Abstract
The RNA polymerase II core promoter is a structurally and functionally diverse transcriptional module. RNAi depletion and overexpression experiments revealed a genetic circuit that controls the balance of transcription from two core promoter motifs, the TATA box and the downstream core promoter element (DPE). In this circuit, TBP activates TATA-dependent transcription and represses DPE-dependent transcription, whereas Mot1 and NC2 block TBP function and thus repress TATA-dependent transcription and activate DPE-dependent transcription. This regulatory circuit is likely to be one means by which biological networks can transmit transcriptional signals, such as those from DPE-specific and TATA-specific enhancers, via distinct pathways.
Keywords: Core promoter, RNA polymerase II, transcription, TBP, Mot1, NC2
The RNA polymerase II core promoter comprises the sequences that direct the initiation of transcription. Although it has often been presumed that the core promoter is a generic entity, current evidence indicates that there is considerable diversity in core promoter structure and function. Hence, the core promoter is a regulatory element (for reviews, see Smale and Kadonaga 2003; Sandelin et al. 2007; Juven-Gershon et al. 2008).
Here, we focus on the relation between two core promoter motifs—the downstream core promoter element (DPE) and the TATA box. The TATA box is the most ancient core promoter motif, as it is conserved from archaebacteria to humans. It has a consensus of TATAWAAR, where the upstream T nucleotide is typically located about −31 or −30 relative to the A + 1 in the Initiator (Inr) element. The DPE appears to be conserved among metazoans. It is strictly located from +28 to +33 relative to the A + 1 in the Inr, and has a consensus of RGWYVT in Drosophila.
Both the TATA box and DPE are binding sites for the TFIID basal transcription factor, but TFIID appears to have distinct modes of binding to the two core promoter motifs. The TBP subunit of TFIID binds to the TATA box, whereas the TAF6 and TAF9 subunits of TFIID are in close proximity to the DPE. In addition, the DNase I footprinting patterns on TATA-containing versus DPE-containing promoters are different (for example, see Burke and Kadonaga 1996). In particular, TFIID footprints of DPE-dependent core promoters exhibit a periodic 10-bp DNase I digestion pattern that suggests an extended, close interaction of TFIID from the Inr through the DPE (Burke and Kadonaga 1996; Kutach and Kadonaga 2000).
There are differences in the functional properties of DPE-dependent versus TATA-dependent core promoters. For instance, an enhancer-trapping analysis in Drosophila revealed the existence of DPE-specific as well as TATA-specific transcriptional enhancers (Butler and Kadonaga 2001). It was also found that a set of factors (TFIIA, TFIIB, TFIID, TFIIE, TFIIF, TFIIH, RNA polymerase II, PC4, and Sp1) that is sufficient for transcription of promoters containing both TATA and DCE (downstream core element; Lee et al. 2005) motifs is not able to transcribe a DPE-dependent promoter (Lewis et al. 2005). In that case, DPE-dependent transcription was additionally found to require casein kinase II (CKII) and Mediator. In other studies, NC2 (also known as Dr1-Drap1), which was originally identified as a repressor of TATA-dependent transcription, was found to activate transcription from five different DPE-dependent core promoters in reactions performed with a nuclear extract (Willy et al. 2000). With a purified transcription system, however, NC2 activation of a DPE-dependent core promoter was not observed (Lewis et al. 2005).
To determine the nature of the factors that promote DPE-dependent versus TATA-dependent transcription, we investigated the properties of key transcription factors by RNAi depletion, overexpression, and chromatin immunoprecipitation (ChIP) analyses with multiple DPE-dependent and TATA-dependent promoters. The new findings reveal a regulatory circuit that controls the balance between DPE-dependent versus TATA-dependent transcription.
Results and Discussion
RNAi depletion of TBP reduces TATA-dependent but not DPE-dependent transcription
In this study, we used cultured Drosophila cells as the experimental system to investigate DPE versus TATA function. We created two sets of reporter constructs that contain either TATA or DPE motifs driving a luciferase reporter gene (Supplemental Fig. 1; Fig. 1). The DPE-dependent and TATA-dependent promoters in each set are identical, except for the sequences at the positions of the DPE and TATA motifs (Supplemental Table 1), and have comparable transcriptional activities (Supplemental Fig. 1).
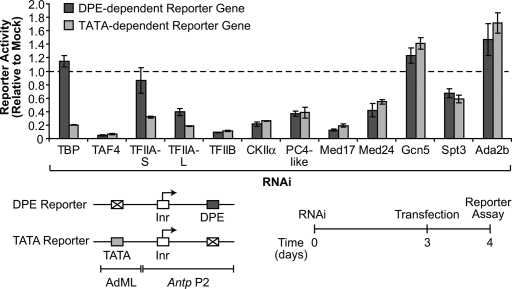
Figure 1.
