Abstract
A body of evidence supports the idea that newly replicated Escherichia coli chromosomes segregate progressively as replication progresses, with spatial separation of sister genetic loci occurring ∼15 min after their replication. We show that the time of this cohesion can be modulated by topoisomerase IV (TopoIV) activity. Impairment of TopoIV prevents segregation of newly replicated sister loci and bulk chromosome segregation, whereas modest increases in TopoIV decrease the cohesion time substantially. Therefore, we propose that precatenanes, which form as replication progresses by interwinding of newly replicated sister chromosomes, are responsible for E. coli sister chromosome cohesion.
Keywords: Sister chromosome cohesion, Escherichia coli, precatenanes, topoisomerase IV
The heritable maintenance of the genetic material over generations requires not only that it be accurately replicated but that newly replicated chromosomes are faithfully transmitted to daughter cells at cell division. Defects in chromosome segregation are associated with genetic disease and cancer. In eukaryotes, newly replicated sister chromosomes remain associated and aligned until the onset of mitosis by dedicated cohesion mechanisms, using SMC (structural maintenance of chromosomes) proteins. Formation and dissolution of cohesion is under strict spatial and temporal control (for review, see Nasmyth and Haering 2005). Similarly, in the archaeon, Sulfolobus sulfataricus, sister cohesion seems to extend from the replicative to the post-replicative phase of the cell cycle (Robinson et al. 2007). In contrast, a growing body of evidence supports the view that newly replicated loci in bacteria segregate progressively as replication proceeds, with a period of sister cohesion (the time between locus replication and separation of the two sister loci) that is substantially less than S phase (Viollier et al. 2004; Neilsen et al. 2006; Reyes-Lamothe et al. 2008). Nevertheless, some work has led to the conclusion that extensive sister cohesion may hold sisters together for a large fraction of S phase (Sunako et al. 2001; Bates and Kleckner 2005).
Topological entanglement of newly replicated sisters has been implicated in facilitating sister chromosome cohesion in bacteria, archaea, and eukaryotes, with precatenanes (interwound sister DNA duplexes present in a replicating chromosome), catenanes (interwound replicated sister duplexes), or hemicatenanes (duplexes interlinked by single-strand interwindings) mediating such entanglement (Lopes et al. 2003; Espeli and Marians 2004; Nasmyth and Haering 2005; Robinson et al. 2007).
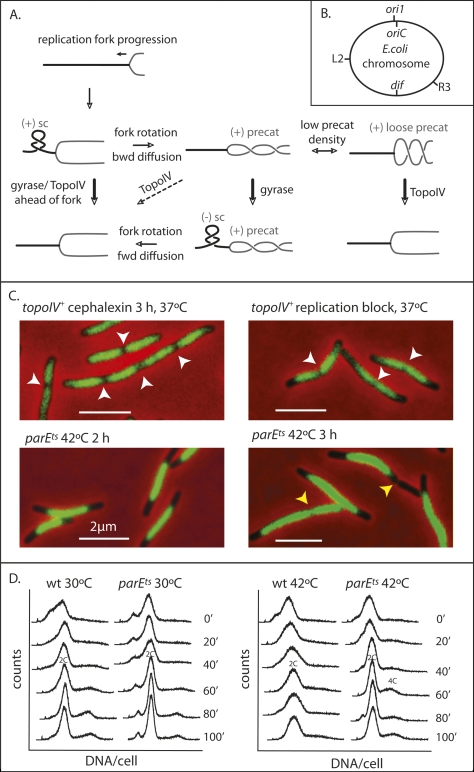
The right-hand (RH) interwinding of the two strands of a DNA duplex (linkage) has to be completely removed in order that newly replicated chromosomes can be segregated to daughter cells. Complete segregation of newly replicated Escherichia coli chromosomes requires the removal of ∼4.2 × 105 links in one generation time or less. RH precatenanes will arise when RH links in duplex DNA ahead of a progressing replication fork diffuse backward to behind the fork, thus interlinking the two newly replicated sisters by a redistribution of total linkage (Fig. 1A; Champoux and Been 1980; Espeli and Marians 2004). In the absence of topoisomerase action, replication progression leads to an increased density of RH links ahead of a replication fork. This takes the form of overtwisted RH duplex, which can adopt (+) supercoils within a left-handed (LH) plectonemic superhelix. The topological torsion arising from this overtwisting can be released by rotation of the fork and replisome, thereby generating RH precatenanes. In principle, precatenane formation should not interfere with replication progression. As a corollary to this, if the DNA ahead of the fork is undertwisted and contains (−) supercoils, precatenanes are expected to be removed by forward diffusion of any duplex interwindings.
Figure 1.
