Abstract
The tryptophans of integral membrane proteins have been suggested to play specific roles as ‘interfacial anchors’, based on their preference for a location near the lipid head groups. Still, the underlying mechanism behind this behavior remains unclear. NMR experiments can provide an important tool to study this interaction in an actual bilayer environment. Here solid-state deuterium nuclear magnetic resonance was used to study the tryptophans in membrane-spanning model peptides from the WALP family (acetyl-GWW(LA)nWWA-ethanolamide with n = 5 and 6.5) in samples of mechanically aligned dimyristoylphosphatidylcholine (DMPC) bilayers. The data indicate that the tryptophans near the C-terminal end of the peptide display a significantly different behavior from those near the N-terminus. This is reflected prominently in a large difference in the motion experienced by the indoles at either end of the peptide, highlighting the directionality of the helix. Nevertheless, our observations indicate high levels of motional freedom for all tryptophans in these membrane spanning domains, that exceed the dynamics for the helix itself. These observations signify that steric and dynamic features of the polypeptide context modulate the tryptophan anchoring in the membrane interface. Measurements of WALP19 in the ether-linked DMPC analog ditetradecylphosphatidylcholine (missing the lipid carbonyls) show very similar Trp dynamics and suggest similar orientations for some or all of the tryptophans. This suggests that the lipid acyl chain carbonyls play at most a minor role in the anchoring interaction between these Trp residues and the DMPC interfacial region.
Traditionally, the interactions of integral membrane proteins with the surrounding lipids have been characterized in terms of the exposure of their hydrophobic transmembrane domains to the lipid acyl chains. It has become apparent in the meantime that the regions directly adjacent to the hydrophobic domains play an important role in this interaction, and are often enriched in specific amino acids. Charged as well as aromatic residues appear to fulfill a special function for membrane proteins, acting as interfacial anchoring residues. In particular, the aromatic tryptophans have drawn attention.
The first indications of special properties come from the distribution of tryptophans in membrane proteins. This amino acid is relatively uncommon in proteins in general, comprising only 1.2 % of all residues in soluble proteins. However, Trp1 is found to be almost three times more common in membrane proteins (1), and then particularly in those regions where the proteins interact with the lipid head groups (2, 3). These kinds of observations together with considerations of the anchoring of gramicidin subunits (4) led to suggestions of a particular function for aromatic residues in transmembrane domains (5). In addition to general and statistical analyses of protein sequences, available membrane protein three-dimensional structures have shown the same pattern of Trp localization, in proteins such as the photosynthetic reaction center (6), the Kcsa potassium channel (7), and several bacterial porins (8).
These observations have naturally resulted in substantial interest in the biophysical characterization of the interfacial anchoring role of Trp residues. Several groups have studied compounds homologous to the tryptophan's indole side chain, which also display a distinct preference for a localization near the head group or interfacial regions of the membrane (9-13). Other recent publications employed experimental or computational simulations to examine the tryptophan-lipid interactions in detail (14-16). Considering its predominantly hydrophobic structure, the Trp indole side chain is expected to have an inherent affinity for the lipid bilayer. Additional physical or chemical characteristics are required to explain its observed distinct (and significant) preference for the membrane interface region. The small-molecule and theoretical studies have yielded a variety of possible contributing factors, without reaching a final consensus. A prominent feature is the presence of a hydrogen bond donor, in the form of the Nε-H bond, but various studies suggest that it is not the main or sole cause of anchoring. Another parameter is the rigid and bulky nature of this aromatic ring system, which might disfavor an environment of highly disordered acyl chains. Recent studies have highlighted a potentially dominant role for electrostatic interactions between the aromatic ring and the electrostatically heterogeneous lipid interface, with a particular focus on cation-π interactions between the lipid headgroups and the indole π-electron distribution (13-16). For a systematic experimental examination of the various interactions, and the roles they play in actual membrane proteins, it is important to closely mimic the lipid bilayer membrane environment and examine (model) systems that can mimic the structural variety of membrane proteins.
This paper reports on solid-state 2H NMR measurements of specifically deuterium-labeled tryptophans in macroscopically oriented peptide/lipid samples. The benefit of planar lipid bilayers is that they emulate the biological membrane more closely than detergents or lipids in non-bilayer arrangements that are often used in x-ray diffraction or solution NMR experiments (17, 18). Macroscopically aligned lipid bilayers have provided an essential means to study membrane proteins, transmembrane peptides and membrane-associated peptides by solid state NMR. Such measurements allow direct determination of molecular orientations relative to the membrane environment, and have yielded unique insights into such issues as hydrophobic matching, transmembrane helix tilt, and peptide-lipid interactions. They include the well-known PISEMA experiments (19, 20), but also a range of other, complementary methods (21-23). These methods have a proven track record, especially when combined with other experimental and theoretical approaches (see e.g. (24)). Here, we employ 2H NMR experiments on macroscopically oriented samples to study the average orientations and dynamics of tryptophans in a well-defined lipid bilayer environment. Such aligned 2H NMR experiments have previously proved successful in the examination of tryptophans in membrane-spanning channels formed by the peptide gramicidin A (25-30). The tryptophans in gramicidin channels were found to be virtually immobile relative to the peptide backbone, and the orientations of their side chains were successfully determined.
In this study we apply solid-state 2H-NMR methods to examine the behavior of tryptophans in α-helical membrane-spanning peptides. The examined peptides are part of the so-called WALP family, which is characterized by an alternating Ala-Leu core flanked by two pairs of tryptophan ‘anchors’ (31). For instance, WALP19 is a 19-residue WALP peptide with the sequence acetyl-GWW(LA)6LWWA-ethanolamide, and WALP16 is acetyl-GWW(LA)5WWA-ethanolamide. Previous studies have shown that WALP16 and WALP19 adopt strictly transmembrane orientations in model dimyristoylphosphatidylcholine (DMPC) bilayers, with the tryptophan residues located near the lipid headgroups (31-34). Those experiments also demonstrated that the DMPC bilayers remain stable in the presence of these particular WALP peptides, in contrast to the situation of other (longer) lipids that are induced to form non-bilayer phases by the presence of these peptides (31, 34, 35). Additionally, WALP16 and WALP19 have been studied in bilayers formed by the corresponding ether-linked lipid, ditetradecyl phosphatidylcholine. Interestingly, the results show little difference in the indole behavior in these two systems. In each lipid, the tryptophans maintain certain preferred (average) orientations, but simultaneously experience significant motion. The results indicate a notable difference between the behavior of tryptophans near the C-terminus compared to those near the N-terminus. The similar dynamics in ester- and ether-linked lipid bilayers suggest that the lipid carbonyl groups are non-essential for the anchoring of the Trp's to the lipid head groups and the resulting stabilization of the membrane spanning peptides in the bilayer.
Materials and Methods
Materials
Dimyristoylphosphatidylcholine (DMPC) and ditetradecylphosphatidylcholine (DTPC) were purchased from Avanti Polar Lipids (Alabaster, AL). Deuterated trifluoroacetic acid (CF3COOD), deuterium depleted water, ring-deuterated tryptophan (Trp-d5), and D2O were purchased from Cambridge Isotope Laboratories (Andover, MA). Unlabeled Fmoc-protected amino acids and peptide resins were from Advanced ChemTech (Louisville, KY) and NovaBiochem (San Diego, CA). 2,2,2-Trifluoroethanol was obtained from J.T. Baker (Phillipsburg, NJ).
Peptide synthesis
The WALP16 (n=5) and WALP19 peptides (n=6.5), of sequence acetyl-GWW(LA)nWWA-ethanolamine, were synthesized by solid-phase FastMoc® methods with a singly labeled at each of the four Trp positions. Fully ring-labeled tryptophans were purchased as Trp-d5, with deuterons on all five available carbons of the indole ring, and were manually protected with an N-terminal Fmoc group (36). Selectively deuterated tryptophans were obtained by 1H-2H exchange of unlabeled Fmoc-Trp with deuterated CF3COOD at 4 °C, as described by Koeppe et al. (28). This results in the predominant deuteration of the indole C2 (70% 2H) and C5 (20% 2H) positions2. A single labeled Fmoc-Trp-dx was incorporated by FastMoc® chemistry as described previously (36, 37), using 2-fold excess of the labeled residue followed by an extra coupling step with 5-fold excess of unlabeled Fmoc-Trp, giving 95% deuteration. The purity of the resulting peptides was checked by reversed phase HPLC, yielding a typical purity of 90%, and their identity was confirmed by electrospray ionization mass spectrometry (see also Supporting Information). The over 95% inclusion of the deuterated Trp-d5 was observed in the mass spectrometry data as a mass gain of 5 compared to the equivalent unlabeled peptide.