Depletion of TBP reduces TATA-dependent but not DPE-dependent transcription. Drosophila S2 cells were depleted of the indicated factors by RNAi, and then transfected with TATA-dependent or DPE-dependent luciferase reporter genes. The activities of the RNAi-depleted extracts are reported as luminescent units per microgram of protein of RNAi-depleted extracts relative to the luminescent units per microgram of protein of mock RNAi-treated control extracts. The experimental scheme and reporter constructs are depicted at the bottom of the figure. “PC4-like” is Ssb-C31a, which is the Drosophila protein that is most closely related to mammalian PC4. The error bars represent the standard deviation.
We investigated the effects of several transcription factors upon DPE versus TATA transcription by RNAi depletion analysis (Fig. 1). The transcription factors were selected on the basis of their fundamental importance as well as their potential role in DPE-dependent transcription. We first carried out RNAi depletion of each target factor (for Western blot data, see Supplemental Figs. 2–4), and then transfected one-half of the cells with the DPE-dependent reporter construct and the other half of the cells with the TATA-dependent reporter. The resulting transcription levels were assessed by measurement of the luciferase activities relative to those in mock RNAi controls.
Depletion of TBP sharply decreases TATA-dependent transcription, but has little effect on DPE-dependent transcription (Fig. 1). This effect was observed with a distinct and independent set of DPE-dependent and TATA-dependent reporter constructs (Supplemental Fig. 5) as well as with a different nonoverlapping dsRNA probe for TBP (Supplemental Fig. 6). Consistent with the ability of TFIIA to promote TBP binding to DNA (for example, see Buratowski et al. 1989; Maldonado et al. 1990), we observed that depletion of TFIIA reduces TATA transcription more than DPE transcription with two different sets of reporter constructs (Fig. 1; Supplemental Fig. 5). In contrast, we did not see differential DPE versus TATA effects upon RNAi depletion of TAF4 (which is essential for the structural integrity of TFIID) (Wright et al. 2006), TFIIB, CKIIα, a PC4-like protein, subunits of Mediator (Med17, Med24), or subunits of the SAGA/TFTC complex (Gcn5, Spt3, Ada2b) (Fig. 1; Supplemental Fig. 5).
Thus, these findings indicate that TBP and, to a lesser extent, TFIIA have a key role in discriminating between DPE- versus TATA-dependent transcription. The stronger effect of TBP relative to TFIIA is consistent with an auxiliary function of TFIIA, such as its ability to increase the binding of TBP to the TATA box. Because depletion of TBP did not adversely affect DPE-dependent transcription, we considered the possibility that DPE-dependent transcription might involve a factor, such as SAGA/TFTC, that lacks TBP (Wieczorek et al. 1998; for review, see Nagy and Tora 2007). We therefore tested the effect of depletion of three SAGA/TFTC subunits (Gcn5, Spt3, and Ada2b), but did not see a substantial decrease in DPE-dependent transcription or any differential DPE versus TATA effects. Thus, it appears unlikely that SAGA/TFTC is important for DPE-dependent transcription. Lastly, upon depletion of CKII, Mediator, PC4-like, TAF4, and TFIIB, we observed a decrease in both DPE-dependent and TATA-dependent transcription. These results are consistent with a more general transcriptional function rather than a DPE-specific or TATA-specific activity for these factors.
NC2 and Mot1 promote DPE-dependent transcription by acting via TBP
NC2 has been previously found to be a DPE-specific transcriptional activator (Willy et al. 2000). With a different biochemical system, however, NC2-mediated enhancement of DPE transcription was not observed (Lewis et al. 2005). We therefore sought to clarify these apparently contrasting results by RNAi analysis of NC2 with our DPE versus TATA reporter gene systems (Fig. 1; Supplemental Fig. 1). NC2 comprises two subunits, NC2α (Drap1) and NC2β (Dr1). Upon RNAi depletion of either NC2α or NC2β (Fig. 2A; Supplemental Fig. 4A), we observed a more substantial decrease in DPE- relative to TATA-dependent transcription with two different sets of reporter genes (Fig. 2B; Supplemental Fig. 5) as well as with two different dsRNAs (Supplemental Fig. 6). These results therefore indicate that NC2 promotes DPE-dependent transcription relative to TATA-dependent transcription in cultured cells.
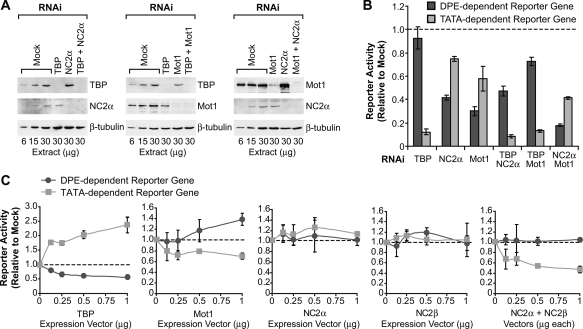
Figure 2.
