Abstract
Actin filament binding by the focal adhesion (FA)-associated protein talin stabilizes cell-substrate adhesions and is thought to be rate-limiting in cell migration. Although F-actin binding by talin is known to be pH-sensitive in vitro, with lower affinity at higher pH, the functional significance of this pH dependence is unknown. Because increased intracellular pH (pHi) promotes cell migration and is a hallmark of metastatic carcinomas, we asked whether it increases FA remodeling through lower-affinity talin–actin binding. Talin contains several actin binding sites, but we found that only the COOH-terminal USH-I/LWEQ module showed pH-dependent actin binding, with lower affinity and decreased maximal binding at higher pH. Molecular dynamics simulations and NMR of this module revealed a structural mechanism for pH-dependent actin binding. A cluster of titratable amino acids with upshifted pKa values, including His-2418, was identified at one end of the five-helix bundle distal from the actin binding site. Protonation of His-2418 induces changes in the conformation and dynamics of the remote actin binding site. Structural analyses of a mutant talin-H2418F at pH 6.0 and 8.0 suggested changes different from the WT protein, and we confirmed that actin binding by talin-H2418F was relatively pH-insensitive. In motile fibroblasts, increasing pHi decreased FA lifetime and increased the migratory rate. However, expression of talin-H2418F increased lifetime 2-fold and decreased the migratory rate. These data identify a molecular mechanism for pH-sensitive actin binding by talin and suggest that FA turnover is pH-dependent and in part mediated by pH-dependent affinity of talin for binding actin.
Keywords: intracellular pH, NHE1, migration
Focal adhesion (FA) remodeling is a rate-limiting determinant in haptokinetic migration of adherent cells. At the leading edge of migrating cells, FAs undergo rapid cycles of assembly and turnover, creating and disrupting, respectively, sites of traction necessary for forward movement of the cell body. Force generation for traction requires linkage among the extracellular matrix, integrin receptors, and actin filaments. Actin filaments do not directly bind to the cytoplasmic domain of integrins but bind to integrin-associated FA proteins such as talin and vinculin. Although several mechanisms contribute to FA remodeling in migrating cells (1), emerging evidence indicates that talin plays a central role in the dynamic linkage between integrins and actin filaments necessary for cell migration (2, 3). Talin functions in distinct albeit complementary mechanisms that promote FA turnover. First is cleavage of talin by the protease calpain, which also modulates adhesion complex composition and likely signaling functions of talin (4). Second is regulated talin binding to actin filaments, which is proposed to act as a clutch to control FA turnover and membrane protrusion dynamics (3, 5–7). How actin binding by talin is dynamically regulated during cell migration, however, remains undetermined.
Previous studies indicate that actin binding by talin in vitro is pH-sensitive, with lower-affinity binding at higher pH (8–10), although the functional significance of this regulation is unknown. In motile cells, an increase in intracellular pH (pHi) promotes FA remodeling (11–13) and velocity (12, 14). We speculated that increased pHi in motile cells might promote dynamic turnover of FAs by lowering the affinity of actin binding by talin. We examined this speculation by using constant-pH molecular dynamics (CpHMD) simulations, pH-dependent NMR structure, and biochemical analysis, which revealed a mechanism for pH-sensitive actin binding by talin. Because talin1 is the likely FA talin, our data refer to this isoform. We tested structural predictions in migrating fibroblasts and found that talin acts as an exquisitely sensitive pH sensor in binding actin filaments and in controlling FA stability and migratory rate. These data suggest that a clutch-like mechanism controlling FA turnover is pH-sensitive.
Results
Decreased pH Increases the Affinity of F-Actin Binding by Talin.
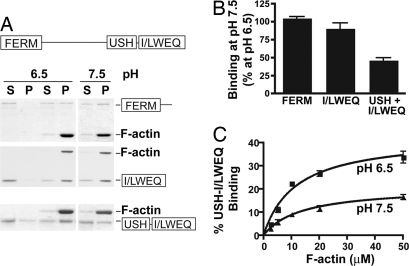
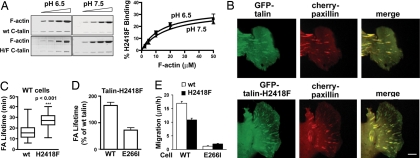
Because the NH2-terminal FERM domain and the COOH-terminal I/LWEQ domain of talin contain actin binding sites that could mediate a clutch-like action for FA remodeling (15), we tested the pH dependence of actin binding by each domain. Using F-actin cosedimentation, we found that actin binding by the FERM domain (1–433) and the I/LWEQ domain including the coiled-coil dimerization segment (2341–2541) was pH-insensitive within the physiological range of pH 6.5–7.5 (Fig. 1 A and B). There also was no change in total F-actin at pH 6.5 compared with pH 7.5. However, maximum F-actin binding (Bmax) by a module containing the I/LWEQ domain and the adjacent upstream helical segment (USH) (2300–2541) was pH-sensitive and >2-fold greater at pH 6.5 (42.7 ± 3.1%) compared with pH 7.5 (20.8 ± 2.0%) (P < 0.01, n = 7) (Fig. 1C). Additionally, at pH 6.5 the Kd of 2.92 ± 0.62 μM was significantly lower compared with a Kd of 8.68 ± 3.91 μM at pH 7.5 (P < 0.01, n = 3). Although the I/LWEQ domain is a conserved actin-binding module also found in the fungal protein Sla2p, Dicty ostelium TalA and TalB, and metazoan proteins Hip1 (Huntington-interacting protein 1) and Hip12, (16), the adjacent USH segment was previously reported to inhibit F-actin binding in vitro by the I/LWEQ domain (17). However, attenuated actin binding by the USH segment was determined at pH 8, which is consistent with our data indicating that the USH segment plays a pivotal role in conferring decreased affinity for F-actin binding by the I/LWEQ domain at higher pH values.