TopoIV impairment prevents sister nucleoid separation. (A) Schematic of topological processing during DNA replication. As a replication fork moves forward, the DNA ahead of the fork becomes overwound and the density of (+) RH linkage increases, forming (+) supercoils (sc) with a LH plectonemic superhelix ahead of the fork. The torsional stress so generated is relieved if rotation of the fork replisome occurs, thereby redistributing the RH linkage. This leads to formation of (+) RH precatenanes behind the fork. These are only good substrates for TopoIV if their density is low (“loose” precat), so their crossing angle can become optimal for TopoIV action (Stone et al. 2003). If precatenane-containing DNA becomes underwound ahead of the fork, generating (−) sc, then diffusion forward of RH precatenanes to remove (−) sc will be favored. It has also been proposed that accumulation of (−) sc ahead of a fork can lead to LH (−) precatenanes in newly replicated DNA, with these also being an in vivo substrate for TopoIV (see Schvartzman and Stasiak 2004). We believe this is unlikely to be a common occurrence, because it would exacerbate chromosome unlinking and be energetically unfavorable because ATP hydrolysis by gyrase ahead of a fork, generating (−) supercoiling, would need to be countered by ATP hydrolysis by TopoIV to remove LH precatenanes. Unreplicated DNA is shown as a thick black line and newly replicated DNA is shown as a gray line. Arrows indicate the direction of reaction. Thick arrows indicate efficient reactions and dashed arrows inefficient reactions. (B) Schematic of the E. coli genetic loci used. (C) Nucleoids in the indicated cells. Replication was blocked by TetR binding to tetO arrays at R3, ∼700 kb anti-clockwise of dif. White arrows indicate positions of nucleoid splitting and yellow arrows show positions of cytokinesis in a bright-field image. Nucleoids were stained with DAPI (0.5 μg/mL). Bars, 2 μm. (D) Flow cytometry (DNA content per cell) of wild-type and parEts cells grown at 30°C and 42°C. Rifampicin, which prevents replication initiation, and cephalexin, which blocks cytokinesis, were added at 0 min, and samples were taken at the indicated times.
Precatenanes, catenanes, and knots are unlinked by type II topoisomerases. Bacterial topoisomerase IV (TopoIV) seems especially adapted for this role, while the other type II topoisomerase, DNA gyrase, is adapted to act on a single duplex to remove (+) supercoiling and to add (−) supercoiling (for review, see Espeli and Marians 2004; Schvartzman and Stasiak 2004). The type I topoisomerase, topoisomerase III (TopoIII), uses single-strand DNA passage reactions to decatenate and remove other DNA entanglements, like hemicatenanes and Holliday junctions (for review, see Wu and Hickson 2006).
Here we report experiments that lead to the conclusion that precatenanes are a major contributor to sister chromosome cohesion in E. coli and that TopoIV acts during the cell cycle to remove precatenanes as they form during replication progression.
Results
Inhibition of TopoIV prevents E. coli nucleoid splitting and genetic locus segregation
Although TopoIV is the major decatenating enzyme in E. coli (Zechiedrich and Cozzarelli 1995; Zechiedrich et al. 1997), it has not been clear whether it acts primarily during replication fork progression, removing any precatenanes that form during replication and possibly removing (+) supercoiling ahead of the replication fork (Khodursky et al. 2000) or whether TopoIV action is targeted primarily to the replication terminus region (ter) to remove precatenanes and catenanes at the completion of replication (Espeli et al. 2003a, b; for review, see Espeli and Marians 2004).
The cytological and flow cytometry observations that TopoIVts strains do not segregate their sister chromosomes normally at restrictive temperature, although they appear to complete replication (Grainge et al. 2007), hinted that TopoIV acts throughout the whole replication cycle to remove precatenanes formed as replication progresses.
In order to explore further the consequences of impairing TopoIV activity, nucleoids derived from TopoIVts cells and TopoIV+ replication-blocked cells were compared with TopoIV+ cells after cephalexin treatment, which permits clear display of segregated sister nucleoids (Wang et al. 2006). All experiments reported here use growth in glycerol-minimal medium, giving a generation time of ∼100 min at 37°C, with most cells initiating DNA replication soon after birth and completing replication 50–65 min later (Wang et al. 2005, 2006; Reyes-Lamothe et al. 2008).
When replication was blocked at locus R3, located 700 kb anti-clockwise of the dif site (Fig. 1B), by TetR-Cfp binding to tetO operator arrays (Possoz et al. 2006), the nucleoid is clearly split into two partially replicated and segregated sister chromosomes, just as in unblocked TopoIV+ conditions (Fig. 1C), an observation consistent with previous demonstrations that nucleoid splitting is initiated midway through S phase (Bates and Kleckner 2005; Wang et al. 2005). However, when parEts cells, temperature-sensitive for the TopoIV ATPase subunit, were grown for 2 h or 3 h at 42°C, the nucleoids appeared smooth, with no sign of splitting (Fig. 1C; Grainge et al. 2007). After 3 h at 42°C, bright-field images showed large unsegregated nucleoids located in the middle of the cell, thereby preventing normal cytokinesis. In a minority of cells, cytokinesis occurred either at DNA-free regions, generating anucleate cells, or over the nucleoid to “cut” the chromosome. These observations suggest strongly that the action of TopoIV is needed for sister nucleoid separation throughout replication, rather than solely at replication termination. Therefore, we infer that precatenanes arise as replication proceeds and are the major substrate for TopoIV action on replicating chromosomes.