Mot1 and NC2 act in opposition to TBP to promote DPE transcription relative to TATA transcription. (A) Mot1, NC2, and TBP are efficiently depleted by RNAi in S2 cells. (B) The ability of Mot1 and NC2 to affect DPE-dependent versus TATA-dependent transcription requires TBP. RNAi depletion analysis of the indicated factors was carried out as in Figure 1. (C) Overexpression of TBP has the opposite effect as overexpression of Mot1 or NC2 upon DPE-dependent versus TATA-dependent transcription. The indicated expression vectors were cotransfected with DPE-dependent or TATA-dependent luciferase reporter genes. In each series of transfections, the total amount of expression vector was maintained at a constant level by the inclusion, where necessary, of a compensatory amount of empty vector (pAc5.1). The reporter gene activities with the expression vectors are given relative to those obtained with the empty vector alone. The error bars represent the standard deviation.
Next, we tested the effects of Mot1 (also known as BTAF1 and Hel89B) on DPE versus TATA transcription. Like NC2, Mot1 antagonizes TBP function. NC2 represses TATA-dependent transcription by blocking the association of TBP with other factors such as TFIIA and TFIIB (for review, see Thomas and Chiang 2006). Mot1 is an ATPase that removes TBP from DNA by an ATP-dependent mechanism (for example, see Auble et al. 1994; Pereira et al. 2003). Genetic studies in Saccharomyces cerevisiae suggest that NC2 and Mot1 have related functions (Prelich 1997; Lemaire et al. 2000). NC2 and Mot1 bind to overlapping regions in the yeast genome and form a complex with TBP and DNA (Darst et al. 2003; van Werven et al. 2008). In addition, although NC2 and Mot1 are often thought to be repressive, a positive function for these factors has been observed in vitro and in vivo (Willy et al. 2000; Andrau et al. 2002; Cang and Prelich 2002; Dasgupta et al. 2002, 2005; Geisberg and Struhl 2004; Albert et al. 2007; van Werven et al. 2008).
We observed that RNAi depletion of Mot1 (Fig. 2A; Supplemental Fig. 2) has a stronger detrimental effect on DPE-dependent than TATA-dependent transcription (Fig. 2B). This effect was seen with two different sets of reporter genes as well as with two independent nonoverlapping dsRNA fragments (Supplemental Figs. 5, 6). Thus, like NC2, Mot1 promotes DPE- relative to TATA-dependent transcription.
To investigate the relationship between TBP, NC2, and Mot1 in the regulation of core promoter activity, we codepleted different combinations of these factors and determined the resulting effects upon DPE versus TATA transcription. Codepletion of both NC2α and Mot1 preferentially decreases DPE relative to TATA transcription to an extent that is similar to that seen upon depletion of either NC2α or Mot1 alone (Fig. 2B). These results suggest that NC2 and Mot1 promote DPE-dependent transcription via the same pathway. In contrast, when we codepleted TBP + Mot1 or TBP + NC2α, we observed nearly the same effect on DPE versus TATA transcription as that seen upon depletion of TBP alone (Fig. 2B). These findings suggest that TBP is downstream from NC2 and Mot1 in the pathway that regulates DPE versus TATA transcription. Thus, NC2 and Mot1 appear to modulate DPE versus TATA transcription by acting via TBP.
Opposing effects of overexpression of TBP versus Mot1 or NC2
To complement the RNAi depletion studies, we investigated the effects of overexpression of TBP, Mot1, or NC2 in S2 cells (Fig. 2C). In these experiments, we cotransfected TBP, Mot1, or NC2 expression vectors along with the DPE-dependent or TATA-dependent reporter constructs. Overexpression of TBP increases TATA-dependent transcription and decreases DPE-dependent transcription. Conversely, overexpression of Mot1 increases DPE-dependent transcription and decreases TATA-dependent transcription. Overexpression of both subunits of NC2 decreases TATA-dependent transcription, but has little effect on DPE-dependent transcription. Consistent with the two NC2 subunits functioning together in a complex, overexpression of NC2α alone or NC2β alone has no effect on DPE-dependent or TATA-dependent transcription. In addition, we carried out a parallel set of overexpression experiments with TBP, Mot1, and NC2 with a different set of DPE-dependent and TATA-dependent reporter genes, and obtained nearly identical results (Supplemental Fig. 7). These findings further demonstrate that TBP favors TATA relative to DPE transcription, whereas Mot1 and NC2 favor DPE relative to TATA transcription.
