under σS control later during entry into stationary phase (Weber et al. 2006; our unpublished data) and regulates the activity of cellulose synthase (Römling et al. 2005; Brombacher et al. 2006). Negative effects of the FlhDC/motility system onto the σS/curli system are highlighted in red, effects by which the σS/curli system down-regulates the FlhDC/motility system are in green. Triangles indicate additional signal input not further specified.
This complex mutual inhibition of the two control cascades provides the basis of a switch mechanism. Throwing the switch from motility to adhesion during transition into stationary phase requires not only σS accumulation but also a precisely timed shut-off of flagellar gene expression, for which a cessation of flhDC expression as well as the ClpXP-mediated proteolysis of the motility master regulator FlhDC are essential. Only then YhjH is reduced sufficiently that its c-di-GMP-reducing PDE activity can be overcome by the increasingly expressed GGDEF proteins YegE and YedQ (Fig. 7).
With FliZ, the flagellar system contains a potent inhibitor for the general stress response and curli expression
FliZ is encoded in a flagellar class 2/3 operon together with the flagellar σ factor FliA, but FliZ is not required for motility (a fliZ derivative of W3110 is even slightly more motile than the parental strain; data not shown), and also in Salmonella a fliZ mutation had a minor modulatory effect only (Kutsukake et al. 1999) or even no effect (Frye et al. 2006) on flagellar gene expression. Here, we show that FliZ is a potent inhibitor for the σS-controlled general stress response (Fig. 1; Supplemental Table S1), with those genes being most affected that are under multiple σS control in feedforward loops (csgB, gadB). As σS levels are not affected (Fig. 1), FliZ reduces σS activity. Interestingly, there is a strong overlap between FliZ-down-regulated and Crl-activated genes (Supplemental Table S1). Crl binds to σS (Bougdour et al. 2004) and thereby stimulates EσS holoenzyme formation during entry into stationary phase (Typas et al. 2007a). During entry into stationary phase, a crl mutant exhibits a delay in the expression of certain σS-dependent genes, which have relatively low-affinity promoters and are therefore most susceptible to lower cellular levels of EσS (Typas et al. 2007a).
Formally, FliZ could be an anti-σ factor for σS. Alternatively, it may modulate the activity of EσS at a subset of promoters (e.g., those with low affinity). Both mechanisms would be consistent with FliZ apparently antagonizing Crl. A simple anti-σS activity, however, seems unlikely as overproduction of FliZ is detrimental for growth also in rapidly growing cells in which σS is hardly present (data not shown); even total inhibition of σS activity would not be expected to inhibit growth (as rpoS knockout mutations do not have such impact). Moreover, FliZ contains a region with similarity to certain phage integrases, which is also present in XerD, a DNA-binding protein and site-specific recombinase involved in chromosome decatenation (Subramanya et al. 1997).
While the precise biochemical function of FliZ remains to be characterized, the physiological function of FliZ is already apparent: By interfering with the activity of σS, which already accumulates during the post-exponential growth phase (Lange and Hengge-Aronis 1994), FliZ transiently gives motility (and therefore a foraging strategy) priority over the general stress response—as long as flagellar gene expression keeps going. FliZ acts as a timing factor for the expression of certain σS-controlled genes, including those involved in curli expression (also thereby antagonizing Crl), as illustrated by the mlrA and ydaM genes. Their timing of activation is precisely controlled by FliZ (Fig. 2) and by Crl (Supplemental Fig. S3). MlrA is most strongly affected and exhibits the latest induction among the three essential activators for csgD transcription (the other ones being σS and YdaM), and therefore can act as a timing device in the expression of the curli regulator CsgD.
Finally, a fliZ mutation resulted in premature expression of various additional σS-dependent genes, which include two EAL genes (actually the majority of GGDEF/EAL genes is under σS control) (our unpublished data). If the encoded c-di-GMP phosphodiesterases become active under these conditions, this would explain why a fliZ mutation can partially suppress the nonmotility phenotype of the yhjH mutant (Girgis et al. 2007; also observed by us).