Fig. 1.
Actin binding by the C terminus of talin is pH-dependent. (A) Actin cosedimentation assays using 10 μg of F-actin and 5 μg of the indicated segments of talin were incubated for 60 min at pH 6.5 and 7.5 and centrifuged to sediment F-actin. Proteins in the pellet and the supernatant were separated by SDS/PAGE and stained with Coomassie. (B) Relative abundance of talin domains cosedimenting with actin was determined by NIH Image software and is expressed as mean ± SEM of four to seven separate assays. (C) Talin–actin binding at pH 6.5 and 7.5 determined with 1 μM of the USH-I/LWEQ domain and the indicated concentrations of F-actin.
Simulations Suggest That Talin–Actin Binding Is Allosterically Regulated by pH.
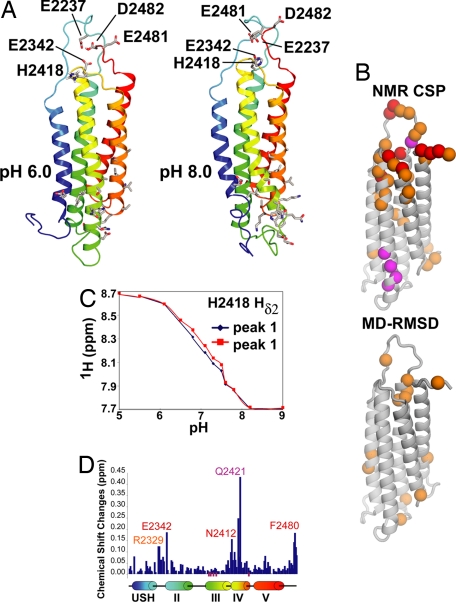
Computer simulations were carried out to investigate possible mechanisms by which F-actin binding by the USH-I/LWEQ module of talin could be regulated by pH. The simulations were originally carried out by using a homology model based on the structure of Hip1R (Hip-1-related). During the course of this study, Critchley and coworkers determined an NMR structure of the talin I/LWEQ domain (18) including the USH segment, but excluding the COOH-terminal dimerization domain (Protein Data Bank ID code 2JSW). Simulations using this NMR structure led to conclusions very similar those of the homology model, and those results are shown here. The I/LWEQ domain contains only one histidine residue, His-2418. Near this residue there are also a number of negatively charged amino acids, and computational pKa prediction using multiconformation continuum electrostatics (19) suggested that Glu-2337, Glu-2342, Glu-2481, and Asp-2482 could have upshifted pKa values. We hypothesized that some or all of these residues form a pH sensor. All five are clustered at one end of the five-helix bundle (Fig. 2A) distal from the putative actin binding site as defined by mutagenesis (20) and are conserved in mammalian talin1 but not in Caenorhabditis elegans or Drosophila.
Fig. 2.
Constant pH computational simulations and NMR reveal pH-dependent conformational changes in talin USH-I/LWEQ. (A) Average structures obtained from the final 5 ns of CpHMD show structural differences between pH 6.0 and 8.0. The different protonation states for His-2418 and Asp-2482 influence the conformation observed for the actin binding site, located ≈40 Å away. The residues Glu-2337, Glu-2342, His-2418, Glu-2481, and Asp-2482, predicted to form the pH sensor, are highlighted for pH 6.0 and 8.0. The actin binding site, formed by residues belonging to helices III and IV, is also depicted for both pH values. The residues His-2418 and Asp-2482 are mostly protonated at pH 6.0 and deprotonated at pH 8.0. The residues Glu-2337, Glu-2481, and Asp-2482 are deprotonated at both pH values. (B) NMR chemical shift perturbations, classified according to chemical shift changes as medium (orange, ΔδTOT = 0.05–0.1 ppm) and large (red, ΔδTOT > 0.1 ppm), where the total chemical shift difference is ΔδTOT = | Δ1H| + |Δ15N| × 0.2. Residues that showed broadening at pH 8.0 relative to pH 6.0 are shown in magenta. The differences between the average structures at pH 6 and pH 8 in the simulations (A) also identified the residues localized near the pH sensor and the actin binding site as the ones with the largest differences. (C) NMR titration curves for His-2418 (observing the histidine Hδ2 proton, which appeared as a doublet in the range between pH 6.5 and 7.5 and coalesced at high/low pH). (D) Chemical shift changes (ΔδTOT = |Δ1H| + |Δ15N| × 0.2) along the linear sequence of USH-I/LWEQ with the positions of the five helices in the NMR model shown below. Magenta bars below the chart indicate residues that broaden at pH 8.0 relative to pH 6.0.
To investigate how the structure and dynamics of the talin USH-I/LWEQ module might be modulated by pH, CpHMD simulations were carried out at several pH values. We report results of the simulations and NMR data at pH 6.0 and 8.0 to emphasize the effect of changing protonation states. Results at intermediate pH values corresponding to those used in the in vivo experiments smoothly interpolate between these two extremes. Between pH 6.0 and 8.0 the side chain of His-2418 showed a large change in its fractional protonation (pH 6.0, 73% protonated; pH 8.0, 2%). The simulations also suggested that Asp-2482 might have a significantly upshifted pKa and titrate in this range as well (pH 6.0, 98% protonated; pH 8.0, 0%). However, this residue is located at the immediate COOH terminus of the construct, and this prediction could be an artifact of the particular monomeric construct used.