Mot1 and NC2 have opposite effects as TBP upon transcription of endogenous genes
To examine the functions of TBP, Mot1, and NC2 in a more natural context, we investigated the effects of RNAi depletion of TBP, Mot1, or NC2 upon transcription of endogenous DPE- or TATA-containing genes in Drosophila Kc cells. In these experiments, we employed secondary/late ecdysone-responsive genes that are activated upon ecdysone induction. In this manner, we were able to characterize the requirements for TBP, Mot1, and NC2 for transcriptional activation.
Many genes in Drosophila are activated by the steroid hormone 20-hydroxyecdysone (20HE) (for review, see King-Jones and Thummel 2005). We obtained a list of genes that are induced by 20HE in Drosophila Kc cells (generous gift of Dr. Lucy Cherbas and Dr. Peter Cherbas, Indiana University) (L. Cherbas and P. Cherbas, unpubl.). From this list, we identified secondary/late-response genes with DPE+Inr motifs (CG9511, CG16876, Glut1) or TATA + Inr motifs (Obp99c, CG4500) in their core promoters. We confirmed the 20HE induction of these genes in Kc cells by using real-time RT–PCR (Supplemental Fig. 8). In addition, we verified the transcription start sites of each of these genes by primer extension analysis of mRNA isolated from Kc cells (Supplemental Fig. 9).
We thus carried out the RNAi analysis of the endogenous secondary/late-response genes as follows (Fig. 3): TBP, TAF4, NC2α, and Mot1 were each individually depleted by RNAi in Kc cells for 4 d, and then the ecdysone-responsive genes were induced with 20HE for 24 h. The total RNA was isolated, and the transcript levels of the selected genes were determined by real-time RT–PCR. We observed that depletion of TBP decreases transcription of the TATA-containing promoters and increases transcription of the DPE-containing promoters. Thus, these results suggest not only that TBP activates TATA-dependent promoters, but also that it represses DPE-dependent promoters. Conversely, we found that depletion of Mot1 or NC2α decreases transcription of DPE-containing promoters and increases transcription of TATA-containing promoters. These findings suggest a positive function of Mot1 and NC2 at DPE-dependent promoters and a negative function at TATA-containing promoters. RNAi depletion of TAF4 causes a substantial decrease in transcription from both DPE-containing and TATA-containing promoters. These results further support the conclusion that TAF4 is required for both DPE-dependent and TATA-dependent transcription.
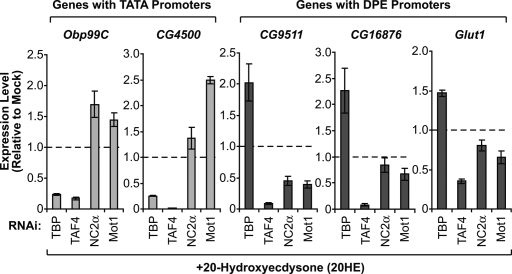
Figure 3.
Mot1 and NC2 have opposite effects as TBP upon transcription of DPE- versus TATA-containing endogenous genes in Drosophila cells. These studies examined secondary/late ecdysone-responsive genes that are activated upon ecdysone induction. Drosophila Kc167 cells were depleted of the indicated factors by RNAi, and then induced with 20-hydroxyecdysone (20HE; 1 μM) for 24 h. The transcription levels of two TATA-containing genes (that lack DPE motifs) and three DPE-containing genes (that lack TATA elements) were determined by real-time RT–PCR. The error bars represent the SEM.
The RNAi depletion analysis with the endogenous genes (Fig. 3) leads to nearly the same conclusions as the experiments with the transfected luciferase reporter genes (Figs. 1, 2B). Both sets of experiments indicate that TBP favors TATA-dependent relative to DPE-dependent transcription, and that Mot1 and NC2 favor DPE-dependent relative to TATA-dependent transcription. However, it is useful to note the two distinctions. First, TBP depletion results in an increase in transcription from endogenous DPE-containing genes, but does not alter transcription from transfected DPE-dependent reporter genes. Second, depletion of Mot1 or NC2α causes an increase in transcription from endogenous TATA-containing genes, but results in a slight decrease in transcription from transfected TATA-dependent reporter genes. The analysis of the endogenous genes is likely to provide a more accurate representation of TBP, Mot1, and NC2 activity than the studies with the transfected genes, because the endogenous genes are in their natural context at the normal copy number and the experiments with the endogenous genes do not involve the extra transfection procedure. Thus, the findings from the analysis of the endogenous genes suggest a repressive function of TBP at DPE-dependent promoters as well as a repressive function of Mot1 and NC2 at TATA-dependent promoters.