Oriented NMR sample preparation
The preparation of highly aligned NMR samples is essential for optimal experimental results(38, 39), and has been optimized for these samples as described previously (34). The samples consist of 4 μmol peptide and 80 μmol DMPC in aligned bilayers distributed among approximately 40 glass cover slides. The samples were hydrated with deuterium-depleted water to achieve 40% (w/w) overall hydration, ensuring fully hydrated bilayers (40, 41), followed by stacking of the glass slides with application of gentle pressure to encourage alignment, and finally packed in a sealed container to prevent dehydration. For optimal alignment the samples were allowed to equilibrate at 40 °C prior to use, for at least 48 hours. In addition to 31P-NMR tests of lipid alignment and integrity (see below), some aligned samples were also checked for chemical integrity using analytical HPLC chromatography and mass spectrometry (see Supporting Information).
NMR measurements
The proper alignment of the lipid bilayers was confirmed using phosphorus NMR experiments as described by Van der Wel et al. (34). Oriented 2H NMR measurements were used to study the tryptophan conformations (25, 34). Both the 31P and the 2H measurements were performed with the samples oriented with the normal to the glass slides (and thus to the membranes) either along the magnetic field direction (β=0°) or perpendicular to it (β=90°), as illustrated in the Supporting Information. Unless indicated otherwise, the NMR experiments were performed at a sample temperature of 40°C, using a Bruker AMX2 300 spectrometer. The experimental temperature and hydration levels were maintained to ensure that the DMPC and DTPC lipids are above their respective gel-to-liquid crystalline transition temperatures of 23.1 and 26.2 °C (42). The 2H measurements involved 1 to 2 million acquisitions and were done using a quadrupolar echo pulse sequence (43) with an echo delay of 30-75 μs, a 3 μs pulse length, and a 30 ms interpulse time. A 100 Hz line broadening was applied to both the 2H and 31P spectra.
Data analysis
The observed 2H NMR doublets reflect the quadrupolar splittings for the C-D bonds, which are correlated to their orientation relative to the magnetic field according to the following equation:
The angles θ and φ specify the orientation of the indole C-D bond within the magnetic field, where θ is the angle between the bond and the magnetic field z-axis and φ the angle between the indole ring plane and the magnetic field (also see ref. (29)). (e2qQ/h) is the quadrupolar coupling constant (QCC), which is 180 kHz for aromatic C-D bonds, in the absence of motion (44). The parameter η is the asymmetry parameter, indicating any deviation from axial symmetry for electric field gradient of the C-D bond. Motion of the indole rings will result in reduction of the observed quadrupolar splittings due to motional averaging. In the case of isotropic motion, this can be represented by a reduction of the order parameter S to a value less than 1. (Note that this is equivalent to using an ‘effective’ quadrupolar coupling constant (QCCeff) that is reduced relative to the ‘static’ value.) Such an isotropic motional model has been used successfully in studies of the tryptophans of gramicidin A (28) and for deuterated alanines in various WALP peptides (34, 45, 46). These studies demonstrated that the method can be very sensitive to the exact orientation and conformation of the deuterated amino acid side chains. Recent work on tryptophan deuterium NMR has yielded a refined indole ring geometry and a calculation of the electric field gradient (EFG) tensor elements for each C-D bond using 3-methyl-indole as a model compound (28, 29). This indole structure showed a number of deviations from previously used idealized indole rings, including a non-90° angle of the C2-H bond with the central ‘bridging’ C8-C9 bond, and a slight deviation of the angle between the C4-2H and C7-2H bonds from 180°. The latter deuterons yield virtually identical quadrupolar splittings that may however appear ‘twinned’ due to their slight deviation from a 180° relative orientation. (The calculated EFG elements also suggested a significant non-zero asymmetery parameter.) Thus, one can expect the deuterons attached to carbons C-2, C-4/7, C-5, and C-6 typically to have four distinct quadrupolar splittings, resulting in four different doublets (25, 30).
The assignment of the peaks to each of the deuterons is here partially facilitated by use of the selectively labeled tryptophans. A computer program was developed, based on extensions of previous methods used for the analysis of the gramicidin tryptophans (25). The analysis relies on the reorientation of the indole ring, combined with a simulation of motion through a reduced isotropic order parameter, to calculate a predicted spectrum and match it to the observed spectrum. Since in many cases we observe significant overlap of signals, resulting in fewer doublets than the number of labeled C-D bonds, the program examines and compares all possible assignment patterns, including multiple bonds assigned to a single doublet. A standard α-helical peptide model as produced by the InsightII (Accelrys Inc., Princeton NJ) molecular modeling program was either aligned with the membrane normal or tilted by 4° to the specific orientation found previously for WALP19 (34). It was found that the InsightII default Trp residues deviated significantly from our determined planar indole geometry. For this reason, the structures were modified to reflect the indole geometry determined previously (28). Based on this peptide model, the orientation of a Trp side chain was defined by the χ1 and χ2 side-chain torsion angles. For each (χ1, χ2) combination, the ring C-D bond orientations were determined, yielding the corresponding quadrupolar splittings (at β=0°) according to equation (1). The differences between the observed and calculated |Δvq|'s are expressed as the root mean square deviation (RMSD), in order to determine the best simulation parameters.
Recent 2H NMR experiments with labeled alanines have shown that WALP19 is tilted in the DMPC bilayer by about 4°, in a specific direction (34). In the calculations presented here we used a structural model that was tilted accordingly. Due to the small tilt angle, one would expect only small deviations in the results. Indeed, reference calculations with an untilted model yielded closely similar results, with virtually identical Trp dynamics and only minor variations in (χ1, χ2) angles corresponding to the suggested orientations (results not shown).
Peptide structural considerations
Two structural considerations affect the range of accessible Trp orientations. First, the indole ring tends to orient itself such that its hydrophobic part is closer to the membrane's hydrophobic center and the Nε-H bond points toward the lipid head groups, where it can form hydrogen bonds. In the particular case of an α-helix, these tendencies affect the tryptophans near the two ends of the helix quite differently, due to the directionality of the helix and the Cα-Cβ bond direction. As illustrated in Figure 1A, different (χ1/ χ2) combinations lead to optimal alignments toward the respective bilayer surfaces. In addition, including the peptide backbone in our simulations allowed us to exclude side chain orientations that would place the ring in steric conflict with the α-helical peptide backbone. Figure 1B and 1C indicate the excluded (χ1, χ2) side chain orientations, showing a difference between the N- and C-terminal tryptophans. The latter were found to experience fewer steric restrictions due to the backbone (a result of the Cα-Cβ bond direction and their close proximity to the end of the peptide). Examinations of existing protein structures also reveal that specific residues tend to exhibit only restricted sets of (χ1, χ2) combinations (or rotamers). Statistical analysis of large numbers of protein structures has yielded rotamer libraries. Lovell et al. (47) found that of 140 ‘α-helical’ tryptophans, 55% had a χ1 value of near −177° (or 183°), and 37% had χ1 near −65° (or 295°). Black dots in Figure 1B and 1C indicate the locations of the backbone-independent rotamers as generated by Dunbrack (48). Backbone-based steric considerations have led to the formation of backbone dependent rotamer libraries (48), which conform to the data shown here. The white areas in Figures 1B and 1C show that one can anticipate the Trp side chain orientations at either end of the peptide to be restricted within distinct subsections of the (χ1/χ2) space.
Figure 1.

Restrictions on Trp orientations. Panel (A) shows the dependence of indole orientation on the χ1 and χ2 angles. The respective areas indicate those side chain orientations in which the indole's Nε-H bond is pointing toward the N-terminus or C-terminus of the α-helix. Panels (B) and (C) show the steric hindrance that the side chain encounters with the backbone, for the Trps at the N-terminus and at the C-terminus, respectively. Dashed areas indicate an unfavorable steric hindrance, defined here as less than 2 Å distance between any pair of atoms. Black dots indicate common rotamer conformations as indicated by Dunbrack (48).