Switching from motility to adhesion requires down-regulation of flagellar gene expression and proteolysis of the flagellar master regulator FlhDC
With FliZ giving priority to motility, down-regulation of the flagellar system is required to allow the activation of σS-dependent genes, including the control cascade for the expression of the adhesive curli fimbriae. Consistently, the expression of the relevant regulatory genes flhDC, fliA, fliZ, and yhjH ceases at an OD578 between 2 and 2.5 in LB-grown cells (actually in a temporal order that reflects the flagellar gene hierarchy) (Fig. 4). The curli regulator CsgD starts to accumulate, and expression of its target gene csgB begins shortly thereafter (at an OD578 of 2.8–3) (Fig. 6).
What is the triggering signal for this down-regulation of the entire flagellar system? Even though the c-di-GMP control system YegE/YhjH modulates motility (Fig. 3; see below), it is not involved in the down-regulation of flagellar gene expression (Fig. 4D). Also, mutations in regulatory systems known to be able to interfere with flhDC expression—i.e., the Rcs and BarA/UvrY two-component systems as well as LrhA (summarized in Prüß et al. 2006)—did not influence the post-exponential shut-down of flhDC expression (S. Busse, A. Kademci, and R.Hengge, unpubl.). Induction of σS itself and its successful competition with σ70 (driven by ppGpp accumulation during entry into stationary phase (Lange et al. 1995; Jishage et al. 2002) may contribute to flhDC down-regulation, as the latter is less pronounced in the rpoS mutant (Fig. 4B).
The observed down-regulation of flagellar gene expression and motility cannot be a consequence of repression of flhDC expression alone. In addition, the activator proteins of the flagellar system already present in the cell have to be inactivated. Dilution by further cell divisions would not be effective, as cells already approach stationary phase when these processes take place. Getting rid of proteins under such conditions often occurs by proteolysis (Jenal and Hengge-Aronis 2003). The transcriptional activators of the motility system—i.e., FlhDC and FliA—are known to be degraded by the ClpXP and Lon proteases, respectively (Tomoyasu et al. 2003; Barembruch and Hengge 2007). Here, we demonstrate that the Clp protease is indeed essential for the precisely timed reduction of the expression and therefore overall cellular activity of the c-di-GMP-degrading PDE YhjH, which has to be overcome to switch to curli induction (Figs. 5, 6). As YhjH is not degraded under these conditions (Supplemental Fig. S6), ClpXP-dependent proteolysis of FlhDC provides the basis for the curli-negative phenotype of the clpP mutant. That disappearance of FlhDC protein is absolutely necessary for curli induction, also finally provides a clear physiological rationale for FlhDC degradation.
Role of GGDEF/EAL proteins YegE, YedQ, YeaJ, and YhjH in the motility-to-adhesion switch
In E. coli, the YhjH protein is crucial for motility in post-exponential cells—i.e., either on nutrient-poor soft agar plates (Fig. 3A) or in liquid LB cultures beyond an OD578 of 3.5—suggesting that it is the dominant c-di-GMP phosphodiesterase (Fig. 3B) under these conditions. This is in contrast to Salmonella, where a yhjH mutation has a minor effect on motility only (Frye et al. 2006; Ryjenkov et al. 2006; Simm et al. 2007). Reduced motility in the E. coli yhjH mutant depends on the presence of several GGDEF proteins, which act in an additive manner, with their relative contributions depending on the actual environmental conditions: YegE is most effective at 28°C, YeaJ is the major player at 37°C (where curli are not expressed), and YedQ plays a minor role at both temperatures (Fig. 3; Supplemental S4). YegE contains a GGDEF domain as well as a degenerate EAL domain; YeaJ and YedQ are GGDEF-only proteins. All three proteins contain intact GGDEF motifs, representing the enzymatically active site (A-site) of diguanylate cyclases (Chan et al. 2004), as well as an intact I-site, which is an allosteric c-di-GMP-binding site imposing product inhibition on these enzymes (Chan et al. 2004; Christen et al. 2006). Taken together, this indicates that YegE, YeaJ, and YedQ are the di-guanylate cyclases that contribute to variable extents to a common pool of c-di-GMP, which can be degraded by YhjH.