TBP ChIP increases upon induction of TATA-containing but not DPE-containing promoters
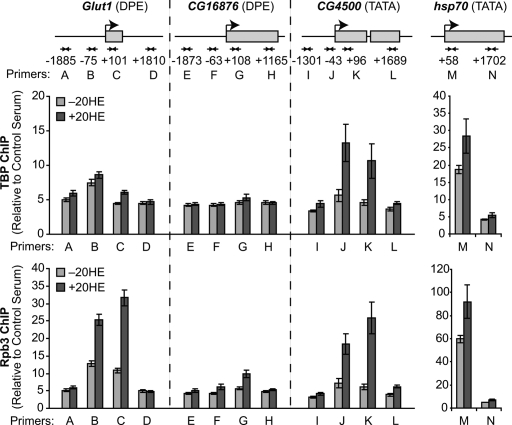
We further characterized the secondary/late ecdysone-responsive genes by ChIP analysis (Fig. 4) with TBP and RNA polymerase II (Rpb3 subunit), for which ChIP-quality antibodies were available. With the TATA-containing CG4500 promoter, there is increased ChIP signal for both TBP and Rpb3 in the promoter region upon 20HE induction. In the control/reference TATA-containing hsp70 promoter, we also observed an increase in ChIP of TBP and Rpb3 in the promoter region (Lebedeva et al. 2005). By comparison, with the DPE-containing Glut1 and CG16876 promoters, there is increased ChIP of Rpb3 in the promoter region upon 20HE induction; however, the ChIP signal for TBP does not increase under the same conditions. The absence of an increased ChIP signal for TBP with the DPE-containing promoters does not necessarily indicate that TBP is not present at the promoter; for instance, it is possible that TBP may be in an altered configuration that masks the accessibility of the antibodies. Yet, whether or not TBP is in close proximity to the DPE-containing promoters, these results show that there are differences in the nature of the interaction of TBP with TATA-containing versus DPE-containing promoters.
Figure 4.
TBP ChIP increases in TATA- but not DPE-containing promoters upon ecdysone induction. ChIP analyses of TBP and RNA polymerase II (Rpb3 subunit) were carried out with Drosophila Kc167 cells in the absence or presence of 20HE. Three ecdysone-responsive genes as well as hsp70, as a reference/control, were analyzed. The amounts of immunoprecipitated DNA with each set of the indicated primers were quantitated by real-time PCR. The enrichments observed with anti-TBP or anti-Rpb3 relative to a control nonimmune serum are shown. The error bars represent the SEM from three independent sets of experiments. The diagrams of the genes are not drawn to scale.
It is also relevant to note that we chose to use secondary/late-response genes in these studies, because secondary/late genes are more likely than primary/early-response genes to be in a naïve state prior to ecdysone induction. To test this notion, we carried out RNAi depletion analyses with two primary/early-response genes, E74A and E75B, both of which contain DPE motifs. With these genes, we did not observe any change in transcription upon RNAi depletion of TBP, TAF4, Mot1, or NC2α (data not shown). Moreover, ChIP analysis further revealed that both TBP and RNA polymerase II (Rpb3 subunit) are present at the promoters prior to ecdysone induction. Therefore, it appears likely that these primary/early-response genes exist in a preactivated state that does not require the subsequent action of factors such as TFIID, Mot1, or NC2.
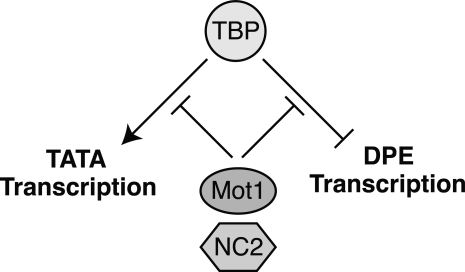
A genetic circuit in which Mot1 and NC2 oppose TBP to control DPE versus TATA transcription
The RNAi depletion and overexpression data reveal a regulatory circuit with the following properties: TBP activates TATA-dependent transcription and represses DPE-transcription; then, Mot1 and NC2 act to block both the activating and repressive functions of TBP (Fig. 5). In this model, there are opposing forces that alter the balance between DPE versus TATA transcription. A decrease in TBP or an increase in Mot1/NC2 favors DPE transcription, whereas an increase in TBP or a decrease in Mot1/NC2 favors TATA transcription. Importantly, the functions of Mot1 and NC2 are dependent on TBP, as seen in Figure 2B. In addition, the proposed circuit is consistent with the known antagonistic relationship between TBP and NC2 as well as between TBP and Mot1.
Figure 5.
Mot1, NC2, and TBP create a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. In this model, TBP activates TATA transcription and represses DPE transcription, and Mot1 and NC2 act to inhibit the function of TBP. Thus, a decrease in TBP and/or an increase in Mot1/NC2 favors DPE transcription, whereas a decrease in Mot1/NC2 and/or an increase in TBP favors TBP transcription.