It has been reported that TopoIV can play an important role in removing (+) supercoils ahead of a replication fork (Khodursky et al. 2000), although other studies have inferred that gyrase is the primary topoisomerase acting there (for review, see Espeli and Marians 2004). To help resolve this uncertainty, we used flow cytometry to assess the time it takes to complete replication by performing a time-course “run-out” experiment in which replication can be completed but not reinitiated in the absence of cell division (Fig. 1D). The replication progression profiles over time for TopoIV+ and parEts strains grown in glycerol minimal medium at 30°C and 42°C were similar, and both strains showed no changes in profile after 60–80 min at 42°C. We conclude that TopoIV inactivation has little influence on replication fork progression, a result confirmed and extended later, when the time from replisome appearance at initiation to disappearance at termination is monitored in time-lapse experiments. We also used flow cytometry to show that parEts strains can reinitiate replication at 42°C (data not shown).
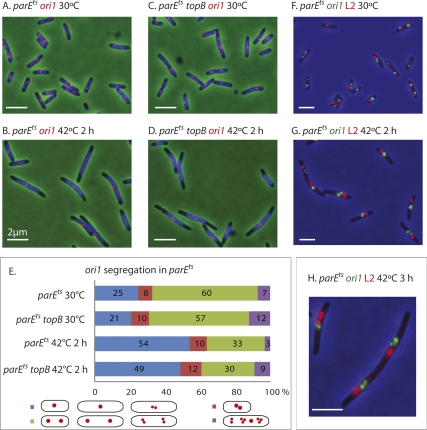
To examine the consequences of TopoIV impairment on segregation of genetic loci, initial experiments examined the segregation of ori1, a locus 15 kb anti-clockwise of oriC (Fig. 1B) (Wang et al. 2006) in a parEts strain at permissive and restrictive temperatures. When cells growing at 30°C were shifted for 2 h to 42°C, the proportion of cells containing a single ori1 focus increased from 25% to 54% (parEts) and from 21% to 49% (parEts topB) (Fig. 2A–E). These initial values for the proportion of cells containing a single ori1 focus are similar to the values reported previously for cells growing exponentially at 37°C (Wang et al. 2005). Since the time at 42°C corresponds to little more than a generation time, this result shows that ori1 segregation is severely impaired when TopoIV activity is compromised, with TopoIII, encoded by the topB, having no observable effect. Sister ori1 segregation occurs normally in TopoIV+ cells grown at 42°C (data not shown).
Figure 2.
Sister ori1 and L2 genetic loci fail to segregate normally when TopoIV is impaired. (A–D) Snapshot images of cells, with ori1 labeled (red). (Blue) DAPI nucleoid staining. (E) Quantitation of the data from the experiments shown in A–D. Cells were classified into the four categories shown in the schematic. (Blue) Cells with one ori1 focus, and longer cells (>4 μm) with two closely spaced ori1 foci (d < 1/5 cell length). (Red) Shorter cells (<4 μm) with two closely spaced foci. (Green) Cells with well-separated foci (d ≥ 1/5 cell length) or two clusters of separated foci. (Purple) Cells with more than two foci or clusters of foci. The cells in the red group were distinguished from those in the blue group because we cannot be sure whether those in the red group have foci that are unable to separate normally or are on the way to normal separation. (F–H) Simultaneous labeling of ori1 (green) and L2 (red) genetic loci. Bars, 2 μm.
The segregation of ori1 and locus L2, located 1 Mb anti-clockwise of oriC (Fig. 1B), were examined simultaneously (Fig. 2F,G). The segregation of L2, like ori1, was inhibited at 42°C. Even when some separation of loci was observed, sister ori1 or L2 loci remained close together and separate from those of the other locus; reinitiation of replication in parEts cells at 42°C is evident from the clusters of more than two sister loci (Fig. 2H). In time-lapse studies of the parEts strain at 42°C we observed that closely spaced sister ori1 foci transiently fuse and reseparate, consistent with them being interwound by precatenanes (see below; data not shown). We believe that proper segregation of other sister loci will be similarly inhibited when TopoIV is impaired. Similar results were obtained with a parCts mutant (data not shown).
Increased TopoIV levels reduce sister locus cohesion time
The demonstration that TopoIV impairment leads to a failure of normal chromosome segregation, despite allowing normal replication progression, supports the view that precatenanes can form as replication proceeds and that failure to remove these with TopoIV prevents normal locus segregation. Nevertheless, these observations do not address whether precatenation is the major or sole source of sister cohesion or whether some other cohesion mechanism holds sisters together for the ∼15-min period of normal cohesion, with precatenanes persisting only long enough to influence cohesion when TopoIV activity is impaired.