By binding to the effector protein YcgR, this c-di-GMP reduces motility (Ryjenkov et al. 2006; Christen et al. 2007). Mutations in all these factors do not affect the expression of the flagellar genes (Fig. 4D), which means that the effector protein YcgR probably interacts with a component of the flagellar basal body and thereby interferes with motor function directly. A similar conclusion was drawn for the activity of DgrA, a c-di-GMP-binding protein and homolog of YcgR, which in Caulobacter crescentus inhibits motility of swarmer cells (Christen et al. 2007). Consistent with a direct effect on motor function, a yhjH mutant has an altered flagellar rotation pattern (Girgis et al. 2007). Taken together, inhibition of motility during entry into stationary phase occurs at all possible levels: Besides a shut-down of flagellar gene expression, which also depends on proteolytic elimination of FlhDC, activity of existing flagella is reduced by the c-di-GMP control module YegE + YedQ + YeaJ/YcgR.
In addition, the antagonists YegE and YhjH are key factors for inversely coordinating motility with curli expression. YegE is required for normal curli expression (Fig. 6A). Also, in Salmonella, YegE (STM2123) plays a positive role in curli formation, which, however, has not been clarified further (Kader et al. 2006). YhjH, on the other hand, is a clear inhibitor of curli expression (Fig. 6A). Interestingly, YegE/YhjH-controlled c-di-GMP affects motility via YcgR, but this effector is not involved in the control of curli expression (Fig. 6B). That c-di-GMP controlled by YegE/YhjH can act via different effector components suggests that this c-di-GMP is freely diffusible. The target of the YegE/YhjH system in curli control is the transcription of the csgD gene (Fig. 6; Supplemental Figs. S7, S8). Taken together, FlhDC-dependent expression of YhjH in combination with σS-dependent induction of its main antagonist YegE generates a switch mechanism based on titration of antagonistic enzymes, which functions as a checkpoint system that “unlocks” csgD and therefore curli expression only after the shut-down of flagellar gene expression during entry into stationary phase has occurred.
At the same time, another DGC/PDE pair, YdaM/YciR, acts strongly and highly specifically on csgD transcription only (Fig. 6C; Weber et al. 2006), but does not affect motility (Fig. 3; Supplemental Fig. S5). At the moment, it is an open question how the two c-di-GMP control systems coordinate their activities at the csgD promoter, but our data suggest that the YdaM/YciR system does not contribute to the overall cellular c-di-GMP level (as YegE/YhjH does), but may act in a more local manner onto the csgD promoter region only. Sequestration of various DGC/PDE systems has previously been suggested based on the existance of sometimes several dozens of GGDEF/EAL proteins in single species (E. coli has 29 such proteins) (Jenal and Malone 2006; Kader et al. 2006; Ryan et al. 2006; Weber et al. 2006) and is certainly an issue that has to be studied more closely in the future.
Conclusions and perspectives
In the present study, we characterized the complex switch mechanism that controls the transition from motility-based foraging to the general stress response and curli-based adhesion that operates in E. coli during entry into stationary phase. Overall, this mechanism relies on mutual interference of two transcription factor cascades, which integrates transcription, proteolysis, and enzymatic activities. The switch operates on two clearly distinct hierarchical levels (Fig. 7). (1) At the top level, the activities of the unstable master regulators FlhDC and σS (and therefore the entire motility and general stress response systems) are inversely controlled by σ factor competition for core RNAP (σ70, σF, and σS) and by the FlhDC-dependent FliZ inhibiting σS activity. (2) At a lower level, the second messenger c-di-GMP, which specifically reduces flagellar activity and stimulates the expression of the adhesive curli fimbriae activator CsgD, is balanced by FlhDC-controlled degradation (via YhjH) and σS-controlled production (via YegE, YedQ).
