Abstract
Background
The objective of the present study was to map candidate loci influencing naturally occurring variation in triacylglycerol (TAG) storage using quantitative complementation procedures in Drosophila melanogaster. Based on our results from Drosophila, we performed a human population-based association study to investigate the effect of natural variation in LAMA5 gene on body composition in humans.
Results
We identified four candidate genes that contributed to differences in TAG storage between two strains of D. melanogaster, including Laminin A (LanA), which is a member of the α subfamily of laminin chains. We confirmed the effects of this gene using a viable LanA mutant and showed that female flies homozygous for the mutation had significantly lower TAG storage, body weight, and total protein content than control flies. Drosophila LanA is closely related to human LAMA5 gene, which maps to the well-replicated obesity-linkage region on chromosome 20q13.2-q13.3. We tested for association between three common single nucleotide polymorphisms (SNPs) in the human LAMA5 gene and variation in body composition and lipid profile traits in a cohort of unrelated women of European American (EA) and African American (AA) descent. In both ethnic groups, we found that SNP rs659822 was associated with weight (EA: P = 0.008; AA: P = 0.05) and lean mass (EA: P= 0.003; AA: P = 0.03). We also found this SNP to be associated with height (P = 0.01), total fat mass (P = 0.01), and HDL-cholesterol (P = 0.003) but only in EA women. Finally, significant associations of SNP rs944895 with serum TAG levels (P = 0.02) and HDL-cholesterol (P = 0.03) were observed in AA women.
Conclusion
Our results suggest an evolutionarily conserved role of a member of the laminin gene family in contributing to variation in weight and body composition.
Background
As the prevalence of obesity and its related co-morbidities continue to increase worldwide [1], there is considerable effort being devoted to identify genetic pathways and mechanisms that control fat storage. To gain insights into the genetic basis of natural variation in fat storage, we have used D. melanogaster as a model system. Like mammals, insects store fat as TAG in neutral lipid droplets that are accumulated in the fat body, the functional equivalent of both mammalian liver and white adipose tissue. Drosophila shares many of the components of TAG biosynthesis, degradation, and regulation with mammals, including many of those implicated in human lipodystrophies, diabetes, and obesity [2]. D. melanogaster has proven to be an important model system to identify genetic loci that contribute to variation in quantitative traits, including lipid metabolism [3,4]. In contrast to rodent and human mapping efforts where high-resolution mapping is constrained by intensive labor demands and expense [5], in Drosophila the transition from chromosomal regions [quantitative trait loci (QTL)] identified by recombination mapping to candidate genes [quantitative trait genes (QTGs)] is made possible through the use of quantitative complementation (QC) tests with deficiency and mutant stocks [4,6]. This approach has been highly effective for identifying genetic loci within QTL that contribute to variation in several Drosophila traits, including low heritability traits such as olfactory behavior and life-span [4,7]. The QC test has been also used in mice to investigate the effect of a mutation of the Rgs2 gene on anxiety behaviors [8]. Recently, Drosophila deficiency mapping has been greatly enhanced by the release and availability of the DrosDel and Exelixis deficiency stocks in which all deficiencies occur in the same genetic background and have molecularly defined breakpoints [9,10]. The availability of Exelixis P and piggyBac stocks with single gene insertions all in the same co-isogenic background [11] has also significantly improved our ability to identify positional candidate genes within refined QTL regions.