Therefore, to test whether precatenanes are the only or major source of cohesion, we set out to determine whether TopoIV overexpression reduces cohesion time. In initial experiments we noted that even modestly increased levels of TopoIV are toxic to cells, with both nucleoid segregation and ori1 focus segregation impaired (data not shown). Nevertheless, the presence of plasmid pWX35, which expresses TopoIV from the arabinose promoter, was able to partially suppress the temperature-sensitive phenotype of parEts cells in medium lacking arabinose, conditions that give basal levels of protein expression (Guzman et al. 1995; X. Wang, R. Reyes-Lamothe, and D.J. Sherratt, unpubl.). Therefore, a basal level of plasmid-encoded TopoIV expression is sufficient for viability and chromosome segregation at 42°C in a parEts strain.
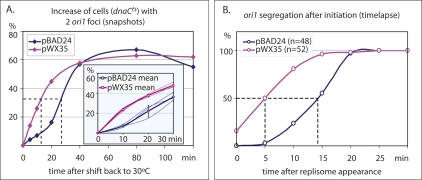
We therefore assessed cohesion time for ori1 in cells with the normal TopoIV complement (estimated to be ∼1000 molecules per cell; Espeli and Marians 2004), plus the basal complement from pWX35 (expected to be <1000 molecules per cell; Guzman et al. 1995). Sister ori1 cohesion time was assessed in two ways. First, initiation of DNA replication was synchronized using ori1-labeled dnaCts cells with pWX35 or the “empty” vector, pBAD24. Cells grew normally at 30°C, and, when shifted to 37°C, finished ongoing rounds of replication and cell division but could not reinitiate replication. When shifted back to 30°C, most cells reinitiate replication within 5 min (Maisnier-Patin et al. 2001; Reyes-Lamothe et al. 2008). Analysis of the increase in cells with two ori1 foci with respect to time showed an additional complement of TopoIV provided by basal expression from pWX35 decreased the time at 30°C needed to reach 50% of maximal increase in two ori1 foci from 28 min to 13 min (Fig. 3A).
Figure 3.
Increased TopoIV decreases sister cohesion time. (A) Snapshot analysis in dnaCts TopoIV+ cells containing either an “empty” plasmid (pBAD24) or a plasmid with basal low-level expression of TopoIV (pWX35). The main plot shows the increase in cells containing two ori1 foci in a single experiment carried out over 120 min at 30°C from replication initiation. The inset shows three experiments carried out for a shorter time, with the mean values shown as thick lines and standard errors as vertical bars. (B) Time-lapse analysis of ori1 segregation in DnaC+ cells from replisome appearance at initiation.
In a second set of experiments, 5-min time-lapse analysis was used to measure the time from replisome appearance at initiation to ori1 segregation in exponentially growing cells in minimal medium at 37°C (generation time ∼100 min), with pWX35 or the parental plasmid, pBAD24. Since replisome appearance marks initiation and ori1 is replicated within a minute of initiation, this gives a direct measure of cohesion time for ori1 in unperturbed cells (Reyes-Lamothe et al. 2008). Fifty percent of ori1 loci had segregated within 14 min of replisome appearance in wild-type cells. This value was reduced to 5 min in cells with the additional complement of TopoIV from pWX35. Taken together, these results provide compelling evidence that precatenanes in wild-type cells are a major contributor to sister cohesion and that this period of cohesion can be reduced by increasing TopoIV. Therefore, normal levels of TopoIV appear to limit the rate at which newly replicated sisters are decatenated.
TopoIV inhibition does not prevent normal outward migration of newly initiated replisomes
We show that sister replisomes originating from a single initiation event at oriC separate into separate cell halves ∼5 min after replication initiation and propose that this separation reflects the tracking of sister replisomes along the compacted DNA of the nucleoid (Reyes-Lamothe et al. 2008). A prediction of this model is that TopoIV impairment should not interfere with sister replisome separation. In contrast, if sister replisome separation is a consequence of chromosome segregation, then replisome separation should be blocked when TopoIV is impaired.