Significant differences in structure and dynamics were observed between pH 6.0 and pH 8.0 mediated by the changing protonation states [Fig. 2A and supporting information (SI) Fig. S1]. The backbone rmsd between the average structures at pH 6.0 and 8.0 is 4.2 Å. Some of the most pronounced differences in the backbone structure and dynamics were distant from the pH sensor, especially the loop connecting helices II and III. Significant conformational changes also were seen in portions of the helices near this loop including residues implicated in actin binding (20). Backbone conformational changes in the pH sensor itself were also significant. The computational simulations thus suggest a model in which protonation of residues in the pH sensor, including His-2418, may modulate conformation and dynamics of distal residues in the actin binding site.
NMR Spectral Perturbations Are Associated with pH Changes.
To test this model, NMR experiments were carried out on the talin USH-I/LWEQ domain lacking the COOH-terminal dimerization helix (2300–2501) at several pH values. Increasing solvent pH from 6.0 to 8.0 was associated with spectral perturbations for a subset of backbone amide resonances in the 2D 15N-HSQC spectrum of talin USH-I/LWEQ; these peaks showed significant alterations in chemical shift, intensity, or both (Fig. 2 B and D and Fig. S2). In agreement with modeling results, the residues showing the largest chemical shift changes and/or the most pronounced line broadening cluster largely to two regions at opposite ends of the USH-I/LWEQ domain: the pole containing the residues we predict to comprise the pH sensor and an area on the opposite pole proximal to the putative actin binding site (20) and the loop joining helices IV and V (Fig. 2B). These spectral perturbations indicate there are significant changes in structure or dynamics and the charge of titrating side-chain groups in the talin USH-I/LWEQ module on increasing pH from 6.0 to 8.0.
Actin binding to I/LWEQ domains requires ≈40 residues at the COOH terminus that are proposed to form a coiled coil responsible for dimerization (20, 21). Using actin cosedimentation, we confirmed previous findings (22) that a COOH-terminal-truncated talin (2341–2501) lacking the dimerization domain does not bind F-actin (data not shown). However, comparison of amide (1HN, 15N) chemical shift changes with pH (6.0 vs. 8.0) for monomeric and dimeric USH-I/LWEQ showed almost identical changes for constructs with and without the C-terminal dimerization helix (Fig. S3). This result suggests that the model of pH allosterically regulating the talin binding site, although based on simulations and NMR experiments using a monomeric construct, may also explain the pH-dependent binding of full-length, dimeric talin to actin.
To probe whether the single histidine residue (His-2418) in the talin USH-I/LWEQ module plays a role in pH sensing, we determined the pKa of His-2418 by NMR (Fig. 2C). pH titrations observing the histidine Hδ2 proton indicated that His-2418 has a pKa of 7.2, which is upshifted from the ≈6.5 pKa of isolated histidine in aqueous buffer (23).
H2418F Mutant Shows Different Spectral Perturbations in Response to pH Change.
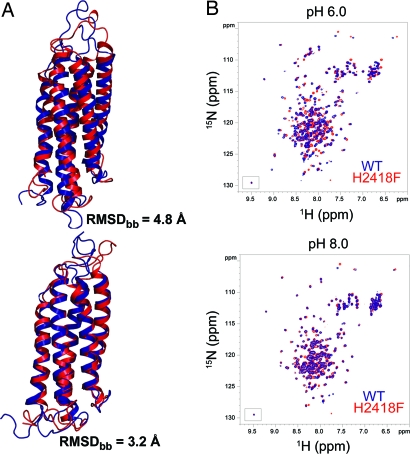
To further investigate the role of the His-2418 in pH sensing, several possible mutants were generated and investigated in silico by using CpHMD simulations, with the goal of identifying a mutant with reduced pH dependence. Of these, the mutant USH-I/LWEQ-H2418F was found to generate the smallest perturbations to the backbone structure and dynamics of the protein at pH 8.0 (3.2-Å backbone rmsd between mutant and WT) and larger changes at pH 6.0 (4.8-Å backbone rmsd). This prediction was confirmed by NMR HSQC experiments. Two-dimensional 15N-HSQC spectra of WT USH-I/LWEQ and the H2418F mutant at pH 8.0 were essentially identical, apart from a few residues around the phenyalanine substitution that showed small chemical shift changes (Fig. 3B), indicating that the backbone structures of the mutant and WT are similar at pH 8.0. Decreasing pH from 8.0 to 6.0 led to chemical shift perturbations in the mutant and WT (Fig. 3 and Figs. S2 and S4). Many of the same residues were affected in the H2418F mutant and WT; however, signals for amino acids often shifted in different directions in 2D 15N-HSQC spectra. Additionally, the patterns of chemical shift changes were dissimilar, indicating that chemical environments and structure in the mutant and WT apparently differ at pH 6.0.
Fig. 3.
Mutation of His-2418 modifies pH-dependent structural properties of talin USH-I/LWEQ. (A) Superimposition of the average structures obtained from the last 5 ns of CpHMD for WT (blue) and H2418F mutant (red). The backbone rmsd values obtained between WT at pH 6.0 and mutant at pH 8.0 is 4.8 Å, whereas the rmsd between WT at pH 8.0 and mutant at pH 6.0 is 3.2 Å. (B) 2D 15N-HSQC spectra of WT (blue) and H2418F mutant (red) USH-I/LWEQ talin at pH 6.0 (Upper) and 8.0 (Lower).