In this system, shutting down flagellar gene expression is not just a strategy to avoid wasting energy for making new flagella that are no longer needed in cells that hardly grow any longer. Rather, it is part of an exquisitely balanced switching mechanism in which precisely timed inactivation of distinct genes under flagellar control (i.e., fliZ and yhjH), which also requires proteolytic removal of existing FlhDC, is a strict prerequisite for turning on the general stress response including curli formation and adhesion.
An interesting question for further studies is the nature of the trigger that actually throws the FlhDC/motility to σS/adhesion switch. Is it just σS accumulation as cells experience increasing nutrient limitation? If so, ppGpp is likely to be an essential trigger (Lange et al. 1995; Jishage et al. 2002). Also, how is the motility/adhesion switch integrated with other regulatory circuits that influence various biofilm functions; e.g., the Rcs or Csr regulatory systems?
Finally, this switch has to be rapidly reversible—at the top level, ClpXP-dependent proteolysis of σS may be essential for reversibility and, in fact, rapid degradation of σS is completely re-established within a couple of minutes upon refeeding of a stationary phase culture (Pruteanu and Hengge-Aronis 2002). At the lower c-di-GMP-controlled level, reversibility would be achieved by YhjH not being degraded, but rather outcompeted by various DGCs when cells enter into stationary phase. Whether the DGC/PDE-dependent c-di-GMP balance switches toward motility or toward curli expression (and other CsgD-dependent biofilm-associated functions) should depend not only on relative expression of the DGCs and PDEs involved, but also on the activities of their not yet characterized sensory input domains.
Materials and methods
Bacterial strains and growth conditions
All strains used in this study are derivatives of the E. coli K-12 strains W3110 (Hayashi et al. 2006) or MC4100 (Casadaban 1976). All strains carry a Δlac deletion. The construction of mutant alleles, plasmids, and single-copy lacZ reporter fusions are described in detail in the Supplemental Material (including relevant references).
Cells were grown in LB medium (Miller 1972) at 28°C under aeration if not otherwise indicated (as this is the appropriate temperature for curli expression). Antibiotics and IPTG were added as detailed in the Supplemental Material. Growth was monitored by measuring the optical density at 578 nm (OD578).
SDS page and immunoblot analysis
Sample preparation for SDS-PAGE and immunoblot analysis were performed as described previously (Lange and Hengge-Aronis 1994). Five micrograms or 10 μg of cellular protein were applied per lane. Polyclonal sera against σS, CsgD, and YhjH (custom-made by Pineda-Antikörper-Service), a goat anti-rabbit IgG alkaline phosphatase conjugate (Sigma), and a chromogenic substrate (BCIP/NBT; Boehringer Mannheim) were used.
Northern blot analysis
For RNA preparation and Northern blot analysis, cells were grown in LB medium at 28°C and harvested at an OD578 of 4.0. The procedure and materials used were exactly as described before (Weber et al. 2006).
c-di-GMP phosphodiesterase activity
Phosphodiesterase activity of purified (see above) C-terminally His6-tagged YhjH and YhjHE48A (0.5 μM) was determined using radiolabeled c-di-GMP prepared with the purified di-guanylate cyclase PleD* exactly as described (Weber et al. 2006).
Determination of β-galactosidase activity
β-galactosidase activity was assayed by use of o-nitrophenyl-β-D-galactopyranoside (ONPG) as a substrate and is reported as micromoles of o-nitrophenol per minute per milligram of cellular protein (Miller 1972). Experiments showing the expression of lacZ fusions along the entire growth cycle were done at least twice, and a representative experiment is shown.
Bacterial motility assay
Motility was tested on swim plates containing 0.5% bacto-tryptone, 0.5% NaCl, and 0.3% agar. Three microliters of overnight cultures (adjusted to an OD578 of 4.0 in their own supernatant) were inocculated into the swim plates, and cells were allowed to grow and swim for 3–5 h at the temperature indicated.
Acknowledgments
Financial support was provided by the Deutsche Forschungsgemeinschaft (He 1556/13-1), the Dr. Hans-Messner-Stiftung, and the Fonds der Chemischen Industrie.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.475808.