We previously mapped multiple QTL responsible for natural variation in TAG storage using a population of recombinant inbred (RI) lines derived from two unrelated Drosophila strains, Oregon R (ORE) and Russian 2b (2b) [3]. In this study we used quantitative deficiency mapping to fine-map two of the TAG QTL, one encompassing the cytological region 27B-30D on chromosome 2 and the other encompassing 63A-65A on chromosome 3. Subsequently, we performed QC tests with single gene mutant stocks to identify four candidate genes influencing TAG levels. One of the genes identified is uncoupling protein 4c (Ucp4c), which encodes a product involved in uncoupling of oxidative phosphorylation in mitochondria [12]. Notably, two mammalian homologues of Ucp4c, ubiquitous UCP2 and skeletal-muscle-specific UCP3, have already been shown to regulate mammalian fatty acid metabolism [13]. In addition, several human population studies have reported a strong association between polymorphic variants in UCPs genes and BMI [14]. The remaining three genes are novel candidate genes affecting fat storage: CG9135, CG1399, and Laminin A (LanA). CG9135 and CG1399 belong to a family of genes of unknown function [12]. LanA encodes a protein belonging to the α subfamily of laminin chains [12]. Laminins are heterotrimeric glycoproteins present in the basement membrane matrix where they play a role in cell-matrix adhesion, migration, growth, and differentiation of various cell types [15]. While in mammals different combinations of five α, four β and three γ chains can assemble into at least 15 diverse laminins [15], Drosophila appears to use only one β, oneγ, and two α chains [12]. The Drosophila laminin A chain has significant sequence homology with mammalian laminin α5 chain [16]. In humans, laminin α5 is encoded by the LAMA5 gene, which spans approximately 78 kb on chromosome 20q13.2-q13.3 [17]. Several genome-wide linkage scans have linked this chromosomal region 20q13.2-q13.3 to variation in body mass index (BMI) and percentage body fat [18]. In addition, QTL affecting body weight and adiposity have been mapped to a region on mouse chromosome 2 that is syntenic with chromosome 20q13.2-q13.3 in humans [18]. Taken together with our results from Drosophila, these observations suggested that polymorphisms in LAMA5 contribute to natural variation in body weight and adiposity in humans. To explore this hypothesis we examined the association between genetic variants in the human LAMA5 gene and phenotypic variation in several anthropometric traits, including those reflecting body composition and lipid profile in a in a cohort of 228 unrelated EA and AA pre-menopausal women. We selected three haplotype-tagging SNPs from the International HapMap project http://www.hapmap.org: rs659822 (T > C) in intron 1, rs2297588 (G > A) in intron 51, and rs944895 (T > C) a non-synonymous SNP in exon 68. Our results imply that genetic variation in the LAMA5 gene affects variation in human body composition and lipid profile. However, additional genetic work and functional studies will be necessary to identify causal associations.
Results
Fine mapping and identification of positional candidate genes for TAG storage in Drosophila
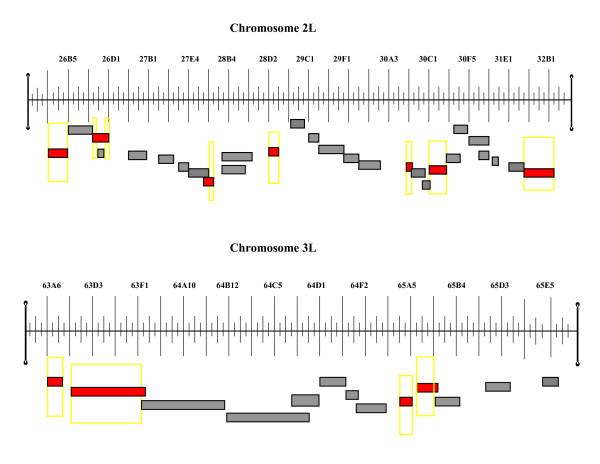
We tested the effects of 38 deficiencies that span the QTL intervals at cytological regions 27B-30D and 63A-65A. After Bonferroni corrections for multiple comparisons, seven deficiencies significantly failed to complement the TAG storage phenotypes of ORE and 2b in the 27B-30D QTL region and four deficiencies in the 63A-65A QTL region (Figure 1). The combined data therefore revealed multiple sub-QTL regions each containing at least one gene affecting variation in TAG storage (Figure 1).
Figure 1.
Quantitative deficiency mapping of D. melanogaster TAG QTL. Long ticks mark sections and short ticks mark subsections of physical maps in the cytological interval 25F5;32B3 on the left (L) arm of chromosome 2 and in the cytological interval 63A6;65E8 on the left arm of chromosome 3. Gray bars represent non-significant deficiencies and red bars correspond to deficiencies with significant failure to complement ORE and 2b QTL for TAG storage. Yellow frames indicate regions where a QTL affecting TAG content between ORE and 2b maps.