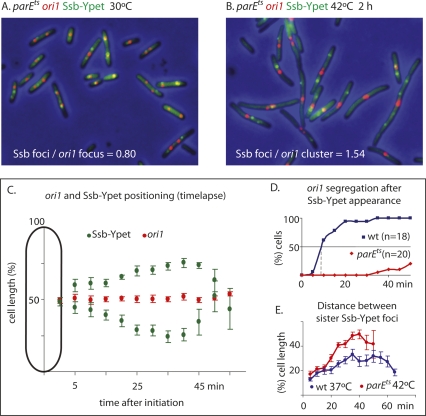
A functional bright fluorescent protein derivative of Ssb, Ssb-Ypet, expressed from the endogenous chromosomal ssb promoter, was used as a replisome marker in a parEts strain containing a marked ori1 locus. Analysis of elongating parEts cells at 42°C showed similar replication phase parameters and initiation times to those of wild-type cells (Fig. 4; data not shown). Examination of the snapshots showed that sister replisomes still migrate to separate cell halves soon after initiation, although sister ori1 foci did not segregate; this is reflected in an increase in the ratio of Ssb foci/ori1 foci after 2 h (Fig. 4A,B). Analysis by time lapse of 10 parEts cells, shifted to 42°C 35 min before commencement of observation, confirmed this behavior (Fig. 4C,D). An ori1 focus or two closely spaced foci remained close to midcell, whereas the sister replisome moved outward for much of S phase. The time from replisome appearance to disappearance fell in the range of 45–60 min (mean 52 min), a value similar to that obtained with wild-type cells examined in control time lapses (data not shown). This confirms that impairment of TopoIV does not interfere with replication progression. Significantly, the separation between the sister replisomes in TopoIV-impaired cells increases progressively up to the last stages of replication, as opposed to the situation in TopoIV+ cells, where the pattern is confounded by the recurrent movement of the two replisomes toward and then away from each other, presumably as a result of the reorganization of DNA to accommodate the growing nucleoids during chromosome segregation; this leads to a reduced outward movement (Fig. 4E; Reyes-Lamothe et al. 2008; data not shown). Therefore, these data consolidate the conclusion that sister replisomes track along DNA.
Figure 4.
Sister replisomes migrate to separate cell halves in the absence of locus segregation. (A,B) Snapshot analysis of ori1 and replisome (Ssb-Ypet) in parEts strain at 30°C and 42°C (2 h). (C) Time-lapse analysis (5 min). Ten parEts cells were switched to 42°C (35 min) and then followed at 42°C by time lapse, and the mean positions of the ori1 foci (either a single focus or transient pair of closely spaced foci) and Ssb-Ypet foci are shown (bars show standard errors). (D) ori1 separation in parEts at 42°C (data from C) compared with ori1 separation in TopoIV+ cells grown at 42°C (not shown). The indicated time intervals are the time from replisome appearance to the last point at which a replisome was present. (E) The separation between sister replication forks after initiation from the data presented in C is compared with a similar experiment using wild-type cells grown at 37°C.
Discussion
The results presented here provide compelling evidence that the normal period of E. coli sister chromosome cohesion (∼15 min after replication of a locus) is mediated by precatenation, presumably arising from the diffusion of RH crossings within a duplex ahead of a fork to interwindings between newly replicated duplex sisters behind the fork. Such backward diffusion requires rotation of the fork and its associated replisome, a process that would seem easier to accommodate when sister replisomes act independently rather than in a coordinated replication factory (Reyes-Lamothe et al. 2008).
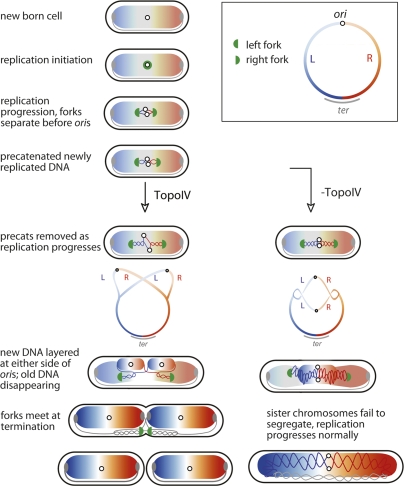
Taken together the data lend strong support to the schematic in Figure 1 and the model in Figure 5. The model proposes that after replication initiation at midcell, DNA gyrase begins to remove most of the linkage between parental duplex strands, by acting on (+) supercoils as they accumulate in front of the progressing sister replication forks; even before (+) supercoils accumulate, gyrase could act to maintain the normal level of (−) supercoiling. Gyrase normally acts throughout replication progression. Although TopoIV can also remove (+) supercoils, the results here show that replication fork progression is not significantly reduced in the absence of TopoIV. Sister replisomes move apart soon after initiation, and any rotation of the forks and their associated replisomes will generate precatenanes, which cohese the newly replicated sisters. TopoIV activity in the region of newly replicated DNA is such that precatenanes persist on average for ∼15 min, the observed period of cohesion between newly replicated sisters. Although TopoIV has evolved to act primarily on duplex interwindings present in catenanes and precatenanes, its action on RH catenanes is expected to be limited to those with a low density of crossings (Fig. 1A). Our observation that once TopoIVts cells have been incubated for 1–2 h at 42°C, normal chromosome or locus segregation does not resume within three to four generations of growth at 30°C (data not shown) could be because increased RH precatenane density prevents the adoption of a suitable crossing angle in the interlinks; thereby preventing effective TopoIV action (Stone et al. 2003). Alternatively, an accumulation of precatenanes might lead to structures that cannot be resolved by TopoIV.