References
- Adler J., Templeton B. The effect of environmental conditions on the motility of Escherichia coli. J. Gen. Microbiol. 1967;46:175–184. doi: 10.1099/00221287-46-2-175. [DOI] [PubMed] [Google Scholar]
- Aldrigde P.D., Karlinsey J.E., Aldridge C., Birchall C., Thompson D., Yagasaki J., Hughes K.T. The flagellar-specific transcription factor, σ28, is the type III secretion chaperone for the flagellar-specific anti-σ28 factor FlgM. Genes & Dev. 2006;20:2315–2326. doi: 10.1101/gad.380406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amsler C.D., Cho M., Matsumura P. Multiple factors underlying the maximum motility of Escherichia coli as cultures enter post-exponential growth. J. Bacteriol. 1993;175:6238–6244. doi: 10.1128/jb.175.19.6238-6244.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barembruch C., Hengge R. Cellular levels and activity of the flagellar σ factor FliA of Escherichia coli are controlled by FlgM-modulated proteolysis. Mol. Microbiol. 2007;65:76–89. doi: 10.1111/j.1365-2958.2007.05770.x. [DOI] [PubMed] [Google Scholar]
- Barker C.S., Prüss B.M., Matsumura P. Increased motility of Escherichia coli by insertion sequence element integration into the regulatory region of the flhD operon. J. Bacteriol. 2004;186:7529–7537. doi: 10.1128/JB.186.22.7529-7537.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnhart M.M., Chapman M.R. Curli biogenesis and function. Annu. Rev. Microbiol. 2006;60:131–147. doi: 10.1146/annurev.micro.60.080805.142106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benach J., Swaminathan S.S., Tamayo R., Handelman S.K., Folta-Stogniew E., Ramos J.E., Forouhar F., Neely H., Seetharaman J., Camilli A., et al. The structural basis of cyclic diguanylate signal transduction by PilZ domains. EMBO J. 2007;26:5153–5166. doi: 10.1038/sj.emboj.7601918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bokranz W., Wang X., Tschape H., Römling U. Expression of cellulose and curli fimbriae by Escherichia coli isolated from the gastrointestinal tract. J. Med. Microbiol. 2005;54:1171–1182. doi: 10.1099/jmm.0.46064-0. [DOI] [PubMed] [Google Scholar]
- Bougdour A., Lelong C., Geiselmann J. Crl, a low temperature-induced protein in Escherichia coli that binds directly to the stationary phase σ subunit of RNA polymerase. J. Biol. Chem. 2004;279:19540–19550. doi: 10.1074/jbc.M314145200. [DOI] [PubMed] [Google Scholar]
- Brombacher E., Baratto A., Dorel C., Landini P. Gene expression regulation by the curli activtor CsgD protein: Modulation of cellulose biosynthesis and control of negative determinants for microbial adhesion. J. Bacteriol. 2006;188:2027–2037. doi: 10.1128/JB.188.6.2027-2037.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown P.K., Dozois C.M., Nickerson C.A., Zuppardo A., Terlonge J., Curtiss R. MlrA, a novel regulator of curli (Agf) and extracellular matrix synthesis by Escherichia coli and Salmonella enterica serovar typhimurium. Mol. Microbiol. 2001;41:349–363. doi: 10.1046/j.1365-2958.2001.02529.x. [DOI] [PubMed] [Google Scholar]
- Casadaban M.J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage λ and μ. J. Mol. Biol. 1976;104:541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Chan C., Paul R., Samoray D., Amiot N., Giese B., Jenal U., Schirmer T. Structural basis of activity and allosteric control of diguanylate cyclase. Proc. Natl. Acad. Sci. 2004;101:17084–17089. doi: 10.1073/pnas.0406134101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chilcott G.S., Hughes K.T. Coupling of flagellar gene expression to flagellar assembly in Salmonella enterica serovar typhimurium and Escherichia coli. Microbiol. Mol. Biol. Rev. 2000;64:694–708. doi: 10.1128/mmbr.64.4.694-708.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christen B., Christen M., Paul R., Schmid F., Folcher M., Jenoe P., Meuwly M., Jenal U. Allosteric control of cyclic di-GMP signaling. J. Biol. Chem. 2006;281:32015–32024. doi: 10.1074/jbc.M603589200. [DOI] [PubMed] [Google Scholar]