To identify candidate genes affecting TAG levels we then performed QC tests using crosses of the ORE and 2b parental strains to mutants of 21 of the genes that map in the refined sub-QTL regions (see Additional File 1: Summary of quantitative complementation tests with mutants of positional candidate genes in D. melanogaster). Four of the genes tested showed a quantitative failure to complement, indicating that allelic differences between ORE and 2b strains at these loci contribute to the differences in TAG storage between the two strains: Ucp4c, CG9135, CG13993, and LanA (see Additional File 1: Summary of quantitative complementation tests with mutants of positional candidate genes in D. melanogaster).
Drosophila LanA influences TAG storage, live weight, and total protein content
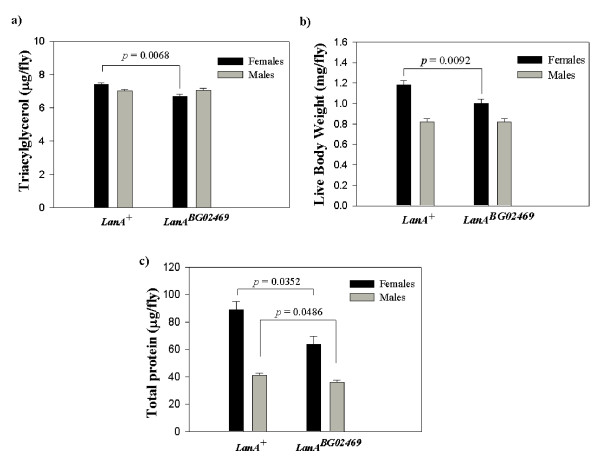
To independently verify the effect of the LanA gene on TAG storage, we measured this trait in flies that were homozygous for the insertional mutation LanABG02469 and non-mutant flies from the co-isogenic control line. We also investigated the effects of the insertion on live body weight and total protein content. For male flies, we found no significant difference between mutant and controls for TAG and live body weight (Figure 2a and 2b). However, LanABG02469 male flies had slightly reduced total protein content (P = 0.0486) compared to controls (Figure 2c). On average, total protein content of LanABG02469 males was 10% lower than that of controls. While the effect on males was minimal, the LanABG02469 mutation had a dramatic effect on female traits. Females with the LanABG02469 mutation had significantly lower TAG storage (P = 0.0068), live body weight (P = 0.0092), and total protein content (P = 0.0352) compared to control flies (Figure 2a–c). The reductions in female TAG storage, live weight, and total protein content relative to control flies were 10%, 16%, and 29%, respectively.
Figure 2.
Effects of Drosophila LanA allele on TAG storage, body weight, and total protein content. Values represent mean ± SEM of TAG storage (panel a), live body weight (panel b), and total protein content (panel c) for n = 9 independent replicates of homozygous LanABG02469 and wild-type male and female flies.
Human LAMA5 variants contribute to variation in anthropometric traits, body composition, and serum lipids
Table 1 summarizes the baseline characteristics of the human cohort stratified by ethnicity. Significant differences in total fat mass (TFM), serum TAG levels, and high density lipoprotein cholesterol (HDL-C) were observed between EA and AA. EA had higher mean TFM and serum TAGs and AA had higher mean HDL-C (Table 1).
Table 1.
Characteristics of the human subjects by ethnicity
| Phenotype (unit of measurement) | European Americans | African Americans |
| (n = 101) | (n = 127) | |
| Age (yr) | 34.66 ± 6.0 | 33.4 ± 5.7 |
| BMI | 27.56 ± 2.18 | 27.50 ± 2.44 |
| Height (cm) | 165.6 ± 0.6 | 163.9 ± 0.6 |
| Weight (kg) | 75.67 ± 9.1 | 73.67 ± 9.0 |
| Total fat mass (kg) | 32.14 ± 6.9* | 30.29 ± 6.7 |
| Lean tissue mass (kg) | 40.22 ± 3.6 | 39.78 ± 4.4 |
| Triacylglycerol (mg/dl) | 115.06 ± 57.3*** | 67.3 ± 25.6 |
| Total cholesterol (mg/dl) | 160.24 ± 31.2 | 155.86 ± 34.1 |
| HDL-cholesterol (mg/dl) | 36.14 ± 9.2*** | 43.21 ± 10.8 |
| LDL-cholesterol (mg/dl) | 101.08 ± 26.7 | 99.18 ± 31.9 |
Values are means ± SD. P values for difference between European Americans and African Americans are obtained using Student's t-test. *p < 0.05 and ***p < 0.001.