Figure 5.
Model for cellular organization of replication and chromosome segregation (see the text for details).
Precatenanes can also be resolved independently of TopoIV if (−) supercoils accumulate in front of a replication fork; this will favor diffusion forward of the interlinks, thereby reducing the level (−) supercoils (Fig. 1A). Therefore, increased (−) supercoiling ahead of a fork will act to prevent RH precatenane formation and to facilitate removal of any precatenanes that arise (Fig. 1A). An accumulation of (−) supercoiling ahead of a fork, accompanied by fork-replisome rotation, could lead to formation of LH precatenanes, which would be removed efficiently by TopoIV (Schvartzman and Stasiak 2004). However, any loss of (−) supercoiling in unreplicated DNA will tend to hinder duplex unlinking and replication fork progression. Our experiments did not identify a role for TopoIII in bulk decatenation.
Once sister loci have become decatenated they segregate to opposite cell halves. This segregation is accompanied by nucleoid remodeling to generate the observed patterns of locus distribution. Toward the end of replication, sister replisomes converge in the midcell region, replisomes dissociate once replication is complete and final decatenation and chromosme dimer resolution is mediated by TopoIV and XerCD-dif dimer resolution (Grainge et al. 2007).
Since impairment of TopoIV activity leads to a dramatic inhibition of locus separation, even when the locus is close to oriC, precatenanes are likely to form as replication progresses and be resolved by TopoIV soon afterward; indeed, TopoIV activity could be associated with replication forks. This observation is not necessarily in contradiction to the proposal that temporal and spatial control of TopoIV activity leads to most of its activity being targeted to ter at the end of replication (Espeli et al. 2003a, b). This is because we also demonstrate that the normal levels of TopoIV activity appear to be limiting for decatenation of at least the ori1 region, despite the relative abundance of TopoIV.
A modest increase in TopoIV activity reduces cohesion time substantially, consistent with precatenanes having a major, and perhaps the sole, role in sister chromosome cohesion in E. coli. Precatenanes inaccessible to TopoIV could still be responsible for the ∼5 min that loci remain spatially unseparated after replication when TopoIV is increased, or perhaps this reflects the time between locus replication and initiation of segregation. Alternatively, other unidentified cohesion processes could be responsible for this short-term cohesion, although the E. coli SMC complex, MukBEF, appears not to be required for sister locus cohesion (Danilova et al. 2007). Our demonstration that sister replisomes still segregate during conditions of TopoIV impairment helps substantiate our proposal that sister replisomes track independently along DNA.
Overexpression of the replisome clamp loader, γ, removes the requirement for TopoIV in bulk replication, although the cell is now dependent on FtsK-facilitated XerCD-dif site-specific recombination to remove final catenation at the end of replication (Levine and Marians 1998; Espeli et al. 2003b; Grainge et al. 2007). Excess γ could act by limiting rotation of the fork or by facilitating gyrase action ahead of the fork, thereby minimizing diffusion backward of linkage and maximizing resolution of any precatenanes by forward diffusion. With regard to this, replisomes still act independently under conditions of γ overexpression (R. Reyes-Lamothe and D.J. Sherratt, unpubl.).
The work reported here provides the most direct evidence for the in vivo generation during replication of precatenanes in chromosomal DNA and for a biological role for such precatenanes in sister cohesion. In previous experiments, precatenanes have been demonstrated in plasmid substrates in bacteria and in eukaryotes, under conditions in which replication is blocked or topoisomerases are inhibited (for review, see Espeli and Marians 2004; Schvartzman and Stasiak 2004). In eukaryotes, precatenation/catenation should occur as a direct consequence of unlinking during replication, but other dedicated cohesion mechanisms, which are under strict temporal and spatial control, ensure that newly replicated sisters remain cohesed and aligned until the biologically appropriate time (for review, see Nasmyth and Haering 2005). In those bacteria, like E. coli, that support overlapping replication cycles and that have chromosomes that complete segregation several generations after initiation, it may be particularly important to ensure that locus segregation occurs progressively soon after replication. It will be interesting to see whether those bacteria and archaea with a strict separation of S phase and cytokinesis have extended periods and lengths of cohesion and whether they use eukaryote cohesin-like mechanisms or appropriately controlled DNA entanglement–disentanglement.
Currently, we have no measure of the level of precatenanes, and indeed whether their density varies in a genetically determined way throughout the chromosome. For example, the level could be determined by the existence of preferential DNA-binding sites for the action of DNA gyrase and TopoIV or by the presence of sites of DNA replication pausing, which might facilitate fork rotation and/or gyrase action ahead of a fork. Temporal and spatial control of topoisomerase activity may also play an important role (Espeli et al. 2003a, b; Espeli and Marians 2004). Our belief is that most linkage is removed by the action of DNA gyrase during replication and that on average <1% of linkage is transferred to precatenanes during bulk replication, corresponding to less than one precatenane per kilobase; under conditions of TopoIV impairment, transient spatial separation of sister loci is still observed. We do not yet know whether there are biological functions that depend on the cohesion introduced by precatenanes For example, does this facilitate recombinational repair between newly replicated sisters, or indeed the sister chromosome segregation process itself? Attempts to answer these questions can be addressed now since it is possible to modulate the periods and extents of cohesion and to control chromosome segregation through precatenation.