Taken together, the simulations and NMR data suggest that the mutant would undergo different structural and dynamical changes as a function of pH compared with WT protein (Fig. 3A and Fig. S5), supporting the model that protonation of His-2418 may modulate the conformation and dynamics of portions of the USH-I/LWEQ domain near the actin binding site, including the solvent exposure of actin binding site residues. Because the H2418F mutant shows significant pH-dependent conformational and dynamical changes, albeit different from those of the WT, we suspect that other residues in the pH sensor could also play a role together with His-2418.
Decreased pHi Decreases FA Turnover.
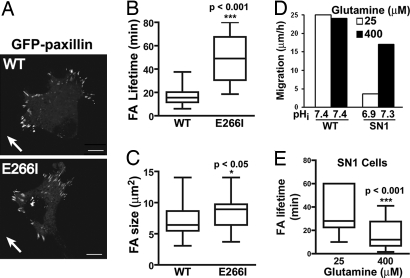
We used two strategies to show that decreased pHi increases FA lifetime. First, we used fibroblasts deficient for the Na-H exchanger NHE1 but stably expressing WT NHE1 (WT cells) or a mutant NHE1 with an E266I substitution that lacks proton translocation (E266I cells) (12, 24). NHE1 plays a central role in regulating pHi by catalyzing an electroneutral exchange of extracellular Na+ for intracellular H+. The pHi of migrating WT and E266I cells is ≈7.5 and ≈7.0, respectively (25), and migratory rate of E266I cells is ≈8-fold slower than WT cells (12). FA turnover in cells migrating at the edge of a wounded monolayer was determined by real-time imaging of GFP-paxillin, which localized to FAs (Fig. 4A and Movies S1 and S2) and had similar expression in WT and E266I cells (Fig. S6). The lifetime of FAs in extending lamellipodia was shorter in WT cells (15.5 ± 1.1 min) compared with E266I cells (37.1 ± 3.2 min) (Fig. 4B) (P < 0.001, n = 80 FA). Additionally, although the total number of FAs in WT and E266I cells was similar (data not shown), the proportion of large (>8 μm2) FAs was greater in E266I cells than in WT cells (Fig. 4C).
Fig. 4.
Decreasing pHi increases FA lifetime. (A) Images from movies of WT and E266I cells expressing GFP-paxillin at the edge of a wounded monolayer. Arrows indicate direction of migration. (Scale bar: 10 μm.) (B and C) Box and whisker plots show lifetime (B) and size (C) of FAs determined by analysis of GFP-paxillin in time-lapse movies from four to seven independent cell preparations. Bars, full range; box, interquartile range; horizontal line, median. (D) Average migration rate and pHi of WT and SN1 cells at the edge of a wounded monolayer determined 6 h after wounding in two independent cell preparations. (E) Box and whisker plot show lifetime of FAs in SN1 cells determined by analysis of GFP-paxillin in time-lapse movies from three independent cell preparations.
Second, we used NHE1-deficient fibroblasts expressing the System N1 transporter (SN1 cells). SN1 is an amino acid transporter that catalyzes the exchange of extracellular glutamine for intracellular H+, and increased extracellular glutamine drives an increase in H+ efflux and pHi (26). We found that decreasing glutamine in the medium from 400 μM to 25 μM decreased pHi from 7.3 to 6.9 and decreased the migratory rate of SN1 cells at the edge of a wounded monolayer (Fig. 4D). Mean FA lifetime in SN1 cells was significantly longer with lower pHi (37 ± 2.2 min) than with higher pHi (16.7 ± 1.5 min) (Fig. 4E) (P < 0.001, n = 67 for 25 μM and n = 54 for 400 μM). These data indicate that FAs are more stable at lower pHi.
Talin-H2418F Has Decreased pH-Sensitive Actin Binding and Modulates FA Lifetime.
To test our predicted structural model of pH sensing by talin, we used F-actin cosedimentation to show that a H2418F substitution in the USH-I/LWEQ segment (H2418F; 2300–2541) had reduced pH-dependent actin binding. At pH 6.5, Bmax (34.8%) was less than WT USH-I/LWEQ but unchanged at pH 7.5 (33.2%) (Fig. 5A). Additionally, dissociation constants for the mutant were relatively pH-insensitive. Kd values at pH 6.5 and 7.5 were 3.99 ± 1.22 μM and 5.95 ± 2.7 μM, respectively, and not significantly different (P > 0.1, n = 3).
Fig. 5.
Talin H2418F has reduced pH-dependent affinity for actin binding and alters FA duration and migration rate. (A) Cosedimentation of 1 μM WT USH-I/LWEQ or USH-I/LWEQ containing an H2418F mutation with increasing actin concentrations after incubation for 60 min at pH 6.5 or pH 7.5. Shown are Coomassie-stained gels of pellet fractions (Left) and kinetics of talin binding as a function of F-actin concentration (Right). (B) Images from movies of WT cells showing colocalization of cherry-paxillin and GFP-talin or GFP-talin-H2418F at the edge of a wounded monolayer 6 h after wounding. (Scale bar: 5 μm.) (C) Lifetime of FAs in WT cells was determined by analysis of cherry-paxillin in 10–15 FAs in movies from five independent cell preparations. (D) Lifetime of FAs in WT and E266I cells expressing full-length talin-H2418F relative to expression of WT talin in cells at the edge of a wounded monolayer. (E) Migration rate of wound-edge WT and E266I cells expressing GFP-talin or GFP-talin-H2418F determined as distance traveled for 10 h after wounding by tracking individual cells. Data are expressed as means ± SEM of transfected cells in movies from three independent cell preparations.