The allele and genotype frequencies for each SNP are shown in Table 2. All genotype groups were in Hardy-Weinberg equilibrium. There was no difference in allele or genotype frequencies between EA and AA for SNP rs2297588. However, there was a difference in genotype frequencies between EA and AA for SNP rs944895 and rs659822 (Table 2). These differences remained when the two populations were tested for pair-wise linkage disequilibrium (LD) between SNPs. In both populations rs2297588 was in weak LD with rs659822 (EA: r2 = 0.16; AA: r2 = 0.14) and was more highly lined with rs944895 (EA: r2 = 0.38; AA: r2 = 0.35), whereas rs944895 was associated with rs659822 in the EA population (r2 = 0.37), but not in the AA population.
Table 2.
Allele and genotype frequencies of LAMA5 rs659822, rs2297588, and rs944895 polymorphisms in the study sample
| European Americans | African Americans | |||||||||||
| (n = 101) | (n = 127) | |||||||||||
| rs659822 | rs2297588 | rs944895 | rs659822 | rs2297588 | rs944895 | |||||||
| Genotype frequency | TT | 0.455b*** | GG | 0.524 | TT | 0.416b** | TT | 0.229 | GG | 0.551 | TT | 0.260 |
| TC | 0.416 | GA | 0.446 | TC | 0.495 | TC | 0.543 | GA | 0.417 | TC | 0.520 | |
| CC | 0.129 | AA | 0.030 | CC | 0.089 | CC | 0.228 | AA | 0.032 | CC | 0.220 | |
| Allele frequency | T | 0.663b* | G | 0.748 | T | 0.664 | T | 0.501 | G | 0.760 | T | 0.520 |
| C | 0.337 | A | 0.252 | C | 0.336 | C | 0.499 | A | 0.240 | C | 0.480 | |
| HWEa | 0.658 | 0.117 | 0.382 | 0.475 | 0.149 | 0.727 | ||||||
aP values of Hardy Weinberg equilibrium (HWE) tests. bComparison between racial groups; *p < 0.05, **p < 0.01, ***p < 0.001.
We next examined whether the SNPs were independently associated with each trait. As significant differences in genotype frequencies were observed for SNP rs944895 and rs659822 between the two populations, we tested the association between SNPs and each trait using the data separately by ethnicity. Age, genetic admixture, and appropriate potential confounding variables were included in the analysis as covariates. While no association was found between SNP rs2297588 and any of the traits (data not shown), we did find significant associations between SNP rs659822 and height, body weight, TFM, and lean tissue mass (LTM) in EA (Table 3) assuming a model of additive effects. When we fit the data to a recessive genetic model we also found a significant association between SNP rs659822 and HDL-C in EA (Table 3). On average, EA women that are homozygous for the C allele had short stature, lower mean body weight, TFM, and LTM than those homozygous for the T allele (Table 3; P < 0.05). EA women that were homozygotes for the C allele also had higher levels of HDL-C than those carrying at least one T allele (Table 3; P < 0.05). The association between SNP rs659822 and variation in weight and LTM was also observed in AA women (Table 3). In this case, however, AA women homozygous for the SNP rs659822 C allele had higher mean weight and LTM than those homozygous for the T allele or heterozygous (Table 3; P < 0.05). Finally, significant associations were observed in AA women between alternative alleles at rs944895 and variation in serum TAG levels and HDL-C assuming the additive and dominant models, respectively (Table 3). On average, AA women homozygous for the T allele had lower TAG levels than those homozygous for the C allele and lower HDL-C than those carrying at least one C allele (Table 3; P < 0.05).
Table 3.