- Christen M., Christen B., Allan M.G., Folcher M., Jenö P., Grzesiek S., Jenal U. DgrA is a member of a new family of cyclic diguanosine monophosphate receptors and controls flagellar motor function in Caulobacter crescentus. Proc. Natl. Acad. Sci. 2007;104:4112–4117. doi: 10.1073/pnas.0607738104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frye J., Karlinsey J.E., Felise H.R., Marzolf B., Dowidar N., McClelland M., Hughes K.T. Identification of new flagellar genes of Salmonella enterica serovar typhimurium. J. Bacteriol. 2006;188:2233–2243. doi: 10.1128/JB.188.6.2233-2243.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel U., Park C., Römling U. Complex regulation of csgD promoter activity by global regulatory proteins. Mol. Microbiol. 2003;49:639–654. doi: 10.1046/j.1365-2958.2003.03594.x. [DOI] [PubMed] [Google Scholar]
- Girgis H.S., Liu Y., Ryu W.S., Tavazoie S. A comprehensive genetic characterization of bacterial motility. PLoS Genet. 2007;3:e154. doi: 10.1371/journal.pgen.0030154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall-Stoodley L.K., Costerton J.W., Stoodley P. Bacterial biofilms: From the natural environment to infectious diseases. Nat. Rev. Microbiol. 2004;2:95–108. doi: 10.1038/nrmicro821. [DOI] [PubMed] [Google Scholar]
- Hayashi K., Morooka N., Yamamoto Y., Fujita K., Isono K., Choi S., Ohtsubo E., Baba T., Wanner B.L., Mori H., Horiuchi T.2006Highly accurate genome sequences of Escherichia coli K-12 strains MG1655 and W3110 Mol. Syst. Biol. 2 :2006.0007. 10.1038/msb4100049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hengge-Aronis R. The general stress response in Escherichia coli. In: Storz G., Hengge-Aronis R., editors. Bacterial stress responses. ASM Press; Washington, DC: 2000. pp. 161–178. [Google Scholar]
- Hengge-Aronis R. Signal transduction and regulatory mechanisms involved in control of the σS subunit of RNA polymerase in Escherichia coli. Microbiol. Mol. Biol. Rev. 2002;66:373–395. doi: 10.1128/MMBR.66.3.373-395.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenal U., Hengge-Aronis R. Regulation by proteolysis in bacterial cells. Curr. Opin. Microbiol. 2003;6:163–172. doi: 10.1016/s1369-5274(03)00029-8. [DOI] [PubMed] [Google Scholar]
- Jenal U., Malone J. Mechanisms of cyclic-di-GMP signaling in bacteria. Annu. Rev. Genet. 2006;40:385–407. doi: 10.1146/annurev.genet.40.110405.090423. [DOI] [PubMed] [Google Scholar]
- Jishage M.L., Ishihama A. Variation in RNA polymerase σ subunit composition within different stocks of Escherichia coli W3110. J. Bacteriol. 1997;179:959–963. doi: 10.1128/jb.179.3.959-963.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jishage M., Kvint K., Shingler V., Nyström T. Regulation of σ factor competition by the alarmone ppGpp. Genes & Dev. 2002;16:1260–1270. doi: 10.1101/gad.227902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kader A., Simm R., Gerstel U., Morr M., Römling U. Hierarchical involvement of various GGDEF domain proteins in rdar morphotype development of Salmonella enterica serovar typhimurium. Mol. Microbiol. 2006;60:602–616. doi: 10.1111/j.1365-2958.2006.05123.x. [DOI] [PubMed] [Google Scholar]