Because actin binding by USH-I/LWEQ-H2418F was relatively pH-insensitive but had an “intermediate” affinity and maximal binding compared with WT at low and high pH, we asked whether it affected FA remodeling, with the prediction that FA lifetime would increase in WT cells but decrease in E266I cells. In cells at the edge of a wounded monolayer, full-length GFP-talin and GFP-talin-H2418F localized with coexpressed cherry-paxillin in FAs (Fig. 5B). Expression of GFP-talin and GFP-talin-H2418F was similar in both cell types, and cleaved fragments were not observed by immunoblotting with antibodies to GFP (Fig. S6). In WT cells, FA lifetime was not significantly different with expression of GFP-talin compared with cells expressing GFP-paxillin only (P > 0.1, n = 60) but was significantly increased with expression of GFP-talin-H2418F (P < 0.001, n = 60) (Fig. 5 C and D and Movies S3 and S4). Additionally, mean FA size was significantly greater in cells expressing GFP-talin-H2418F (9.1 ± 0.5 μm2) than in cells expressing WT GFP-talin (7.6 ± 0.4 μm2; P < 0.05, n = 60). Consistent with increased lifetime of FAs, the migration velocity of WT cells expressing GFP-talin-H2418F was significantly slower compared with cells expressing WT GFP-talin (P < 0.002, n = 20 cells) (Fig. 5E). In contrast, expression of GFP-talin-H2418F in E266I cells significantly decreased FA lifetime (P < 0.01, n = 58) (Fig. 5D and Movies S5 and S6) and increased migratory velocity compared with E266I cells expressing GFP-paxillin only (data not shown) or GFP-talin (Fig. 5E). In E266I cells, the increase in migratory velocity with GFP-talin-H2418F was significant (P < 0.01, n = 20) but smaller in magnitude compared with the decrease in WT cells, most likely because absence of NHE1 activity in motile cells also impairs polarity (25) and actin filament assembly (14). These data indicate that substitution of H2418F in talin changes FA stability, attenuating turnover in WT cells with pHi of 7.5 but increasing turnover in E266I cells with pHi of 7.0. Our data also suggest that His-2418 in talin is a key residue in sensing intracellular pH changes to regulate FA stability and cell migration rate.
Discussion
Increased pHi was previously suggested to promote FA remodeling (12, 13), and we now show that talin acts as a pH sensor in regulating the stability of FAs. Through a combination of computational simulations, NMR, and functional studies, we demonstrate that talin His-2418 plays a key role in pH sensing and pH-dependent actin binding. Computational data provide a structural model for pH sensing in a monomeric USH-I/LWEQ construct, suggesting that protonation of His-2418 modulates conformation and dynamics at the remote actin binding site. NMR chemical shift mapping revealed pH-dependent changes both around the cluster of residues in the vicinity of His-2418, which has an upshifted pKa value, and the actin binding site. The NMR data and computational simulations suggest a model where residues in the talin pH sensor and actin binding site are conformationally coupled through residues in the helices of the I/LWEQ module, although additional work is necessary to identify the mechanism of this coupling and to determine whether modulation is allosteric.
Our data also suggest that regulated actin binding by the COOH terminus of talin may be an important determinant in FA remodeling. Although we showed that the NH2-terminal FERM domain of talin binds F-actin, as previously reported (15), binding is pH-insensitive. Whether actin binding by the FERM domain has functional significance in cells is unclear. The FERM domain is not sufficient for force generation at FAs. Impaired force generation in talin-null cells is restored by expression of full-length talin but not by a truncated talin lacking the COOH terminus but retaining the FERM domain (27). Additionally, a mutant talin lacking the FERM domain but retaining the COOH terminus targets to FAs (22). Although the FERM domain binds the cytoplasmic domain of β integrins (28), the COOH terminus of talin1 contains a second integrin binding site (amino acids 1984–2113) (2).
FA remodeling in motile cells is regulated by multiple mechanisms that likely vary with cell type, substratum, and migratory cue. Kinases, including FA kinase (FAK), Src, extracellular signal-regulated kinase (ERK), and myosin light chain kinase, and adaptor proteins, including paxillin and p130CAS, regulate FA turnover in migrating cells (1). Additionally, three mechanisms could account for dynamic changes in talin-mediated linkage of actin filaments to FAs. First is talin binding to integrins, although whether this is dynamically regulated in motile cells has not been reported. Second is calpain cleavage of talin, which also disassembles other FA components, including paxillin, vinculin, and zyxin (4). Third is regulated talin affinity for F-actin, which our current data and previous findings (8–10) show is pH-dependent. Because integrin activation (11, 29), growth factors (11, 25, 30), and monolayer wounding (25) stimulate NHE1 activity and increase pHi, pH-dependent FA remodeling could be regulated by different migratory cues. Our current study suggests that pH-regulated FA remodeling by dynamic changes in talin affinity for F-actin may be one of several complementary mechanisms controlling FA turnover.