How might TBP repress DPE-dependent transcription? Two possible explanations are as follows. First, in the absence of a TATA box, TBP might interfere with the proper assembly of the transcription initiation complex. Second, there may be an essential DPE-directed transcription factor that is inhibited by TBP. It is possible that DPE-mediated transcription does not directly involve TBP, as there is substantial evidence of RNA polymerase II-mediated transcription occurring in the absence of TBP (for example, see Veenstra et al. 2000; Müller et al. 2001; Martianov et al. 2002; Paulson et al. 2002; Deato and Tjian 2007; Ferg et al. 2007).
We also considered whether either of the TBP-related factors, TRF1 and TRF2, are used instead of TBP at DPE-containing promoters. To this end, we examined the effect of depleting TRF1 or TRF2 upon the expression of DPE-containing versus TATA-containing endogenous genes (Supplemental Fig. 10). TRF1, which is largely involved in RNA polymerase III transcription in Drosophila (Takada et al. 2000; Isogai et al. 2007b), has little or no effect on transcription of DPE-containing or TATA-containing genes. TRF2 is important for both DPE-mediated and TATA-mediated transcription. The effect of TRF2 is similar to that of TAF4, which appears to contribute to both DPE-depentend and TATA-dependent transcription. Neither TRF1 nor TRF2 exhibit an opposite effect on DPE-mediated versus TATA-mediated transcription as do TBP, Mot1, and NC2 (Fig. 3). In addition, a genome-wide ChIP analysis of TRF2 did not reveal an association of TRF2 with DPE-containing genes (Isogai et al. 2007a). Thus, at the present time, there is no evidence suggesting a specific link between either TRF1 or TRF2 and DPE-mdidated or TATA-mediated transcription.
In conclusion, the analysis of TBP, Mot1, and NC2 in the context of DPE-containing versus TATA-containing promoters has revealed a regulatory circuit that controls the balance between DPE-mediated versus TATA-mediated transcription. This circuit may be a key means by which DPE or TATA specificity of transcriptional enhancers is achieved. In the future, it will be interesting and important to build upon this core circuit to identify the connections and mechanisms by which biological networks use DPE and TATA specificity to increase the number of pathways by which signals can be transmitted.
Materials and methods
RNAi and overexpression analyses
For RNAi-coupled reporter assays, cells were treated with dsRNA for 3 d and then transfected with the firefly luciferase reporter (0.2 pmol) and the pol III-Renilla control plasmid (50 ng) with Effectene (Qiagen). For overexpression experiments, the indicated amounts of expression vector were combined, as necessary, with empty expression vector (pAc5.1) to give a total of 1 μg of expression vector, and then cotransfected with the firefly luciferase reporter (0.2 pmol) and the pol III-Renilla control plasmid (50 ng) with Transfectol (Gene Choice). Twenty-four hours after transfection, cells were washed with PBS and then lysed with 1× RLB (Promega). The firefly and Renilla luciferase activities were measured by using reagents from the Dual-luciferase reporter assay systems (Promega). The protein concentration of cell lyastes was measured with the BCA reagent (Pierce). For ecdysone treatment during RNAi, Kc167 cells were treated with dsRNA for 3 or 4 d and then incubated with 1 μM of 20-hydroxyecdysone (20HE) for 24 h. Additional Materials and Methods are available in the Supplemental Material.
Acknowledgments
We thank David Auble, Timur Yusufzai, Russell Darst, Barbara Rattner, and Chin Yan Lim for critical reading of the manuscript. We are especially grateful to Dr. Lucy Cherbas and Dr. Peter Cherbas (Indiana University) for the unpublished information on ecdysone-responsive genes in Kc cells. We thank Biraj Shah for assistance in the preparation of the Drosophila Mot1 antibodies; Drs. Frank Furnari and Webster Cavenee (Ludwig Institute for Cancer Research; UCSD) for the use of their luminometer; Dr. Norbert Perrimon (Harvard Medical School) for the Renilla luciferase control plasmid; Dr. Jerry Workman (Stowers Institute), Dr. John T. Lis (Cornell University), Dr. Michael E. Dahmus (University of California, Davis), and Dr. Sofia G. Georgieva (Russian Academy of Sciences) for antibodies; and Dr. Carl Thummel (University of Utah) for advice on ecdysone-responsive genes. J.-Y.H. was supported by a Helen Hay Whitney Fellowship. This work was supported by a grant from the National Institutes of Health (GM041249) to J.T.K.
Footnotes
Supplemental material is available at http://www.genesdev.org.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.1681808.