Results
NMR experiments
The alignment of the lipids in the NMR samples was checked using 31P NMR. The sample preparation method employed (34) yields lipids in well-aligned bilayers between the glass supports, with approximately 90% of the lipids in an aligned conformation. The presence of 5 mole-% WALP16 and WALP19 peptides did not disturb the alignment, yielding 31P spectra indicative of highly aligned lipid bilayers (see ref. (34) and the Supporting Information). Optimal alignment was generally accompanied by a visual transparency of the stacked samples.
Subsequently, deuterium NMR spectra were recorded to examine the 2H labeled peptides imbedded in the aligned DMPC bilayers. Figure 2 shows the spectra obtained for WALP19 peptides containing a deuterium (d5) labeled tryptophan at position 2, 3, 17, or 18, measured at sample orientations of β=0° (left column in figure) and 90° (right column). These experiments were recorded at a temperature of 40°C, where the lipids are in the liquid crystalline phase. Under these conditions, the peptides undergo rapid axial reorientation, resulting in a 50% reduction in the quadrupolar splittings at β=90° compared to β=0°, as was seen previously (34). The expected four (or under optimal conditions, five) splittings cannot be distinguished in all of the spectra. This can be due to overlap of multiple peaks, or to low intensities for certain signals with large splittings. The former problem is most severe at β=90°, whereas the latter mostly affects measurements performed at β=0°. Measuring at both orientations therefore provides complementary information. Samples were also prepared using the somewhat shorter WALP16 peptide. The spectra for WALP16 are illustrated in Figure 3. The observed quadrupolar splittings for both peptides are listed in Table 1.
Figure 2.
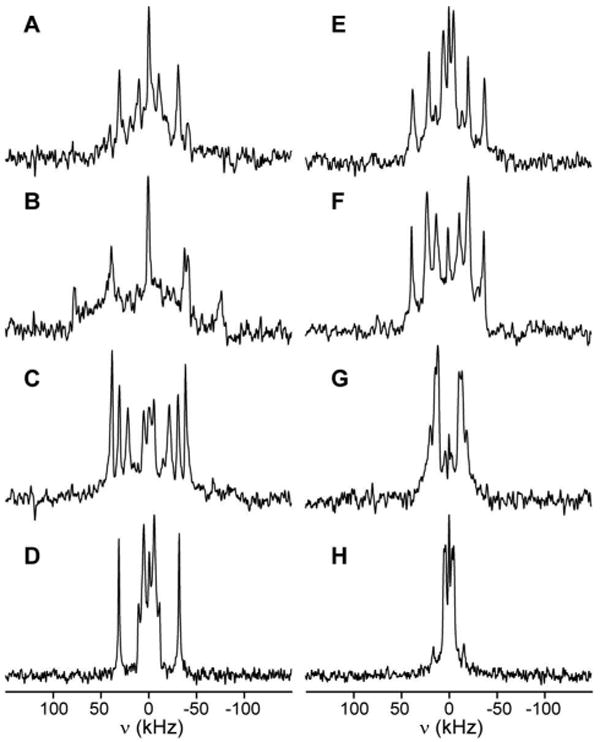
Deuterium NMR spectra for Trp-d5 labeled peptides WALP19-Wx-d5 in oriented DMPC bilayers (1:20 peptide/lipid ratio). Measurements were done at 40% (w/w) hydration and 40 °C experimental temperature. Spectra obtained at β=0° and 90° are shown in the left and right columns, respectively. The label positions are, from top to bottom, W2 in spectra A,E; W3 in spectra B,F; W17 in spectra C,G; and W18 in spectra D,H.
Figure 3.

Deuterium NMR spectra for Trp-d5 labeled peptides WALP16-Wx-d5 in oriented DMPC bilayers (1:20 peptide/lipid ratio). Measurements were done at 40% (w/w) hydration and 40 °C experimental temperature. Spectra obtained at β=0° and 90° are shown in the left and right columns, respectively. The label positions are, from top to bottom, W2 in spectra A,E; W3 in spectra B,F; W14 in spectra C,G; and W15 in spectra D,H.
Table 1.
Quadrupolar splittings, in kHz, of Trp-d5 labeled WALP16 and WALP19 in model membranes at β=0° and 90°, at 40 °C. WALP16 was studied in DMPC and WALP19 in DMPC and its ester-linked analog DTPC. The experimental errors in the measured quadrupolar splittings range from 1-2 kHz.
| labeled residue | WALP16/DMPC | WALP19/DMPC | WALP19/DTPC | |||
|---|---|---|---|---|---|---|
| β = 0° | β = 90° | β = 0° | β = 90° | β = 0° | β = 90° | |
| Trp-2 | 74 | 155 | 75 | 71 | ||
| 80 | 41 | 76 | 38 | 69 | 36 | |
| 60 | 28 | 45 | 28 | 14 | ||
| 26 | 38 | 11 | ||||
|
| ||||||
| Trp-3 | 68 | 155 | 78 | 140 | 70 | |
| 117 | 59 | 85 | 41 | 87 | 42 | |
| 97 | 53 | 26 | 42 | 21 | ||
| 56 | 27 | 20 | ||||
| 27 | 14 | |||||
|
| ||||||
| Trp-14/17 | 70 | 77 | 38 | 72 | 34 | |
| 62 | 29 | 62 | 28 | 42 | 21 | |
| 52 | 44 | 23 | 29 | |||
| 28 | 13 | 11 | 6 | |||
|
| ||||||
| Trp-15/18 | 73 | 36 | 63 | 32 | 67 | 33 |
| 35 | 22 | 10 | 24 | 12 | ||
| 21 | 8 | 11 | 7 | 13 | 6 | |
| 4 | ||||||
Interestingly, in both WALP16 and WALP19 the spectra of the Trps near the N-terminus are systematically different from those near the C-terminus, both in signal intensity and in the magnitudes of the observed quadrupolar splittings. Under identical conditions, Trps 2 and 3 exhibit consistently lower intensity signals. Such intensity differences could be a result of dynamics in the motional regime where the motional exchange reduces the deuterium signal intensity. To evaluate this possibility, the same samples were also measured at elevated temperatures. As the temperature increases, we observed significant increases in signal intensity for the N-terminal residues. The spectra for Trp2 in both peptides are shown in Figure 4. The observed changes in intensity from 40°C to 60°C are consistent with a range of motional exchange of the C-D bond of 105-108 s-1 in this temperature range. The increase in motion at higher temperatures also results in a consistent reduction of all observed quadrupolar splittings, as shown in Table 2 for Trps 2 and 3 of WALP19. This is consistent with an increase in the range of motion of the indole rings as the temperature increases.
Figure 4.
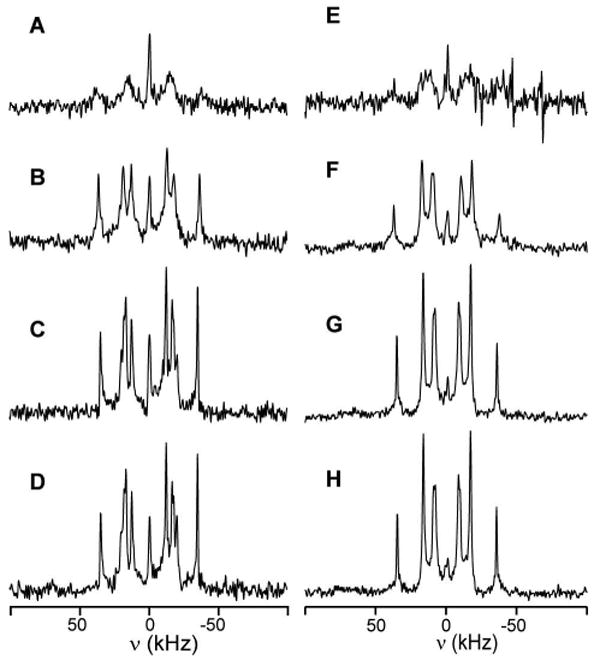
Temperature dependence of the 2H NMR spectra of WALP16-W2-d5 (left) and WALP19-W2-d5 (right). The experimental temperatures are 40, 50, 60, and 65°C, from top to bottom. All spectra are based on 800,000 accumulated free-induction decays, measured at β=90°. The central peaks indicate a residual amount of 2H-containing water molecules in these samples.
Table 2.