An intriguing aspect of pH sensitivity is the ability to regulate multiple proteins in unison. Increased pHi is an evolutionarily conserved signal necessary for several stages of cell migration in addition to FA remodeling, including polarity (12, 14, 25) and the assembly of cytoskeletal filaments (14, 31). Moreover, in addition to talin a number of proteins have pH-dependent actin binding, including HIP1 (17), cofilin (32), and villin (33). Of notable significance is that increased pHi is a hallmark of most metastatic cancers, regardless of the tissue origin or genetic background (34). Hence, the higher pHi of metastatic cells compared with normal cells could increase FA turnover to promote migratory capacity. Moreover, our study underscores the importance of bridging structural and cellular biology to elucidate how physiological changes in pHi regulate cell behaviors.
Materials and Methods
Cell Culture and DNA Constructs.
Materials and methods for cell culture (11, 24, 25) and pHi measurements (24) were as previously described. Transient expression of proteins was obtained by transfection with FuGENE 6 (Roche). GFP-talin was provided by Anna Huttenlocher (University of Wisconsin, Madison), and GFP-paxillin and cherry-paxillin were obtained from Christopher Turner (State University of New York, Syracuse) and Torsten Wittmann (University of California, San Francisco), respectively. GST fusions of mouse talin1 were subcloned into pGEX6P2 and expressed and purified as previously described (24). GST was cleaved with PreScission Protease (GE Healthcare Biosciences). Site-directed mutagenesis was performed to obtain H2418F mutants of talin in pGEX6P2 (talin 2300–2541, 2300–2501) or in pEGFP (full length GFP-talin) vectors.
F-Actin Cosedimentation.
Nonmuscle G-actin (β-actin; Cytoskeleton) was polymerized in 50 mM KCl, 2 mM MgCl2, and 1 mM ATP for 60 min at 32°C. F-actin (1.5 μM) was incubated with the indicated talin fragment in buffer containing 10 mM imidazole (pH 6.5 or 7.5), 1 mM ATP, 1 mM 2-mercaptoethanol, 1 mM EGTA, 0.1 mM CaCl2, and 2 mM MgCl2. The final pH of samples was monitored with a micro-pH meter and adjusted by addition of 0.1 M HCl or KOH. Samples were incubated for 60 min at 32°C and then centrifuged at 100,000 × g for 20 min at 24°C. Proteins in supernatant and pellet fractions were separated by SDS/PAGE and stained with Coomassie. pH-dependent dissociation constants were calculated from transformation of binding curves by using GraphPad Prism 5 software.
MD Simulations.
All CpHMD simulations were carried out by using the AMBER 8 suite of programs (35). The AMBER parm99 force field (36) and generalized Born solvation model (37) were used. Detailed procedures are provided in SI Materials and Methods. Analyses were carried out for the final 5 ns of each simulation, primarily with the ptraj program, and Pymol (38) and VMD (39) were used for visualization purposes.
NMR.
For protein expression, transformed BL21 cells were grown in minimal medium [Na2HPO4, KH2PO4, NaCl, MgSO4, CaCl2, MEM vitamin mix (Gibco), 15NH4Cl, 13C glucose, Isogrow 15N 13C, biotin, and FeCl2] with ampicillin. GST fusion proteins with an intervening PreScission protease recognition site were expressed and purified as described (24), cleaved on-column with PreScission protease, eluted, and concentrated to 1 mM.
Experiments were performed on Bruker DRX 500 and Avance 800 MHz spectrometers. Initial 1HN, 15N, 13Cα, 13Cβ, and 13C′ backbone resonance assignments of a shorter talin USH-I/LWEQ construct lacking the dimerization helix (2300–2482) were made at 318 K and pH 6.0 using standard 3D triple-resonance experiments (18). Resonance assignments obtained at 318 K were transferred to spectra of a longer talin USH-I/LWEQ construct (2300–2501) at 305 K using standard 3D triple-resonance experiments (40). NMR chemical shift perturbations in spectra recorded at different pH values were classified according to chemical shift changes as medium (orange, ΔδTOT = 0.05–0.1 ppm) and large (red, ΔδTOT > 0.1 ppm), were weighted according to ΔδTOT = |Δ1H| + |Δ15N| × 0.2 (41).
Fluorescence Imaging and FA Dynamics.
Extending lamellipodia in cells expressing fluorescent-tagged proteins were selected in the mid-stack of time-lapse movies (Movies S1–S6) to capture their assembly and disassembly. Image J software was used to outline FAs, and an equal area juxtaposed was selected for background fluorescence intensity. The duration of a FA was measured as the time from frame of background-subtracted adhesion (fluorescence signal) formation to the frame of its disappearance. For each movie measurements were obtained for 10–15 individual adhesions on four to six cells. The area outlining FAs was recorded as the size. Statistical analysis was performed using GraphPad Prism 5 software.
Supplementary Material
Acknowledgments.
We thank Alice Goodwin, Dinah Zaghi, and Pascal Wassam for technical assistance and Torsten Wittmann for technical advice and suggestions. This work was supported by National Science Foundation Grant MCB-0346399 (to M.P.J.), National Institutes of Health Grant GM58642 (to D.L.B.), and a grant from the Sandler Family Foundation (to M.J.S.K., M.P.J., and D.L.B.). Work by J.S. and D.L.B. was conducted in a facility constructed with support from Research Facilities Improvement Program Grant C06 RR16490 from the National Center for Research Resources, National Institutes of Health. Work by A.R.G., B.T.G., and D.R.C. was funded by the Wellcome Trust, Cancer Research UK, and National Institutes of Health Grant GM064346 (to the Cell Migration Consortium).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0805163105/DCSupplemental.