Mean ± SEM for anthropometric measures, body composition, and serum lipid profile of study subjects stratified according to LAMA5 rs659822 or rs944895 genotype and ethnicity.
| European American | African American | |||||||
| rs659822 | C/C | C/T | T/T | P* | C/C | C/T | T/T | P |
| n | 13 | 42 | 46 | 29 | 69 | 29 | ||
| BMI | 26.8 ± 0.5 | 27.6 ± 0.3 | 27.7 ± 0.3 | 0.1 | 28.2 ± 0.4 | 27.2 ± 0.3 | 27.4 ± 0.4 | 0.3 |
| Height (cm) | 162.6 ± 1.2 | 164.6 ± 0.8 | 167.3 ± 1.1 | 0.02 | 164.9 ± 1.2 | 164.1 ± 0.8 | 162.5 ± 1.4 | 0.2 |
| Weight (kg) | 69.8 ± 2.3 | 74.7 ± 1.3 | 78.2 ± 1.4 | 0.008 | 76.5 ± 1.7 | 73.2 ± 1.1 | 71.9 ± 1.5 | 0.05 |
| Total fat mass (kg) | 28.8 ± 1.9 | 31.5 ± 1.0 | 33.7 ± 1.0 | 0.01 | 31.3 ± 1.2 | 30.3 ± 0.9 | 29.4 ± 1.1 | 0.3 |
| Lean tissue mass (kg) | 37.6 ± 0.8 | 39.9 ± 0.5 | 41.2 ± 0.5 | 0.003 | 41.4 ± 0.8 | 39.5 ± 0.5 | 38.7 ± 0.8 | 0.03 |
| Triacylglycerol (mg/dl) | 104.9 ± 11.5 | 120.5 ± 11.4 | 112.9 ± 6.3 | 0.8 | 68.1 ± 4.7 | 65.8 ± 3.2 | 70.3 ± 4.3 | 0.6 |
| Total cholesterol (mg/dl) | 153.4 ± 6.3 | 163.0 ± 4.9 | 159.6 ± 4.9 | 0.8 | 162.3 ± 6.5 | 150.1 ± 3.8 | 163.1 ± 6.9 | 0.6 |
| HDL-cholesterol (mg/dl) | 42.4 ± 2.4 | 36.0 ± 1.6 | 34.5 ± 1.1 | 0.003a | 46.1 ± 1.8 | 41.2 ± 1.3 | 45.2 ± 2.1 | 0.07a |
| LDL-cholesterol (mg/dl) | 90.0 ± 5.8 | 102.9 ± 4.1 | 102.5 ± 4.1 | 0.5 | 102.7 ± 6.2 | 95.8 ± 3.6 | 103.8 ± 6.4 | 0.9 |
| rs944895 | ||||||||
| n | 9 | 50 | 42 | 28 | 66 | 33 | ||
| BMI | 26.6 ± 0.7 | 27.5 ± 0.3 | 27.8 ± 0.3 | 0.3 | 27.8 ± 0.4 | 27.6 ± 0.3 | 27.1 ± 0.4 | 0.5 |
| Height (cm) | 163.1 ± 1.2 | 165.6 ± 1.0 | 166.0 ± 0.9 | 0.3 | 163.9 ± 1.4 | 164.4 ± 0.8 | 163.1 ± 1.2 | 0.6 |
| Weight (kg) | 70.9 ± 2.8 | 75.5 ± 1.3 | 76.9 ± 1.3 | 0.09 | 73.5 ± 1.4 | 74.5 ± 1.2 | 72.1 ± 1.6 | 0.7 |
| Total fat mass (kg) | 28.2 ± 2.1 | 31.7 ± 1.0 | 33.3 ± 1.0 | 0.1 | 29.9 ± 1.1 | 30.7 ± 0.9 | 29.7 ± 1.2 | 0.5 |
| Lean tissue mass (kg) | 38.5 ± 1.1 | 40.5 ± 0.6 | 40.2 ± 0.5 | 0.7 | 40.0 ± 0.8 | 40.2 ± 0.5 | 38.8 ± 0.9 | 0.4 |
| Triacylglycerol (mg/dl) | 101.8 ± 8.6 | 117.3 ± 9.2 | 115.2 ± 8.1 | 0.9 | 78.9 ± 5.1 | 65.7 ± 3.1 | 60.9 ± 4.0 | 0.02 |
| Total cholesterol (mg/dl) | 159.8 ± 7.5 | 161.2 ± 4.6 | 159.2 ± 4.9 | 0.7 | 156.5 ± 5.6 | 159.4 ± 4.2 | 148.3 ± 6.5 | 0.7 |
| HDL-cholesterol (mg/dl) | 41.2 ± 3.1 | 35.1 ± 1.4 | 36.3 ± 1.3 | 0.5b | 43.4 ± 2.1 | 45.2 ± 1.4 | 39.1 ± 1.5 | 0.03b |
| LDL-cholesterol (mg/dl) | 98.2 ± 8.6 | 102.7 ± 3.8 | 99.8 ± 4.1 | 0.7 | 97.4 ± 5.1 | 101.1 ± 4.0 | 97.0 ± 6.1 | 1 |
* P values represent the significance of the comparison among genotypes. P values without subscript were calculated assuming additive models. P values with subscripts (a) and (b) were calculated assuming recessive and dominant models, respectively. P values significant after permutations tests to correct for multiple comparisons are highlighted in bold case
Except for the association between SNP rs659822 and weight in AA women, all the single-marker associations remained significant at an experiment-wise P = 0.05 after allowing for multiple testing by permutation analysis [19]. Pair-wise haplotype-based association analyses did not increase the power of these associations (data not shown).