Overview of quadrupolar splittings of WALP19-W2-d5 and WALP19-W3-d5 in DMPC at varying temperatures. Sample orientations are at β=0°.
| Temperature (°C) | Trp-2 | Trp-3 | |||||
|---|---|---|---|---|---|---|---|
| 40 | 76 | 45 | 153 | 88 | 53 | ||
| 50 | 149 | 72 | 43 | 37 | 146 | 84 | 51 |
| 60 | 142 | 69 | 38 | 33 | 143 | 82 | 48 |
| 65 | 142 | 68 | 38 | 33 | -a | -a | -a |
This sample was not measured at 65°C.
Similarly, the difference in motion between the C-terminal and N-terminal Trps is also reflected in very different overall widths of the 2H spectra. To express this difference, we calculated the maximum observed quadrupolar splitting for each residue. Figure 5 compares the maximum splittings observed for the different tryptophans studied here, together with the results of previous studies on highly mobile deuterated indole rings in lipid bilayers (12, 13), and on highly restricted tryptophans of gramicidin A (25, 27). The “free” indole rings—not linked to any peptide—show spectra with a relatively small maximum quadrupolar splitting, indicating a residual spontaneous orientation of these highly mobile rings within the membrane. At the other extreme of motional restriction, the tryptophans of gramicidin A channels have significantly larger maximum quadrupolar splittings. Interestingly, Trp9 of gramicidin A is closest to the bilayer center and exhibits a slightly smaller maximum quadrupolar splitting compared to the other gramicidin Trps. Remarkably, Trp9 becomes slightly more motionally restricted when a fatty acyl chain is esterified to the C-terminal aminoethanol group (Figure 5), presumably reflecting increased steric interactions as a result of the acylation. Within this framework, the Trps in the WALP16 and WALP19 peptides seem to experience intermediate motion between the extremes of unattached indole and gramicidin A. Striking is also the clear and consistent difference between the ‘N-terminal’ and ‘C-terminal’ Trps of the two WALP peptides.
Figure 5.
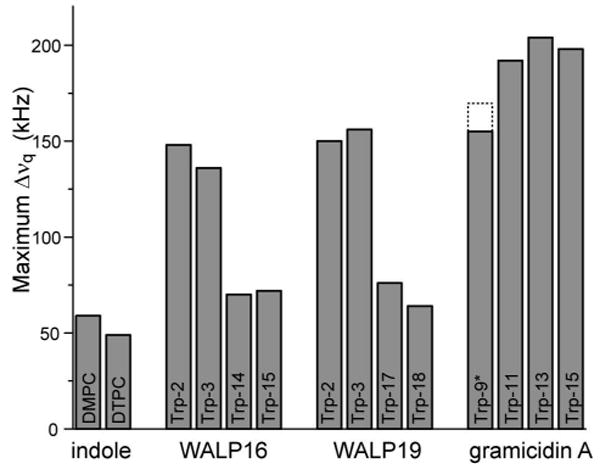
Maximum quadrupolar splittings observed for different indoles. From left to right: ‘free’ indole in DMPC and DTPC (from ref. (12)); the tryptophans of WALP16, WALP19, and gramicidin A, respectively. (*) The dotted lines indicate the decreased motion of Trp9 that accompanies the acylation of gramicidin A (26).
Ether-linked lipids
To investigate the role of the lipid head group structure, WALP19 was also incorporated into bilayers of ditetradecyl phosphatidylcholine (DTPC), the ether-linked analog of DMPC. 31P-NMR spectra showed that these lipids form well-aligned bilayers similar to the DMPC samples. Remarkably, the 2H NMR spectra of Trp-d5 labeled WALP19 peptides in this lipid are very similar to those obtained in DMPC (Figure 6, Table 1). This finding indicates that at least the dynamics of the indoles is largely unaffected by the change in the lipid head group. Due to the extent of overlap it is hard to establish with certainty how the assignments of the observed splittings relate to one another, and therefore how the average orientations of the indole rings may change. We have examined the minimal orientational change required to account for the changed peak positions, if we assume the peak assignments to remain the same. Using estimated motional averaging based on the WALP19/DMPC results (see below), the changes in bond orientations are found to approximate the variation due to experimental error (a Δνq change of 1-2 kHz, equivalent to typical angular deviations of 0.5-4 degrees) for at least some of the residues (see Supporting Information). These results show that the indole ring dynamics (and possibly orientation) are remarkably insensitive to the removal of the carbonyl groups from the lipid headgroups, despite their apparent proximity to this part of the membrane (32).
Figure 6.

2H NMR spectra of ether-linked DTPC bilayers containing WALP19-Wx-d5 peptides. Measurements were done at 40°C, and 1/20 peptide/lipid ratio. Sample orientations of β=0° and 90° are shown in the left and right columns, respectively. The label positions are, from top to bottom, W2 in spectra A,E; W3 in spectra B,F; W17 in spectra C,G; and W18 in spectra D,H.
Orientational analysis
In Figure 5 we already referred to the similar studies on gramicidin A tryptophans and free indoles. In gramicidin A, the tryptophans were found to experience relatively little motion, which allowed accurate simulations of the 2H NMR data using a fairly simple motional model. Conversely, in the case of the membrane-imbedded free indoles, such a quantitative analysis of the indole ring alignment is impossible due to the unknown nature of the ring motion. The present results suggest that motion of the tryptophans in the α-helical WALP peptides is in an intermediate regime. It is unclear whether the approximation of isotropic motional averaging (reasonably successful for the gA tryptophans, which exhibit little motion) will suffice in cases of more pronounced motion of both the peptide and its Trp side chains, as indicated for the WALP tryptophans. In the next sections, we attempt to apply similar methods to WALP19. For this peptide we have the benefit of detailed information on its orientation within the bilayer (34), and the availability of a slightly ‘better’ data set with more identifiable peaks.
Selective labeling
To aid in the peak assignment required for a more quantitative analysis, we selectively labeled tryptophan side chains. 2H NMR measurements of four WALP19 peptides containing a selectively labeled Trp are shown in Figure 7, at β=90° sample orientation. Based on the temperature-dependent data shown above, these peptides with the label at sequence position 2 or 3 were measured at higher temperatures to improve the signal:noise ratio. In each case, we observe one dominant doublet, for which the quadrupolar splitting is attributed to a deuteron at the ring C-2 position. In three cases, a weaker second doublet is seen, due to the partial incorporation of a deuteron at the C-5 position. These data allowed partial assignment of the observed doublets.
Figure 7.
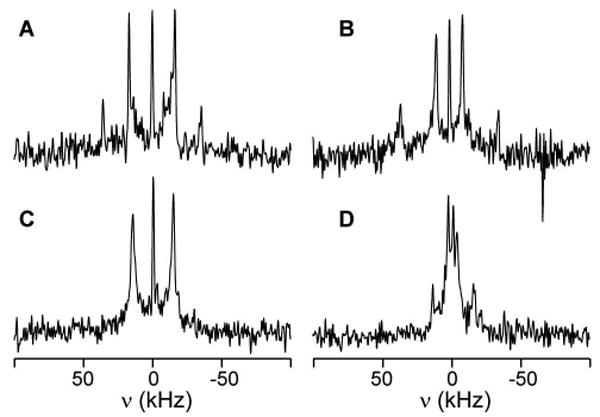
Deuterium NMR spectra for peptides with selectively labeled tryptophans, which are deuterated primarily at indole positions 2 and 5. The samples consist of WALP19 in DMPC, with a selectively labeled Trp at sequence position 2 (A), 3 (B), 17 (C), or 18 (D). Spectra shown were obtained at β=90° and a temperature of 40°C (C&D) or 60°C (A&B).
Motion of the tryptophans
Using assignments for those peaks which could be identified by selective labeling, we performed a series of calculations based on a model which assumes isotropic averaging. This procedure involved systematic calculations of the deuterium quadrupolar splittings at all possible indole ring orientations. Due to the small number of available data points for each indole ring, molecular motion was—of necessity—treated in a highly approximate manner. For the initial modeling, we assumed a simplistic partial isotropic averaging, expressed as an order parameter of less than 1.0. Within this scheme, the calculated sets of quadrupolar splittings were compared to the observed values, evaluating all possible assignment patterns consistent with the selective labeling experiments. The minimum error that could be achieved for each Trp as a function of the isotropic order parameter is shown in Figure 8. At 40°C, the minima for the tryptophans at position 2 and 3 were found to be near an order parameter of 0.75, while tryptophans 17 and 18 had order parameters between 0.5 and 0.6. In spite of the simplified motion assumptions, these relative motion parameters are consistent with the qualitative observations discussed above, indicating that all of the WALP indole rings experience significant amounts of motion, and that those near the C-terminus are most mobile. The effect of higher temperatures is shown in panels C and D, indicating a decrease of the optimal order parameter to 0.60 for Trp-2 at 65°C, and to 0.68 for Trp-3 at 60°C.