Materials and methods
Bacterial strain, growth, and plasmids
Unless otherwise stated, E. coli K12 AB1157 was used (Wang et al. 2005). parEts, parCts, and parental MG1655 strains were a gift from L. Zechiedrich (Grainge et al. 2007). A ΔtopB strain was kindly provided by “NBRP E. coli Strain” (Baba et al. 2006).
Cells were growing at 30°C for parEts, parCts, and dnaC2ts strains and shifted to 42°C, 42°C, and 37°C, respectively, to obtain the temperature-sensitive phenotypes.
Chromosomal genetic loci were visualized using fluorescent repressor–operator systems (Wang et al. 2005, 2006). A lacO array of 240 copies was inserted into ori1, L2 and a 240 copy tetO array into R3 (Fig. 1B) (Wang et al. 2006). LacI-mCherry and TetR-mCerulean were expressed from the chromosomal leuB and galK loci, respectively, regulated by the lac promoter (Reyes-Lamothe et al. 2008). A Ssb-Ypet fusion protein was expressed from the ssb endogenous chromosomal promoter and used as a marker for the replisome (Reyes-Lamothe et al. 2008). Replication blockage at R3 tetO arrays by tightly bound TetR was achieved by removal of anhydrotetracycline (Possoz et al. 2006).
TopoIV was cloned from the pBR322 derivative pLexParEParC (a kind gift of K.J. Marians) to pBAD24, where the cloned genes were under the control of the arabinose promoter (Guzman et al. 1995).
Flow cytometry and fluorescence microscopy
Flow cytometry was performed as in Reyes-Lamothe et al. (2008). Cells for microscopy were grown in minimal glycerol supplemented with required nutrients (Wang et al. 2005). Snapshot images were taken from cells in exponential phase (A600 0.05–0.2). Cells were mounted on 1% agarose in minimal medium at required temperature during time-lapse analysis (5-min intervals). Cells were visualized with a 100× objective on a Nikon Eclipse TE2000-U microscope, equipped with either a Photometrics Cool-Snap HQ CCD or a QuantEM camera, and a temperature-controlled incubation chamber. The images were taken, analyzed, and processed by MetaMorph 6.2.
Acknowledgments
We are grateful to K.J. Marians for supplying a TopoIV expression plasmid. We thank I. Grainge and K. Nasmyth for helpful discussions. The research was funded by the Wellcome Trust. R.R.-L. was supported by a University of Oxford Clarendon post-graduate award and Conacyt.
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.487508.
References
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knock-out mutants: The Keio collection. Mol. Syst. Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bates D., Kleckner N. Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Champoux J.J., Been M.D. Topoisomerases and the swivel problem. In: Alberts B., editor. Mechanistic studies of DNA replication and recombination: ICN-UCLA symposia on molecular and cellular biology. Academic Press; New York: 1980. pp. 809–815. [Google Scholar]
- Danilova O., Reyes-Lamothe R., Pinskaya M., Sherratt D.J., Possoz C. MukB colocalizes with the oriC region and is required for organization of the two Escherichia coli chromosome arms into separate cell halves. Mol. Microbiol. 2007;65:1485–1492. doi: 10.1111/j.1365-2958.2007.05881.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espeli O., Marians K.J. Untangling intracellular DNA topology. Mol. Microbiol. 2004;52:925–931. doi: 10.1111/j.1365-2958.2004.04047.x. [DOI] [PubMed] [Google Scholar]
- Espeli O., Lee C., Marians K.J. A physical and functional interaction between Escherichia coli FtsK and topoisomerase IV. J. Biol. Chem. 2003a;278:44639–44644. doi: 10.1074/jbc.M308926200. [DOI] [PubMed] [Google Scholar]
- Espeli O., Levine C., Hassing H., Marians K.J. Temporal regulation of topoisomerase IV activity in E. coli. Mol. Cell. 2003b;11:189–201. doi: 10.1016/s1097-2765(03)00013-3. [DOI] [PubMed] [Google Scholar]