- Karlinsey J.E., Tanaka S., Bettenworth B., Yamaguchi S., Boos W., Aizawa S.-I., Hughes K.T. Completion of the hook-basal body complex of the Salmonella typhimurium flagellum is coupled to FlgM secretion and fliC transcription. Mol. Microbiol. 2000;37:1220–1231. doi: 10.1046/j.1365-2958.2000.02081.x. [DOI] [PubMed] [Google Scholar]
- Klausen M., Gjermansen M., Kreft J.U., Tolker-Nielsen T. Dynamics of development and dispersal in sessile microbial communities: Examples form Pseudomonas aeruginosa and Pseudomonas putida model biofilms. FEMS Microbiol. Lett. 2006;261:1–11. doi: 10.1111/j.1574-6968.2006.00280.x. [DOI] [PubMed] [Google Scholar]
- Ko M., Park C. Two novel flagellar components and H-NS are involved in the motor function of Escherichia coli. J. Mol. Biol. 2000;303:371–382. doi: 10.1006/jmbi.2000.4147. [DOI] [PubMed] [Google Scholar]
- Kutsukake K., Ikebe T., Yamamoto S. Two novel regulatory genes, fliT and fliZ, in the flagellar regulon of Salmonella. Genes Genet. Syst. 1999;74:287–292. doi: 10.1266/ggs.74.287. [DOI] [PubMed] [Google Scholar]
- Lange R., Hengge-Aronis R. The cellular concentration of the σS subunit of RNA-polymerase in Escherichia coli is controlled at the levels of transcription, translation and protein stability. Genes & Dev. 1994;8:1600–1612. doi: 10.1101/gad.8.13.1600. [DOI] [PubMed] [Google Scholar]
- Lange R., Fischer D., Hengge-Aronis R. Identification of transcriptional start sites and the role of ppGpp in the expression of rpoS, the structural gene for the σS subunit of RNA-polymerase in Escherichia coli. J. Bacteriol. 1995;177:4676–4680. doi: 10.1128/jb.177.16.4676-4680.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Matsumura P. The FlhD/FlhC complex, a transcriptional activator of the Escherichia coli flagellar class II operons. J. Bacteriol. 1994;176:7345–7351. doi: 10.1128/jb.176.23.7345-7351.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller J.H. Experiments in molecular genetics. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1972. [Google Scholar]
- Olsén A., Jonsson A., Normark S. Fibronectin binding mediated by a novel class of surface organelles on Escherichia coli. Nature. 1989;338:652–655. doi: 10.1038/338652a0. [DOI] [PubMed] [Google Scholar]
- O’Toole G.A., Kaplan H.B., Kolter R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000;54:49–79. doi: 10.1146/annurev.micro.54.1.49. [DOI] [PubMed] [Google Scholar]
- Prüß B.M., Besemann C., Denton A., Wolfe A.J. A complex transcription network controls the early stages of biofilm development in Escherichia coli. J. Bacteriol. 2006;188:3731–3739. doi: 10.1128/JB.01780-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruteanu M., Hengge-Aronis R. The cellular level of the recognition factor RssB is rate-limiting for σS proteolysis: Implications for RssB regulation and signal transduction in σS turnover in Escherichia coli. Mol. Microbiol. 2002;45:1701–1714. doi: 10.1046/j.1365-2958.2002.03123.x. [DOI] [PubMed] [Google Scholar]
- Römling U., Rohde M., Olsén A., Normark S., Reinköster J. AgfD, the checkpoint of multicellular and aggregative behaviour in Salmonella typhimurium regulates at least two independent pathways. Mol. Microbiol. 2000;36:10–23. doi: 10.1046/j.1365-2958.2000.01822.x. [DOI] [PubMed] [Google Scholar]
- Römling U., Gomelsky M., Galperin M.Y. C-di-GMP: The dawning of a novel bacterial signalling system. Mol. Microbiol. 2005;57:629–639. doi: 10.1111/j.1365-2958.2005.04697.x. [DOI] [PubMed] [Google Scholar]