Figure 8.
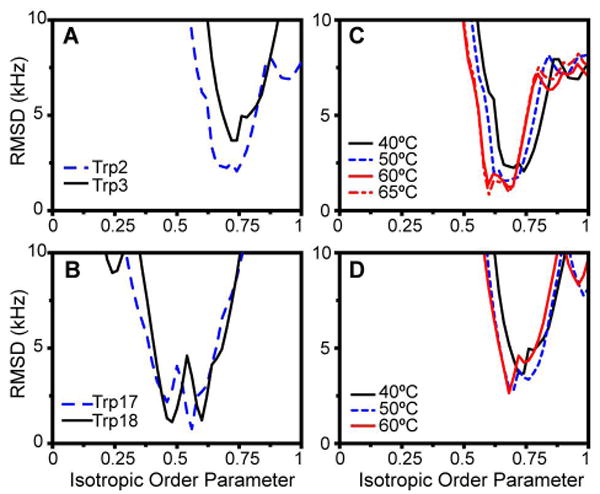
Variation of RMSD error with the isotropic order parameter used in the fitting procedure for WALP19 in DMPC. The figure shows the RMSD associated with the best fit for each order parameter value, for Trp-2 (blue) and Trp-3 (black) (panel A), and Trp-17 (blue) and Trp-18 (black). Panels (C) and (D) show the decrease in order parameter at (50-65°C) on Trp-2 and -3.
Table 3 lists the estimated average angles that the various assigned C-D bonds would have based on the assumed optimal motion averaging. When Trp-2 and Trp-3 near the N-terminus are compared, the corresponding values are very similar (within 5 degrees). The residues near the C-terminus seem more distinctly different from each other. Finding the side chain orientations (χ1,χ2) that are consistent with these angles (or more precisely, with the observed quadrupolar splittings) is perhaps more insightful (especially considering the inherent degeneracy of the single C-D bond orientations listed in Table 3). At each of the optimal isotropic order parameters, one can map out the RMSD for the (χ1,χ2) combinations, to locate the side chain orientations providing the minimum RMSD values. For each of the minima in the charts in Figure 8, such a contour plot is shown in Figure 9. Areas known to exhibit steric hindrance due to the backbone are excluded (see also Figure 1). Those orientations that are both consistent with proper alignment of the indole ring N-H group and have an equivalently low RMSD are indicated with arrows. Typical RMSD values for these minima are 1.0 to 3.4 kHz. Note that the experimental errors in the measured splittings are 1-2 kHz in value (at any given temperature). In many cases the optimal fits include deviations of one or more particular quadrupolar splittings that significantly exceed the experimental error. The latter finding contrasts with the high sensitivity and accuracy of similar previous deuterium-based experiments (28, 29, 34). An additional problem is the presence of various equivalently good solutions that cannot be sufficiently distinguished based on the present data. The available data combined with the isotropic motion model turn out to be insufficient to reliably and accurately determine the behavior of the Trp indole rings in WALP peptides. We suggest that this is predominantly due to a lack of detailed knowledge of the peptide dynamics. If this is indeed the case, the actual averaged angular values (and their associated errors) could deviate from the values estimated from the basic isotropic model (e.g. those listed in Table 3). We have explored a number of different, more advanced models with more explicit simulations of the Trp side chain dynamics, including modest excursions of the side chain around a single χ1/χ2 orientation and calculations of the averaged splittings that would result from larger jumps between the accessible rotamer orientations for the Trp side chain (instead of the assumption of an isotropic reduction of the splittings). Unfortunately, none of these approaches provided significant improvement to the fits between the calculated and measured data. In the absence of further experimental information, the application of complex models would also be hard to verify.
Table 3.
Assigned quadrupolar splittings from WALP19 peptides in DMPC at 40°C, based on results for selectively labeled tryptophans. The last column lists the averaged θ angles between the C-D bond and the membrane normal (assuming isotropic order parameters of 0.75 (Trp-2&3) and 0.55 (Trp-17&18), along with the error associated with an estimated experimental error of 2 kHz in Δνq.
| Labeled Trp | Bond C-2 | Bond C-5 | ||
|---|---|---|---|---|
| Δνq (kHz) | θ (°) | Δνq (kHz) | θ (°) | |
| Trp-2 | 76 | 40.2 ± 0.4; 73.2 ± 0.7 | 155 | 23.3 ± 0.5 |
| Trp-3 | 53 | 44.6 ± 0.4; 66.5 ± 0.5 | 155 | 23.3 ± 0.5 |
| Trp-17 | 62 | 38.5 ± 0.5; 76.4 ± 1.1 | a | a |
| Trp-18 | 11 | 51.8 ± 0.5, 57.8 ± 0.6 | 63 | 38.3 ± 0.5; 77.0 ± 1.2 |
No weaker C-5 peak could be identified for Trp-17.
Figure 9.
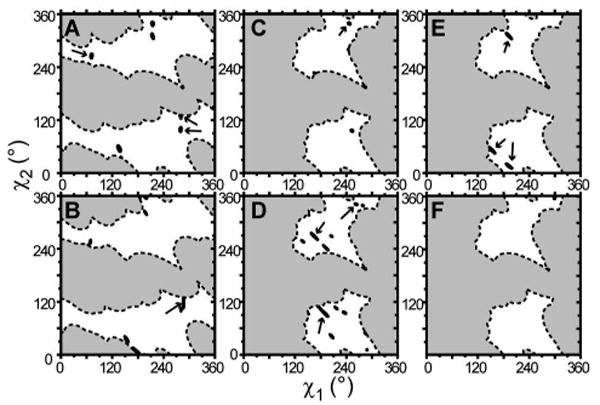
Contour plots from data-fit of WALP19 in DMPC. The panels show the error (RMSD) over the χ1- χ2 range accessible to each Trp, at the optimum isotropic order parameters S=0.74 for Trp-2 (A), 0.73 for Trp-3 (B), 0.46 and 0.56 for Trp-17 (C,D), and 0.48 and 0.60 for Trp-18 (E,F). Arrows indicate orientations with the lowest RMSD (below 2.5 kHz) and having the proper Nε-H orientation (for Trp-3 the best solution with RMSD = 3.4 kHz is indicated).
Discussion
Solid-state deuterium NMR spectroscopy was used to examine the behavior of the tryptophan interfacial anchors of α-helical peptides spanning model lipid bilayer membranes. Similar to what was observed previously in the β-helical peptide gramicidin A, each of the tryptophans was found to be have distinct spectral characteristics. The narrowness of the peaks indicates that the ring motions must be fast on the NMR time scale. The overall magnitudes of the quadrupolar splittings for all WALP Trp residues are intermediate between what was observed for the cases of the essential rigidity of the gramicidin tryptophans and the weakly aligned state of unattached indoles or indole analogs in lipid bilayers (Figure 5). In addition to its inherently larger motion as a whole, the α-helical backbone allows more mobility for the individual side chains than the gramicidin β6.3 helix. Still, the Trp ring motions in these WALP peptides are highly reduced relative to the unconnected (peptide-free) indoles, especially for the residues near the N-terminus consistent with a much more specific alignment of the indoles relative to the lipid bilayer environment.
Helix direction
Under most conditions we find narrow peaks, indicating fast ring dynamics on the NMR time scale. However, the reduced intensities for the N-terminal Trps near 40 °C suggest that their motions are near the intermediate exchange regime (near 105-106 s-1). This is the case for both the WALP16 and WALP19 tryptophans. Another indication of different dynamics between tryptophans at the respective ends of these helices is found in their average quadrupolar splittings. The larger average splittings for the N-terminal residues suggest that these Trp indole rings are experiencing a smaller extent of motion than their counterparts at the opposite end of the peptide.
The observed difference in spectral properties between Trps at the N- and C-terminal ends of an α-helix is not completely unexpected. Even though these WALP peptides have primary sequences that appear quite ‘symmetric,’ the structures of amino acids and of the assembled α-helix are inherently and distinctly directional. The directional feature is for instance indicated by the helix dipole moment and is also seen in the “arrowhead-like” orientations of the side chains along the helix axis. The backbone structure of α-helical peptides dictates that the Cα-Cβ side chain bonds make an angle of ∼58° with the helix axis, measured with respect to the N-terminus (34). This Cα-Cβ bond orientation causes the Trps closer to the C-terminus to require different χ1 and χ2 angles, compared to the Trps near the N-terminus (Figure 1A), in order to direct each of the indole N-H bonds toward a water/membrane interface.