References
- 1.Webb DJ, et al. FAK-Src signalling through paxillin, ERK and MLCK regulates adhesion disassembly. Nat Cell Biol. 2004;6:154–161. doi: 10.1038/ncb1094. [DOI] [PubMed] [Google Scholar]
- 2.Critchley DR. Cytoskeletal proteins talin and vinculin in integrin-mediated adhesion. Biochem Soc Trans. 2004;32:831–836. doi: 10.1042/BST0320831. [DOI] [PubMed] [Google Scholar]
- 3.Nayal A, Webb DJ, Horwitz AF. Talin: An emerging focal point of adhesion dynamics. Curr Opin Cell Biol. 2004;16:94–98. doi: 10.1016/j.ceb.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Franco SJ, et al. Calpain-mediated proteolysis of talin regulates adhesion dynamics. Nat Cell Biol. 2004;6:977–983. doi: 10.1038/ncb1175. [DOI] [PubMed] [Google Scholar]
- 5.Jay DG. The clutch hypothesis revisited: Ascribing the roles of actin-associated proteins in filopodial protrusion in the nerve growth cone. J Neurobiol. 2000;44:114–125. doi: 10.1002/1097-4695(200008)44:2<114::aid-neu3>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 6.Brown CM, et al. Probing the integrin-actin linkage using high-resolution protein velocity mapping. J Cell Sci. 2006;119:5204–5214. doi: 10.1242/jcs.03321. [DOI] [PubMed] [Google Scholar]
- 7.Hu K, Ji L, Applegate KT, Danuser G, Waterman-Storer CM. Differential transmission of actin motion within focal adhesions. Science. 2007;315:111–115. doi: 10.1126/science.1135085. [DOI] [PubMed] [Google Scholar]
- 8.Schmidt JM, Zhang J, Lee HS, Stromer MH, Robson RM. Interaction of talin with actin: Sensitive modulation of filament crosslinking activity. Arch Biochem Biophys. 1999;366:139–150. doi: 10.1006/abbi.1999.1204. [DOI] [PubMed] [Google Scholar]
- 9.Goldmann WH, Hess D, Isenberg G. The effect of intact talin and talin tail fragment on actin filament dynamics and structure depends on pH and ionic strength. Eur J Biochem. 1999;260:439–445. doi: 10.1046/j.1432-1327.1999.00177.x. [DOI] [PubMed] [Google Scholar]
- 10.Lee HS, et al. Characterization of an actin-binding site within the talin FERM domain. J Mol Biol. 2004;343:771–784. doi: 10.1016/j.jmb.2004.08.069. [DOI] [PubMed] [Google Scholar]
- 11.Tominaga T, Barber DL. Na-H exchange acts downstream of RhoA to regulate integrin-induced cell adhesion and spreading. Mol Biol Cell. 1998;9:2287–2303. doi: 10.1091/mbc.9.8.2287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denker SP, Barber DL. Cell migration requires both ion translocation and cytoskeletal anchoring by the Na-H exchanger NHE1. J Cell Biol. 2002;159:1087–1096. doi: 10.1083/jcb.200208050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stock C, et al. Migration of human melanoma cells depends on extracellular pH and Na+/H+ exchange. J Physiol. 2005;567:225–238. doi: 10.1113/jphysiol.2005.088344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patel H, Barber DL. A developmentally regulated Na-H exchanger in Dictyostelium discoideum is necessary for cell polarity during chemotaxis. J Cell Biol. 2005;169:321–329. doi: 10.1083/jcb.200412145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hemmings L, et al. Talin contains three actin-binding sites each of which is adjacent to a vinculin-binding site. J Cell Sci. 1996;109:2715–2726. doi: 10.1242/jcs.109.11.2715. [DOI] [PubMed] [Google Scholar]
- 16.McCann RO, Craig SW. The I/LWEQ module: A conserved sequence that signifies F-actin binding in functionally diverse proteins from yeast to mammals. Proc Natl Acad Sci USA. 1997;94:5679–5684. doi: 10.1073/pnas.94.11.5679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Senetar MA, Foster SJ, McCann RO. Intrasteric inhibition mediates the interaction of the I/LWEQ module proteins Talin1, Talin2, Hip1, and Hip12 with actin. Biochemistry. 2004;43:15418–15428. doi: 10.1021/bi0487239. [DOI] [PubMed] [Google Scholar]
- 18.Gingras AR, et al. The structure of the C-terminal actin-binding domain of talin. EMBO J. 2008;27:458–469. doi: 10.1038/sj.emboj.7601965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Georgescu RE, Alexov EG, Gunner MR. Combining conformational flexibility and continuum electrostatics for calculating pKa's in proteins. Biophys J. 2002;83:1731–1748. doi: 10.1016/S0006-3495(02)73940-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brett TJ, Legendre-Guillemin V, McPherson PS, Fremont DH. Structural definition of the F-actin-binding THATCH domain from HIP1R. Nat Struct Mol Biol. 2006;13:121–130. doi: 10.1038/nsmb1043. [DOI] [PubMed] [Google Scholar]
- 21.Smith SJ, McCann RO. A C-terminal dimerization motif is required for focal adhesion targeting of Talin1 and the interaction of the Talin1 I/LWEQ module with F-actin. Biochemistry. 2007;46:10886–10898. doi: 10.1021/bi700637a. [DOI] [PubMed] [Google Scholar]