- Ryan R.P., Fouhy Y., Lucey J.F., Dow J.M. Cyclic di-GMP signaling in bacteria: Recent advances and new puzzles. J. Bacteriol. 2006;188:8327–8334. doi: 10.1128/JB.01079-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rychlik I., Martin G., Methner U., Lovell M., Cardova L., Sebkova A., Sevcik M., Damborsky J., Barrow P.A. Identification of Salmonella enterica serovar Typhimurium genes associated with growth suppression in stationary-phase nutrient broth cultures and in the chicken intestine. Arch. Microbiol. 2002;178:411–420. doi: 10.1007/s00203-002-0458-7. [DOI] [PubMed] [Google Scholar]
- Ryjenkov D.A., Simm R., Römling U., Gomelsky M. The PilZ domain is a receptor for the second messenger c-di-GMP: The PilZ protein YcgR controls motility in enterobacteria. J. Biol. Chem. 2006;281:30310–30314. doi: 10.1074/jbc.C600179200. [DOI] [PubMed] [Google Scholar]
- Simm R., Lusch A., Kader A., Andersson M., Römling U. Role of EAL-containing proteins in multicellular behavior of Salmonella enterica serovar typhimurium. J. Bacteriol. 2007;189:3613–3623. doi: 10.1128/JB.01719-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanya H.S., Arciszweska L.K., Baker R.A., Bird L.E., Sherratt D.J., Wiglex D.B. Crystal structure of the site-specific recombinase, XerD. EMBO J. 1997;16:5178–5187. doi: 10.1093/emboj/16.17.5178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamayo R., Pratt J.T., Camilli A. Roles of cyclic diguanylate in the regulation of bacterial pathogenesis. Annu. Rev. Microbiol. 2007;61:131–148. doi: 10.1146/annurev.micro.61.080706.093426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomoyasu T., Takaya A., Isogai E., Yamamoto T. Turnover of FlhD and FlhC, master regulator proteins for Salmonella flagellum biogenesis, by the ATP-dependent ClpXP protease. Mol. Microbiol. 2003;48:443–452. doi: 10.1046/j.1365-2958.2003.03437.x. [DOI] [PubMed] [Google Scholar]
- Typas A., Barembruch C., Hengge R. Stationary phase reorganisation of the E. coli transcription machinery by Crl protein, a fine-tuner of σS activity and levels. EMBO J. 2007a;26:1569–1578. doi: 10.1038/sj.emboj.7601629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Typas A., Becker G., Hengge R. The molecular basis of selective promoter activation by the σS subunit of RNA polymerase. Mol. Microbiol. 2007b;63:1296–1306. doi: 10.1111/j.1365-2958.2007.05601.x. [DOI] [PubMed] [Google Scholar]
- Wang S., Fleming R.T., Westbrook E.M., Matsumura P., McKay D.B. Structure of the Escherichia coli FlhDC complex, a prokaryotic heteromeric regulator of transcription. J. Mol. Biol. 2006;355:798–808. doi: 10.1016/j.jmb.2005.11.020. [DOI] [PubMed] [Google Scholar]
- Weber H., Polen T., Heuveling J., Wendisch V., Hengge R. Genome-wide analysis of the general stress response network in Escherichia coli: σS-dependent genes, promoters and σ factor selectivity. J. Bacteriol. 2005;187:1591–1603. doi: 10.1128/JB.187.5.1591-1603.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber H., Pesavento C., Possling A., Tischendorf G., Hengge R. Cyclic-di-GMP-mediated signaling within the σS network of Escherichia coli. Mol. Microbiol. 2006;62:1014–1034. doi: 10.1111/j.1365-2958.2006.05440.x. [DOI] [PubMed] [Google Scholar]
- Zhao K., Liu M., Burgess R.R. Adaptation in bacterial flagellar and motility systems: From regulon members to “foraging”-like behavior in E. coli. Nucleic Acids Res. 2007;35:4441–4452. doi: 10.1093/nar/gkm456. [DOI] [PMC free article] [PubMed] [Google Scholar]