Another consequence of the inequivalence of the two termini is reflected in the level of steric hindrance that the indole rings experience (Figure 1). It seems somewhat paradoxical that the Trps at positions 2 and 3 are less restricted by their peptide environment, yet experience smaller and slower motions compared to the tryptophans near the C-terminus. Apparently the respective (χ1, χ2) angles and the indole/lipid anchoring interactions are compatible with more motional freedom for C-terminal Trps than for N-terminal Trps in these α-helices. The lipid interactions and anchoring roles of these residues could be responsible for the motional differences.
Motional complexity
Despite the partial labeling and extensive modeling of the system and the data, it is not yet possible to predict a single, averaged ring orientation. We feel this situation is due predominantly to uncertainty concerning the precise extent and type of motions that the tryptophans undergo. Within the highly dynamic lipid bilayer, these residues experience combinations of simultaneous motions, including an overall ‘wobble’ of the peptide and rotations about the χ1 and χ2 side chain torsion angles. Considering the anchoring role of the tryptophans, there is likely to be intricate interplay between these various types of motion within the peptide, which could very well be linked to the dynamics of the lipids.
Disentangling the diverse modes of motion for each labeled tryptophan is not trivial. The existing data set could be enhanced by the availability of other experimental parameters. In the case of gramicidin, 15N NMR data proved useful for determining the indole orientations and were for instance able to indicate that the (smaller) librational motions of those tryptophans occur mostly about χ2 rather than about χ1 (27). Compared to the gA analysis, the present situation is also complicated by the extensive overlap of the more averaged quadrupolar splittings, making an analysis in the absence of complete assignments difficult.
Our results highlight the structural differences between the steric environments of the β-helical gramicidin channel and the α-helical WALP transmembrane segments. Extrapolation to the general membrane-spanning domains of α-helical membrane proteins should be done with caution since the tryptophans near the N-terminus could be influenced by their close proximity to the end of the peptide. Figure 1B and C show that, compared to tryptophans in the center of an α-helix (or near the C-terminus), the side chains of both tryptophans near the N-termini of these WALP peptides experience much less steric hindrance from the rest of the peptide. In a longer peptide, or a complete membrane protein, the helix would typically continue beyond the bilayer, resulting in a more restricted local environment. It is possible that such changes in environment would change the observed Trp orientations, in particular for the N-terminal residues.
Considerations for interfacial anchoring
The main purpose for this study has been to examine the functioning of tryptophans as membrane anchors, as reflected in their orientations and motion. Molecular dynamics (MD) simulations can provide useful insights into the physical principles involved, especially when complementing experimental NMR results (24). Recent MD simulations of membrane-imbedded ‘KALP’ peptides have examined the tilting and interfacial anchoring of analogous α-helical peptides with lysine residues as interfacial anchors (49, 50). These studies highlighted the flexible nature of the Lys anchors and suggested the presence of extensive motions of the α-helical peptides as a whole. It was suggested that the ‘snorkeling’ lysines may interact specifically with the lipids, thereby restricting the direction of tilt but still allowing fluctuations in the extent of tilt (49). At the same time, the Lys side chains appeared to ‘follow’ the fluctuations of the highly mobile surrounding lipids.
For the more bulky and more rigid tryptophans, the (χ1, χ2) space that avoids steric clashes with the rest of the α-helix is significantly more restricted than for lysine. The steric restrictions should result in a decreased Trp mobility, consistent with earlier MD simulations on WALP peptides (51). A lower Trp mobility was also indicated by earlier experiments where Trp interfacial anchors instilled a peptide with a more precisely defined anchoring depth than the (more flexible) Lys counterparts (52). In contrast with previous gramicidin data, we observe significant mobility of Trp interfacial anchors in the WALP peptides. The gramicidin A tryptophans experience restrictions due to steric interactions with nearby residues (53) as well as a very immobile backbone structure (54). This situation contrasts with the WALP peptides, where the Trps might have a ‘buffering’ effect on the motions of the peptides within the highly dynamic membrane environment. In a fashion analogous to the observations on the Lys-anchored peptides, the anchoring Trp residues could partially ‘follow’ the fluctuations of the lipid headgroup component to which they are anchoring. Unfortunately, these complex, non-isotropic motions do prevent a detailed analysis of the reported NMR spectral results in terms of specific side chain orientations, as was possible for the more rigid gramicidin channels. Nevertheless, a simplified approximation of the motion does provide an initial estimate of the dynamics.
Within these limitations, we have found that 2H-NMR measurements do provide a convenient and sensitive probe of the tryptophan-lipid interaction, as demonstrated by the comparison of DMPC and DTPC membranes, the latter lacking the lipid carbonyl groups. Despite the significant chemical difference between the ester and ether lipids, it has been found that bilayers comprised of DTPC are structurally similar to DMPC bilayers. One notable difference is that the dipole moment across the head group region is 100 mV smaller in ether lipid bilayers compared to DMPC (55). Our data suggest that hydrogen bonding of the Trp Nε–H groups to the lipid carbonyls is unlikely to constitute a dominant factor for the indole ring behavior, despite the fact that indoles are typically found in close proximity to the carbonyl groups (11, 32, 56). The hydrogen bonding acceptors for the indole Nε-H bonds could alternatively be the lipid's phosphate oxygens and/or the water molecules that hydrate the head groups. It also appears that the change in dipole moment does not seriously affect the tryptophan behavior, arguing against a central role for specific dipole-dipole interactions between the indole rings and the lipid head groups. Our data correspond well to the previously published study of ‘free’ indoles in DMPC and DTPC, where the Nε–H hydrogen bonding represented only a small contribution to the molecular ordering observed for the aromatic rings (12). One notable difference with this previous study is that with peptide-attached indoles we do not observe a reduction in molecular ordering upon removal of the lipid carbonyl groups. This finding is likely related directly to the covalent connection to the ‘fixed’ peptide backbone and the concomitant restriction on the conformational space available to the indole ring (Figure 1). Our observations are consistent with the recent studies (13-16) suggesting that electrostatic interactions (including cation-π interactions) could be among the prominent anchoring mechanisms. Contributions from Nε-H hydrogen bonding would appear to involve partners farther from the bilayer center than the acyl chain ester groups, consistent with the observed indole ring location within the interface (32).
Overall, we conclude that the interfacial Trp residues have a strong tendency to adopt specific (averaged) orientations that are different at the N- and C-terminal ends of WALP19 and WALP16. Presumably, the Trp indole rings fulfill specific interactions with the lipid interfacial region, but their behavior is strongly modulated by the local peptide context. Interestingly, while the steric interactions suggest a somewhat decreased steric hindrance from the local peptide backbone, the Trp side chains near the N-terminus appear to be less mobile than those near the C-terminus. Comparing the 16-residue and 19-residue peptides, indoles at comparable positions relative to the membrane surface adopt similar average orientations and have similar mobilities. This feature might reflect the inherent directional structure of an α-helix, in which the Cα-Cβ bonds are fixed in an orientation that points toward the N-terminus, in an angle of ∼58° (34). This bond orientation inherently directs the indole rings near the N-terminal end of a transmembrane helix in the direction of the lipid bilayer ‘surface.’ By contrast, indoles near the C-terminal must reorient to achieve optimal orientations of the Nε-H groups. The proper alignment of the indole rings appears to be the basis for the anchoring characteristics of tryptophans in membrane spanning peptides and proteins. The present results highlight how the protein environment can modulate the behavior of the tryptophans and may tweak the interplay between various interfacial anchoring mechanisms. Clearly, for a detailed understanding of their functioning in complex membrane proteins and the surrounding lipid environment, it is important also to examine these interactions in the proper polypeptide context. Important aspects to be addressed in more detail could include the effect of further protein folds and associated changes in motional flexibility, and also issues such as lipid headgroup identity and hydrophobic matching effects.
Supplementary Material
(1) Illustration of the macroscopic sample alignment; (2) HPLC data showing peptide purity; (3) Data demonstrating chemical integrity of aligned NMR samples; (4) Table of estimated changes in Trp orientation for the WALP19 peptide in the ether-linked lipid DTPC. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgments
We thank Anna E. Daily for help with the measurements and Sigrid E. Hayden-Schmutzer for valuable comments on the manuscript.