Discussion
We performed quantitative deficiency mapping to dissect two previously identified QTL regions influencing variation in TAG storage among a set of RI lines established from two strains of D. melanogaster, ORE and 2b [3]. The fine mapping revealed that the two QTL broke down into multiple sub-QTL regions (Fig. 1). This indicates that the number of loci influencing variation in TAG levels among these RI lines is much greater than the number suggested by the initial QTL mapping. This finding is consistent with other studies that have fine-mapped QTL for several quantitative traits in Drosophila and mice [4,20,21] and corroborates the complexity of the genetic architecture of quantitative traits.
Several QTL have been associated with BMI, body weight, fat mass, and fat-free mass in human linkage studies [18]. If the complexity observed in model systems turns out to be a common phenomenon also in human traits, then the identification of the genes underlying variation in these traits will remain a challenge. There is increasing evidence that genome-wide association (GWA) studies are a powerful method for identifying genes involved in human complex traits [22]. Taking advantage of the block-like patterns of LD that characterize the human genome [23], these studies rely on the use of hundreds of thousands of "tagging" markers that can capture a significant proportion of the genetic variation and provide power to detect associations. However, one limitation of this approach is that linkage of the markers with variants in a number of genes in the block can make it difficult if not impossible to identify the casual variant affecting the trait. In addition, because of the well-known context dependency of allelic effects of QTL on quantitative traits (e.g. epistasis and genotype by environment interaction) [4], association studies in controlled environments and defined genetic background will potentially allow a more detailed picture of the complexity of the genetic architecture of quantitative traits than that provided by human studies. Studies using D. melanogaster and other model systems will continue to play an important role in pinpointing potential candidate loci affecting quantitative traits.
Using QC tests to mutants of positional genes, we identified Ucp4c, CG9135, CG13993, and LanA as candidate loci that influence variation in TAG storage between ORE and 2b. Notably, three of the implicated loci, Ucp4c, CG9135, and CG13993, are tightly linked, with CG9135 and CG13993 being only 9 kb apart [12]. These however represent only a fraction of the genes underlying the QTL effects identified in this study. Together, there are 286 genes currently mapped in the refined QTL regions. Mutant stocks for 99 of these genes are available from the Drosophila stock center. Many of these mutants are in different genetic backgrounds making it difficult to distinguish allelic effects on a trait at the tested locus from epistatic effects with genetic background. In this study we therefore chose to focus only on loci with mutations in the same genetic background of their controls. QC tests to all available mutations of the genes mapping within the refined regions are underway.