References
- Albert T.K., Grote K., Boeing S., Stelzer G., Schepers A., Meisterernst M. Global distribution of negative cofactor 2 subunit-α on human promoters. Proc. Natl. Acad. Sci. 2007;104:10000–10005. doi: 10.1073/pnas.0703490104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrau J.C., Van Oevelen C.J., Van Teeffelen H.A., Weil P.A., Holstege F.C., Timmers H.T.M. Mot1p is essential for TBP recruitment to selected promoters during in vivo gene activation. EMBO J. 2002;21:5173–5183. doi: 10.1093/emboj/cdf485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Auble D.T., Hansen K.E., Mueller C.G.F., Lane W.S., Thorner J., Hahn S. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes & Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- Buratowski S., Hahn S., Guarente L., Sharp P.A. Five intermediate complexes in transcription initiation by RNA polymerase II. Cell. 1989;56:549–561. doi: 10.1016/0092-8674(89)90578-3. [DOI] [PubMed] [Google Scholar]
- Burke T.W., Kadonaga J.T. Drosophila TFIID binds to a conserved downstream basal promoter element that is present in many TATA-box-deficient promoters. Genes & Dev. 1996;10:711–724. doi: 10.1101/gad.10.6.711. [DOI] [PubMed] [Google Scholar]
- Butler J.E.F., Kadonaga J.T. Enhancer-promoter specificity mediated by DPE or TATA core promoter motifs. Genes & Dev. 2001;15:2515–2519. doi: 10.1101/gad.924301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cang Y., Prelich G. Direct stimulation of transcription by negative cofactor 2 (NC2) through TATA-binding protein (TBP) Proc. Natl. Acad. Sci. 2002;99:12727–12732. doi: 10.1073/pnas.202236699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darst R.P., Dasgupta A., Zhu C., Hsu J.Y., Vroom A., Muldrow T., Auble D.T. Mot1 regulates the DNA binding activity of free TATA-binding protein in an ATP-dependent manner. J. Biol. Chem. 2003;278:13216–13226. doi: 10.1074/jbc.M211445200. [DOI] [PubMed] [Google Scholar]
- Dasgupta A., Darst R.P., Martin K.J., Afshari C.A., Auble D.T. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc. Natl. Acad. Sci. 2002;99:2666–2671. doi: 10.1073/pnas.052397899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dasgupta A., Juedes S.A., Sprouse R.O., Auble D.T. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 2005;24:1717–1729. doi: 10.1038/sj.emboj.7600646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deato M.D., Tjian R. Switching of the core transcription machinery during myogenesis. Genes & Dev. 2007;21:2137–2149. doi: 10.1101/gad.1583407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferg M., Sanges R., Gehrig J., Kiss J., Bauer M., Lovas A., Szabo M., Yang L., Straehle U., Pankratz M.J., et al. The TATA-binding protein regulates maternal mRNA degradation and differential zygotic transcription in zebrafish. EMBO J. 2007;26:3945–3956. doi: 10.1038/sj.emboj.7601821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geisberg J.V., Struhl K. Cellular stress alters the transcriptional properties of promoter-bound Mot1–TBP complexes. Mol. Cell. 2004;14:479–489. doi: 10.1016/j.molcel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- Isogai Y., Keles S., Prestel M., Hochheimer A., Tjian R. Transcription of histone gene cluster by differential core-promoter factors. Genes & Dev. 2007a;21:2936–2949. doi: 10.1101/gad.1608807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai Y., Takada S., Tjian R., Keles S. Novel TRF1/BRF target genes revealed by genome-wide analysis of Drosophila Pol III transcription. EMBO J. 2007b;26:79–89. doi: 10.1038/sj.emboj.7601448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutach A.K., Kadonaga J.T. The downstream promoter element DPE appears to be as widely used as the TATA box in Drosophila core promoters. Mol. Cell. Biol. 2000;20:4754–4764. doi: 10.1128/mcb.20.13.4754-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juven-Gershon T., Hsu J.-Y., Theisen J.W., Kadonaga J.T. The RNA polymerase II core promoter—The gateway to transcription. Curr. Opin. Cell Biol. 2008;20:253–259. doi: 10.1016/j.ceb.2008.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King-Jones K., Thummel C.S. Nuclear receptors—A perspective from Drosophila. Nat. Rev. Genet. 2005;6:311–323. doi: 10.1038/nrg1581. [DOI] [PubMed] [Google Scholar]
- Lemaire M., Xie J., Meisterernst M., Collart M.A. The NC2 repressor is dispensable in yeast mutated for the Sin4p component of the holoenzyme and plays roles similar to Mot1p in vivo. Mol. Microbiol. 2000;36:163–173. doi: 10.1046/j.1365-2958.2000.01839.x. [DOI] [PubMed] [Google Scholar]