Footnotes
This work was supported in part by NIH grants RR15569 and GM70971.
Abbreviations: DMPC, 1,2-dimyristoylphosphatidylcholine; DTPC, 1,2-ditetradecylphosphatidylcholine; gA, gramicidin A; NMR, nuclear magnetic resonance, QCC, quadrupolar coupling constant; TFE, 2,2,2-trifluoroethanol; Trp, tryptophan.
Indole ring positions C2-C7 correspond to Trp atoms named Cδ1, Cε2, Cε3, Cζ3, Cη2, and Cζ2, in the IUPAC naming scheme.
References
- 1.von Heijne G. The distribution of positively charged residues in bacterial inner membrane proteins correlates with the transmembrane topology. EMBO J. 1986;5:3021–3027. doi: 10.1002/j.1460-2075.1986.tb04601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landolt-Marticorena C, Williams KA, Deber CM, Reithmeier RAF. Non-random distribution of amino acids in the transmembrane segments of human type I single span membrane proteins. J Mol Biol. 1993;229:602–608. doi: 10.1006/jmbi.1993.1066. [DOI] [PubMed] [Google Scholar]
- 3.Wallin E, Tsukihara T, Yoshikawa S, von Heijne G, Elofsson A. Architecture of helix bundle membrane proteins: an analysis of cytochrome c oxidase from bovine mitochondria. Protein Sci. 1997;6:808–815. doi: 10.1002/pro.5560060407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.O'Connell AM, Koeppe RE, II, Andersen OS. Kinetics of gramicidin channel formation in lipid bilayers: transmembrane monomer association. Science. 1990;250:1256–9. doi: 10.1126/science.1700867. [DOI] [PubMed] [Google Scholar]
- 5.Schiffer M, Chang CH, Stevens FJ. The functions of tryptophan residues in membrane proteins. Protein Engineering. 1992;5:213–214. doi: 10.1093/protein/5.3.213. [DOI] [PubMed] [Google Scholar]
- 6.Deisenhofer J, Michel H. The photosynthetic reaction center from the purple bacterium Rhodopseudomonas viridis. Science. 1989;245:1463–1473. doi: 10.1126/science.245.4925.1463. [DOI] [PubMed] [Google Scholar]
- 7.Doyle DA, Cabral JM, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R. The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science. 1998;280:69–77. doi: 10.1126/science.280.5360.69. [DOI] [PubMed] [Google Scholar]
- 8.Cowan SW, Schirmer T, Rummel G, Steiert M, Ghosh R, Pauptit RA, Jansonius JN, Rosenbusch JP. Crystal structures explain functional properties of two E. coli porins. Nature. 1992;358:727–733. doi: 10.1038/358727a0. [DOI] [PubMed] [Google Scholar]
- 9.Wimley WC, White SH. Membrane partitioning: Distinguishing bilayer effects from the hydrophobic effect. Biochemistry. 1993;32:6307–6312. doi: 10.1021/bi00076a001. [DOI] [PubMed] [Google Scholar]
- 10.Wimley WC, White SH. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol. 1996;3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 11.Yau WM, Wimley WC, Gawrisch K, White SH. The preference of tryptophan for membrane interfaces. Biochemistry. 1998;37:14713–14718. doi: 10.1021/bi980809c. [DOI] [PubMed] [Google Scholar]
- 12.Persson S, Killian JA, Lindblom G. Molecular ordering of interfacially localized tryptophan analogs in ester- and ether-lipid bilayers studied by 2H-NMR. Biophys J. 1998;75:1365–1371. doi: 10.1016/s0006-3495(98)74054-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaede HC, Yau WM, Gawrisch K. Electrostatic Contributions to Indole-Lipid Interactions. J Phys Chem B. 2005;109:13014–13023. doi: 10.1021/jp0511000. [DOI] [PubMed] [Google Scholar]
- 14.Norman KE, Nymeyer H. Indole Localization in Lipid Membranes Revealed by Molecular Simulation. Biophys J. 2006;91:2046–2054. doi: 10.1529/biophysj.105.080275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petersen FNR, Jensen MO, Nielsen CH. Interfacial Tryptophan Residues: A Role for the Cation-{pi} Effect? Biophys J. 2005;89:3985–3996. doi: 10.1529/biophysj.105.061804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sanderson JM, Whelan EJ. Characterisation of the interactions of aromatic amino acids with diacetyl phosphatidylcholine. Phys Chem Chem Phys. 2004;6:1012–1017. [Google Scholar]
- 17.Chiu ML, Nollert P, Loewen MC, Belrhali H, Pebay-Peyroula E, Rosenbusch JP, Landau EM. Crystallization in cubo: general applicability to membrane proteins. Acta Crystallogr D Biol Crystallogr. 2000;56(Pt 6):781–4. doi: 10.1107/s0907444900004716. [DOI] [PubMed] [Google Scholar]
- 18.Nollert P. Membrane protein crystallization in amphiphile phases: practical and theoretical considerations. Progress in Biophysics and Molecular Biology. 2005;88:339–357. doi: 10.1016/j.pbiomolbio.2004.07.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu CH, Ramamoorthy A, Opella SJ. Threedimensional solid-state NMR experiment that correlates the chemical shift and dipolar coupling frequencies of two heteronuclei. J Magn Res. 1994;109:270–272. doi: 10.1006/jmrb.1995.1063. [DOI] [PubMed] [Google Scholar]
- 20.Ramamoorthy A, Wei YF, Lee DK. PISEMA Solid-State NMR Spectroscopy. Annu Rep NMR Spectrosc. 2004;52:1–52. [Google Scholar]
- 21.Opella SJ, Marassi FM. Structure determination of membrane proteins by NMR spectroscopy. Chemical Reviews. 2004;104:3587–3606. doi: 10.1021/cr0304121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Strandberg E, Ulrich AS. NMR methods for studying membrane-active antimicrobial peptides. Concepts In Magnetic Resonance Part A. 2004;23A:89–120. [Google Scholar]
- 23.Bechinger B, Aisenbrey C, Bertani P. The alignment, structure and dynamics of membrane-associated polypeptides by solid-state NMR spectroscopy. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2004;1666:190. doi: 10.1016/j.bbamem.2004.08.008. [DOI] [PubMed] [Google Scholar]
- 24.Ramamoorthy A, Kandasamy SK, Lee DK, Kidambi S, Larson RG. Structure, Topology, and Tilt of Cell-Signaling Peptides Containing Nuclear Localization Sequences in Membrane Bilayers Determined by Solid-State NMR and Molecular Dynamics Simulation Studies. Biochemistry. 2007;46:965–975. doi: 10.1021/bi061895g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koeppe RE, II, Killian JA, Greathouse DV. Orientations of the Tryptophan 9 and 11 Side Chains of the Gramicidin Channel Based on Deuterium Nuclear Magnetic Resonance Spectroscopy. Biophys J. 1994;66:14–24. doi: 10.1016/S0006-3495(94)80748-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Koeppe RE, II, Killian JA, Vogt TCB, de Kruijff B, Taylor MJ, Mattice GL, Greathouse DV. Palmitoylation-Induced Conformational Changes of Specific Side Chains in the Gramicidin Transmembrane Channel. Biochemistry. 1995;34:9299–9306. doi: 10.1021/bi00029a004. [DOI] [PubMed] [Google Scholar]
- 27.Hu W, Lazo ND, Cross TA. Tryptophan dynamics and structural refinement in a lipid bilayer environment: solid state NMR of the gramicidin channel. Biochemistry. 1995;34:14138–14146. doi: 10.1021/bi00043a019. [DOI] [PubMed] [Google Scholar]
- 28.Koeppe RE, II, Sun H, van der Wel PCA, Scherer EM, Pulay P, Greathouse DV. Combined experimental/theoretical approach to indole ring geometry using deuterium magnetic resonance and ab initio calculations. J Am Chem Soc. 2003;125:12268–12276. doi: 10.1021/ja035052d. [DOI] [PubMed] [Google Scholar]