- Grainge I., Bregu M., Vazquez M., Sivanathan V., Ip S.C., Sherratt D.J. Unlinking chromosome catenanes in vivo by site-specific recombination. EMBO J. 2007;26:4228–4238. doi: 10.1038/sj.emboj.7601849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guzman L.M., Belin D., Carson M.J., Beckwith J. Tight regulation, modulation, and high-level expression by vectors containing the arabinose PBAD promoter. J. Bacteriol. 1995;177:4121–4130. doi: 10.1128/jb.177.14.4121-4130.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodursky A.B., Peter B.J., Schmid M.B., DeRisi J., Botstein D., Brown P.O., Cozzarelli N.R. Analysis of topoisomerase function in bacterial replication fork movement: Use of DNA microarrays. Proc. Natl. Acad. Sci. 2000;97:9419–9424. doi: 10.1073/pnas.97.17.9419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levine C., Marians K.J. Identification of dnaX as a high-copy suppressor of the conditional lethal and partition phenotypes of the parE10 allele. J. Bacteriol. 1998;180:1232–1240. doi: 10.1128/jb.180.5.1232-1240.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes M., Cotta-Ramusino C., Liberi G., Foiani M. Branch migrating sister chromatid junctions form at replication origins through Rad51/Rad52-independent mechanisms. Mol. Cell. 2003;6:1499–1510. doi: 10.1016/s1097-2765(03)00473-8. [DOI] [PubMed] [Google Scholar]
- Maisnier-Patin S., Nordstrom K., Dasgupta S. Replication arrests during a single round of replication of the Escherichia coli chromosome in the absence of DnaC activity. Mol. Microbiol. 2001;42:1371–1382. doi: 10.1046/j.1365-2958.2001.02718.x. [DOI] [PubMed] [Google Scholar]
- Nasmyth K., Haering C.H. The structure and function of SMC and kleisin complexes. Annu. Rev. Biochem. 2005;74:595–648. doi: 10.1146/annurev.biochem.74.082803.133219. [DOI] [PubMed] [Google Scholar]
- Nielsen H.J., Li Y., Youngren B., Hansen F.G., Austin S. Progressive segregation of the Escherichia coli chromosome. Mol. Microbiol. 2006;61:383–393. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- Possoz C., Filipe S.R., Grainge I., Sherratt D.J. Tracking of controlled Escherichia coli replication fork stalling and restart at repressor-bound DNA in vivo. EMBO J. 2006;25:2596–2604. doi: 10.1038/sj.emboj.7601155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes-Lamothe R., Possoz C., Danilova O., Sherratt D.J. Independent positioning and action of Escherichia coli replisomes in live cells. Cell. 2008;133:90–102. doi: 10.1016/j.cell.2008.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson N.P., Blood K.A., McCallum S.A., Edwards P.A.W., Bell S.D. Sister chromatid junctions in the hyperthermophilic archaeon Sulfolobus solfataricus. EMBO J. 2007;26:816–824. doi: 10.1038/sj.emboj.7601529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schvartzman J.B., Stasiak A. A topological view of the replicon. EMBO Rep. 2004;5:256–261. doi: 10.1038/sj.embor.7400101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone M.D., Bryant Z., Crisona N., Smith S.B., Vologodskii A., Bustamente C., Cozzarelli N.R. Chirality sensing by Escherichia coli topoisomerase IV and the mechanism of type II topoisomerases. Proc. Natl. Acad. Sci. 2003;100:8654–8659. doi: 10.1073/pnas.1133178100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sunako Y., Onogi T., Hiraga S. Sister chromosome cohesion of Escherichia coli. Mol. Microbiol. 2001;42:1233–1241. doi: 10.1046/j.1365-2958.2001.02680.x. [DOI] [PubMed] [Google Scholar]
- Viollier P.H., Thanbichler M., McGrath P.T., West L., Meewan M., McAdams H.H., Shapiro L. Rapid and sequential movement of individual chromosomal loci to specific subcellular locations during bacterial DNA replication. Proc. Natl. Acad. Sci. 2004;101:9257–9262. doi: 10.1073/pnas.0402606101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Possoz C., Sherratt D.J. Dancing around the divisome: Asymmetric chromosome segregation in Escherichia coli. Genes & Dev. 2005;19:2367–2377. doi: 10.1101/gad.345305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Liu X., Possoz C., Sherratt D.J. The two Escherichia coli chromosome arms locate to separate cell halves. Genes & Dev. 2006;20:1727–1731. doi: 10.1101/gad.388406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L., Hickson I.D. DNA helicases required for homologous recombination and repair of damaged replication forks. Annu. Rev. Genet. 2006;40:279–306. doi: 10.1146/annurev.genet.40.110405.090636. [DOI] [PubMed] [Google Scholar]
- Zechiedrich E.L., Cozzarelli N.R. Roles of topoisomerase IV and DNA gyrase in DNA unlinking during replication in Escherichia coli. Genes & Dev. 1995;9:2859–2869. doi: 10.1101/gad.9.22.2859. [DOI] [PubMed] [Google Scholar]
- Zechiedrich E.L., Khodursky A.B., Cozzarelli N.R. Topoisomerase IV, not gyrase, decatenates products of site-specific recombination in Escherichia coli. Genes & Dev. 1997;11:2580–2592. doi: 10.1101/gad.11.19.2580. [DOI] [PMC free article] [PubMed] [Google Scholar]