- Lebedeva L.A., Nabirochkina E.N., Kurshakova M.M., Robert F., Krasnov A.N., Evgen’ev M.B., Kadonaga J.T., Georgieva S.G., Tora L. Occupancy of the Drosophila hsp70 promoter by a specific subset of basal transcription factors diminishes upon transcriptional activation. Proc. Natl. Acad. Sci. 2005;102:18087–18092. doi: 10.1073/pnas.0509063102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee D.H., Gershenzon N., Gupta M., Ioshikhes I.P., Reinberg D., Lewis B.A. Functional characterization of core promoter elements: The downstream core element is recognized by TAF1. Mol. Cell. Biol. 2005;25:9674–9686. doi: 10.1128/MCB.25.21.9674-9686.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B.A., Sims R.J., Lane W.S., Reinberg D. Functional characterization of core promoter elements: DPE-specific transcription requires the protein kinase CK2 and the PC4 coactivator. Mol. Cell. 2005;18:471–481. doi: 10.1016/j.molcel.2005.04.005. [DOI] [PubMed] [Google Scholar]
- Maldonado E., Ha I., Cortes P., Weis L., Reinberg D. Factors involved in specific transcription by mammalian RNA polymerase II: Role of transcription factors IIA, IID, and IIB during formation of a transcription-competent complex. Mol. Cell. Biol. 1990;10:6335–6347. doi: 10.1128/mcb.10.12.6335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martianov I., Viville S., Davidson I. RNA polymerase II transcription in murine cells lacking the TATA binding protein. Science. 2002;298:1036–1039. doi: 10.1126/science.1076327. [DOI] [PubMed] [Google Scholar]
- Müller F., Lakatos L., Dantonel J., Strähle U., Tora L. TBP is not universally required for zygotic RNA polymerase II transcription in zebrafish. Curr. Biol. 2001;11:282–287. doi: 10.1016/s0960-9822(01)00076-8. [DOI] [PubMed] [Google Scholar]
- Nagy L., Tora L. Distinct GCN5/PCAF-containing complexes function as co-activators and are involved in transcription factor and global histone acetylation. Oncogene. 2007;26:5341–5357. doi: 10.1038/sj.onc.1210604. [DOI] [PubMed] [Google Scholar]
- Paulson M., Press C., Smith E., Tanese N., Levy D.E. IFN-stimulated transcription through a TBP-free acetyltransferase complex escapes viral shutoff. Nat. Cell Biol. 2002;4:140–147. doi: 10.1038/ncb747. [DOI] [PubMed] [Google Scholar]
- Pereira L.A., Klejman M.P., Timmers H.T.M. Roles for BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription. Gene. 2003;315:1–13. doi: 10.1016/s0378-1119(03)00714-5. [DOI] [PubMed] [Google Scholar]
- Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol. Cell. Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandelin A., Carninci P., Lenhard B., Ponjavic J., Hayashizaki Y., Hume D.A. Mammalian RNA polymerase II core promoters: Insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- Smale S.T., Kadonaga J.T. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- Takada S., Lis J.T., Zhou S., Tjian R. A TRF1:BRF complex directs Drosophila RNA polymerase III transcription. Cell. 2000;101:459–469. doi: 10.1016/s0092-8674(00)80857-0. [DOI] [PubMed] [Google Scholar]
- Thomas M.C., Chiang C.M. The general transcription machinery and general cofactors. Crit. Rev. Biochem. Mol. Biol. 2006;41:105–178. doi: 10.1080/10409230600648736. [DOI] [PubMed] [Google Scholar]
- van Werven F.J., van Bakel H., van Teeffelen H.A.A.M., Altelaar A.F.M., Koerkamp M.G., Heck A.J.R., Holstege F.C.P., Timmers H.Th.M. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes & Dev. 2008 doi: 10.1101/gad.1682308. (this issue) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenstra G.J., Weeks D.L., Wolffe A.P. Distinct roles for TBP and TBP-like factor in early embryonic gene transcription in Xenopus. Science. 2000;290:2312–2315. doi: 10.1126/science.290.5500.2312. [DOI] [PubMed] [Google Scholar]
- Wieczorek E., Brand M., Jacq X., Tora L. Function of TAF(II)-containing complex without TBP in transcription by RNA polymerase II. Nature. 1998;393:187–191. doi: 10.1038/30283. [DOI] [PubMed] [Google Scholar]
- Willy P.J., Kobayashi R., Kadonaga J.T. A basal transcription factor that activates or represses transcription. Science. 2000;290:982–985. doi: 10.1126/science.290.5493.982. [DOI] [PubMed] [Google Scholar]
- Wright K.J., Marr M.T., Tjian R. TAF4 nucleates a core subcomplex of TFIID and mediates activated transcription from a TATA-less promoter. Proc. Natl. Acad. Sci. 2006;103:12347–12352. doi: 10.1073/pnas.0605499103. [DOI] [PMC free article] [PubMed] [Google Scholar]