- 29.Pulay P, Scherer EM, van der Wel PCA, Koeppe RE., II Importance of tensor asymmetry for the analysis of 2H-NMR spectra from deuterated aromatic rings. J Am Chem Soc. 2005;127:17488–17493. doi: 10.1021/ja054935x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu W, Lee KC, Cross TA. Tryptophans in membrane proteins: indole ring orientations and functional implications in the gramicidin channel. Biochemistry. 1993;32:7035–47. doi: 10.1021/bi00078a032. [DOI] [PubMed] [Google Scholar]
- 31.Killian JA, Salemink I, de Planque M, Lindblom G, Koeppe RE, II, Greathouse DV. Induction of non-bilayer structures in diacylphosphatidylcholine model membranes by transmembrane α-helical peptides. Importance of hydrophobic mismatch and proposed role of tryptophans. Biochemistry. 1996;35:1037–1045. doi: 10.1021/bi9519258. [DOI] [PubMed] [Google Scholar]
- 32.de Planque MRR, Bonev BB, Demmers JAA, Greathouse DV, Koeppe RE, Separovic F, Watts A, Killian JA. Interfacial Anchor Properties of Tryptophan Residues in Transmembrane Peptides Can Dominate over Hydrophobic Matching Effects in Peptide-Lipid Interactions. Biochemistry. 2003;42:5341–5348. doi: 10.1021/bi027000r. [DOI] [PubMed] [Google Scholar]
- 33.Weiss TM, van der Wel PCA, Killian JA, Koeppe RE, Huang HW. Hydrophobic mismatch between helices and lipid bilayers. Biophys J. 2003;84:379–385. doi: 10.1016/S0006-3495(03)74858-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.van der Wel PCA, Strandberg E, Killian JA, Koeppe RE., II Geometry and intrinsic tilt of a tryptophan anchored transmembrane alpha-helix determined by 2H NMR. Biophys J. 2002;83:1479–1488. doi: 10.1016/S0006-3495(02)73918-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van der Wel PCA, Pott T, Morein S, Greathouse DV, Koeppe RE, II, Killian JA. Tryptophan-anchored transmembrane peptides promote formation of nonlamellar phases in phosphatidylethanolamine model membranes in a mismatch-dependent manner. Biochemistry. 2000;39:3124–3133. doi: 10.1021/bi9922594. [DOI] [PubMed] [Google Scholar]
- 36.Greathouse DV, Koeppe RE, II, Providence LL, Shobana S, Andersen OS. Design and characterization of gramicidin channels. Methods Enzymol. 1999;294:525–550. doi: 10.1016/s0076-6879(99)94031-4. [DOI] [PubMed] [Google Scholar]
- 37.Greathouse DV, Goforth RL, Crawford T, van der Wel PCA, Killian JA. Optimized aminolysis conditions for cleavage of N-protected hydrophobic peptides from solid-phase resins. J Pept Res. 2001;57:519–527. doi: 10.1034/j.1399-3011.2001.00849.x. [DOI] [PubMed] [Google Scholar]
- 38.Hallock KJ, Henzler Wildam K, Lee DK, Ramamoorthy A. An innovative procedure using a sublimable solid to align lipid bilayers for solid-state NMR studies. Biophys J. 2002;82:2499–2503. doi: 10.1016/S0006-3495(02)75592-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rainey JK, Sykes BD. Optimizing Oriented Planar-Supported Lipid Samples for Solid-State Protein NMR. Biophys J. 2005;89:2792–2805. doi: 10.1529/biophysj.105.063800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bechinger B, Seelig J. Conformational changes of the phosphatidylcholine headgroup due to membrane dehydration. A 2H-NMR study. Chemistry and Physics of Lipids. 1991;58:1–5. doi: 10.1016/0009-3084(91)90105-k. [DOI] [PubMed] [Google Scholar]
- 41.Dvinskikh SV, Castro V, Sandstrom D. Probing segmental order in lipid bilayers at variable hydration levels by amplitude- and phase-modulated cross-polarization NMR. Phys Chem Chem Phys. 2005;7:3255–7. doi: 10.1039/b508190d. [DOI] [PubMed] [Google Scholar]
- 42.McKeone BJ, Pownall HJ, Massey JB. Ether phosphatidylcholines: comparison of miscibility with ester phosphatidylcholines and sphingomyelin, vesicle fusion, and association with apolipoprotein A-I. Biochemistry. 1986;25:7711–7716. doi: 10.1021/bi00371a064. [DOI] [PubMed] [Google Scholar]
- 43.Davis JH, Jeffrey KR, Bloom M, Valic MI, Higgs TP. Quadrupolar echo deuteron magnetic resonance spectroscopy in ordered hydrocarbon chains. Chem Phys Lett. 1976;42:390–394. [Google Scholar]
- 44.Gall CM, DiVerdi JA, Opella SJ. Phenylalanine ring dynamics by solid-state 2H NMR. J Am Chem Soc. 1981;103:5039–5043. [Google Scholar]
- 45.Strandberg E, Ozdirekcan S, Rijkers DTS, van der Wel PCA, Koeppe RE, II, Liskamp RMJ, Killian JA. Tilt Angles of Transmembrane Model Peptides in Oriented and Non-Oriented Lipid Bilayers as Determined by 2H Solid-State NMR. Biophys J. 2004;86:3709–3721. doi: 10.1529/biophysj.103.035402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ozdirekcan S, Rijkers D, Liskamp R, Killian J. Influence of flanking residues on tilt and rotation angles of transmembrane peptides in lipid bilayers. A solid-state 2H NMR study. Biochemistry. 2005;44:1004–1012. doi: 10.1021/bi0481242. [DOI] [PubMed] [Google Scholar]
- 47.Lovell SC, Word JM, Richardson JS, Richardson DC. The penultimate rotamer library. Proteins. 2000;40:389–408. [PubMed] [Google Scholar]
- 48.Dunbrack JRL. Rotamer Libraries in the 21st Century. Curr Opin Struct Biol. 2002;12:431–440. doi: 10.1016/s0959-440x(02)00344-5. [DOI] [PubMed] [Google Scholar]
- 49.Goodyear DJ, Sharpe S, Grant CW, Morrow MR. Molecular dynamics simulation of transmembrane polypeptide orientational fluctuations. Biophys J. 2005;88:105–17. doi: 10.1529/biophysj.104.047506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kandasamy SK, Larson RG. Molecular Dynamics Simulations of Model Trans-Membrane Peptides in Lipid Bilayers: A Systematic Investigation of Hydrophobic Mismatch. Biophys J. 2006;90:2326–2343. doi: 10.1529/biophysj.105.073395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Petrache HI, Zuckerman DM, Sachs JN, Killian JA, Koeppe RE, II, Woolf TB. Hydrophobic matching mechanism investigated by molecular dynamics simulations. Langmuir. 2002;18:1340–1351. [Google Scholar]
- 52.de Planque MRR, Kruijtzer JAW, Liskamp RMJ, Marsh D, Greathouse DV, Koeppe RE, II, de Kruijff B, Killian JA. Different membrane anchoring positions of tryptophan and lysine in synthetic transmembrane α-helical peptides. J Biol Chem. 1999;274:20839–20846. doi: 10.1074/jbc.274.30.20839. [DOI] [PubMed] [Google Scholar]
- 53.Koeppe RE, II, Hatchett J, Jude AR, Providence LL, Andersen OS, Greathouse DV. Neighboring Aliphatic/Aromatic Side Chain Interactions between Residues 9 and 10 in Gramicidin Channels. Biochemistry. 2000;39:2235–2242. doi: 10.1021/bi9920679. [DOI] [PubMed] [Google Scholar]
- 54.Separovic F, Pax R, Cornell B. NMR order parameter analysis of a peptide plane aligned in a lyotropic liquid crystal. Molecular Physics. 1993;78:357–369. [Google Scholar]
- 55.Gawrisch K, Ruston D, Zimmerberg J, Parsegian VA, Rand RP, Fuller N. Membrane dipole potentials, hydration forces, and the ordering of water at membrane surfaces. Biophys J. 1992;61:1213–1223. doi: 10.1016/S0006-3495(92)81931-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Providence LL, Andersen OS, Greathouse DV, Koeppe RE, II, Bittman R. Gramicidin Channel Function Does Not Depend on Phospholipid Chirality. Biochemistry. 1995;34:16404–16411. doi: 10.1021/bi00050a022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(1) Illustration of the macroscopic sample alignment; (2) HPLC data showing peptide purity; (3) Data demonstrating chemical integrity of aligned NMR samples; (4) Table of estimated changes in Trp orientation for the WALP19 peptide in the ether-linked lipid DTPC. This material is available free of charge via the Internet at http://pubs.acs.org.