- 22.Franco SJ, Senetar MA, Simonson WT, Huttenlocher A, McCann RO. The conserved C-terminal I/LWEQ module targets Talin1 to focal adhesions. Cell Motil Cytoskeleton. 2006;63:563–581. doi: 10.1002/cm.20145. [DOI] [PubMed] [Google Scholar]
- 23.Tanokura M. 1H-NMR study on the tautomerism of the imidazole ring of histidine residues. I. Microscopic pK values and molar ratios of tautomers in histidine-containing peptides. Biochim Biophys Acta. 1983;742:576–585. doi: 10.1016/0167-4838(83)90276-5. [DOI] [PubMed] [Google Scholar]
- 24.Denker SP, Huang DC, Orlowski J, Furthmayr H, Barber DL. Direct binding of the Na–H exchanger NHE1 to ERM proteins regulates the cortical cytoskeleton and cell shape independently of H(+) translocation. Mol Cell. 2000;6:1425–1436. doi: 10.1016/s1097-2765(00)00139-8. [DOI] [PubMed] [Google Scholar]
- 25.Frantz C, Karydis A, Nalbant P, Hahn K, Barber DL. Positive feedback between Cdc42 activity and H+ efflux by the Na-H exchanger NHE1 for polarity of migrating cells. J Cell Biol. 2007;179:403–410. doi: 10.1083/jcb.200704169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chaudhry FA, et al. Molecular analysis of system N suggests novel physiological roles in nitrogen metabolism and synaptic transmission. Cell. 1999;99:769–780. doi: 10.1016/s0092-8674(00)81674-8. [DOI] [PubMed] [Google Scholar]
- 27.Jiang G, Giannone G, Critchley DR, Fukumoto E, Sheetz MP. Two-piconewton slip bond between fibronectin and the cytoskeleton depends on talin. Nature. 2003;424:334–337. doi: 10.1038/nature01805. [DOI] [PubMed] [Google Scholar]
- 28.Calderwood DA, et al. The phosphotyrosine binding-like domain of talin activates integrins. J Biol Chem. 2002;277:21749–21758. doi: 10.1074/jbc.M111996200. [DOI] [PubMed] [Google Scholar]
- 29.Schwartz MA, Ingber DE, Lawrence M, Springer TA, Lechene C. Multiple integrins share the ability to induce elevation of intracellular pH. Exp Cell Res. 1991;195:533–535. doi: 10.1016/0014-4827(91)90407-l. [DOI] [PubMed] [Google Scholar]
- 30.Yan W, Nehrke K, Choi J, Barber DL. The Nck-interacting kinase (NIK) phosphorylates the Na+-H+ exchanger NHE1 and regulates NHE1 activation by platelet-derived growth factor. J Biol Chem. 2001;276:31349–31356. doi: 10.1074/jbc.M102679200. [DOI] [PubMed] [Google Scholar]
- 31.King KL, Essig J, Roberts TM, Moerland TS. Regulation of the Ascaris major sperm protein (MSP) cytoskeleton by intracellular pH. Cell Motil Cytoskeleton. 1994;27:193–205. doi: 10.1002/cm.970270302. [DOI] [PubMed] [Google Scholar]
- 32.Pope BJ, Zierler-Gould KM, Kühne R, Weeds AG, Ball LJ. Solution structure of human cofilin: Actin binding, pH sensitivity, and relationship to actin-depolymerizing factor. J Biol Chem. 2004;279:4840–4848. doi: 10.1074/jbc.M310148200. [DOI] [PubMed] [Google Scholar]
- 33.Grey MJ, et al. Characterizing a partially folded intermediate of the villin headpiece domain under non-denaturing conditions: Contribution of His41 to the pH-dependent stability of the N-terminal subdomain. J Mol Biol. 2006;355:1078–1094. doi: 10.1016/j.jmb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 34.Harguindey S, Orive G, Luis Pedraz J, Paradiso A, Reshkin SJ. The role of pH dynamics and the Na+/H+ antiporter in the etiopathogenesis and treatment of cancer. Two faces of the same coin—one single nature. Biochim Biophys Acta. 2005;1756:1–24. doi: 10.1016/j.bbcan.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 35.Mongan J, Case DA, McCammon JA. Constant pH molecular dynamics in generalized Born implicit solvent. J Comput Chem. 2004;16:2038–2048. doi: 10.1002/jcc.20139. [DOI] [PubMed] [Google Scholar]
- 36.Wang J, Cieplak P, Kollman P. How well does a RESP (restrained electrostatic potential) model do in calculating the conformational energies of organic and biological molecules? J Comp Chem. 2000;21:1049. [Google Scholar]
- 37.Onufriev A, Bashford D, Case DA. Exploring protein native states and large-scale conformational changes with a modified generalized born model. Proteins. 2004;55:383–394. doi: 10.1002/prot.20033. [DOI] [PubMed] [Google Scholar]
- 38.DeLano W. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. [Google Scholar]
- 39.Humphrey W, Dalke A, Schulten K. VMD: Visual molecular dynamics. J Mol Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 40.Sattler M, Schleucher J, Griesinger C. Heteronuclear multidimensional NMR experiments for the structure determination of proteins in solution employing pulsed field gradients. Prog Nuclear Magn Reson Spectrosc. 1999;34:93–158. [Google Scholar]
- 41.Hajduk PJ, et al. NMR-based discovery of lead inhibitors that block DNA binding of the human papillomavirus E2 protein. J Med Chem. 1997;40:3144–3150. doi: 10.1021/jm9703404. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.