Our studies in flies further suggest that the effect of Drosophila LanA is not limited to TAG storage, but it extends to body weight and whole-body protein content. The observed result on body weight is interesting since three-week old mice homozygous for a hypomorphic mutation in the LAMA5 gene, the mammalian homolog of LanA, have smaller size than their controls [24]. Here we report that natural variation in this gene may contribute to the underlying variation in these traits in human populations. We identified a significant association between a T/C variant in the human LAMA 5 intron 1 (rs659822) and height, body weight, TFM, LTM, and HDL-C in EA women. EA women homozygous for the less frequent variant (CC) on average had lower body weight, TFM, and LTM than those homozygous for the T allele. The effect of SNP rs659822 on weight and LTM was also observed in AA women. In this case, however, AA women that were homozygous for CC at this SNP had higher weight and LTM than women homozygous TT. The opposite effect of rs659822 genotypes on body weight and LTM in the two ethnic groups is intriguing and might be explained by the complexity of the processes that determine variation in these traits, including allelic epistatic interactions within the LAMA5 gene and interactions with other genes and with the environment. In this regard it is important to point out that the genotype frequencies of SNP rs659822 were significantly different across these two groups, with the frequency of the genotype CC being significantly lower in EA women than in AA (Table 3). This sensitivity of the allelic effects of SNPs on phenotypic traits has also been observed in disease-marker studies [25] and is implied in QTL studies in plants [26], Drosophila [27], and mice [28] that show significant differences in allelic effects on phenotypes depending on the genetic background in which they occur. Lin et al. used theoretical modeling to demonstrate that such "flip-flop" associations can occur because the lack of consideration of other genetic loci or environmental factors that influence complex traits [25]. They argue that this is particularly important when a non-causal genetic variant that is linked with the causal polymorphism is investigated [25]. Because genotypes of all polymorphic sites in the LAMA5 gene were not determined and SNP rs659822 is located in an intron, it is possible that rs659822 is not itself the causal polymorphism, but is in LD with the true causal polymorphism somewhere else in this gene. In our results SNP rs659822 was in weak LD with both rs2297588 (r2 = 0.16) and rs944895 (r2 = 0.37) in EA and both SNPs were not associated with any of the traits in this ethnic group. This observation suggests that SNP rs659822 is the site with the largest association with the true causal polymorphism. An overview of the pattern of linkage disequilibrium across the LAMA5 gene established by the HapMap Project in the CEU population of northern and western European ancestry from Utah showed that SNP rs659822 is in strong LD (r2 = 0.73) with a non-synonymous A to G variant in exon 47 (rs2274934) that could be the responsible polymorphism. Notably, this SNP converts the neutral amino-acid asparagine to the negatively charged amino-acid aspartate in one of the laminin EGF-like domains, which have been suggested to act as signals for cellular growth and differentiation [29]. A change in the amino acid structure of this laminin EGF-like domain might explain our finding that variation in LAMA5 associates with a pleiotropic effect on both anthropometric traits and body composition.
We also identified a significant association between a non-synonymous T to C variant in the exon 68 (rs944895) that converts a tryptophan to an arginine in the laminin G (LG)-like 2 domain and variation in serum TAG levels and HDL-C and in AA subjects. AA women homozygous for the less frequent variant (CC) on average had lower serum TAG and HDL-C levels than those homozygous for the T allele. Laminin LG modules have been implicated in interactions with cellular receptors and other extracellular ligands, such as heparan sulfate proteoglycans (HSPGs) [30]. Interestingly, consistent evidence exists that HSPGs play a role in the turnover of lipoproteins, including the uptake of HDL-C in liver [31]. Moreover, cell surface HSPGs contribute to intracellular TAG accumulation in adipocytes [32]. Studies examining the functional effect of rs944895 polymorphism in lipoprotein metabolism will be necessary to understand the mechanisms underlying our findings.
One limitation of our study is that the human association component involved a fairly small sample size and was restricted only to women. This cohort was chosen because measurements of genetic admixture were available for each individual, which allowed us to adjust for ancestry within ethnic groups and, therefore, limit false-positive results [33]. The human data set was also chosen because it provided detailed measurements of body composition for each individual. Clearly, replication of the results in other human cohorts is necessary [34], but the consistency in genetic effects of this member of the laminin gene family in both the fly and humans supports a generally conserved role for this gene in regulating traits reflecting body composition. This is particularly evident for the effect of the gene on lean tissue mass, which was not only observed in both EA and AA women, but also in male and female Drosophila.
Conclusion
Over the past few years, the number of chromosomal regions that contain one or more genes affecting obesity traits in humans and in mammalian models has dramatically increased [18]. Results from our study indicate that D. melanogaster may be a good model to pinpoint those genes with evolutionarily conserved effects on body composition that fall within the large chromosomal regions identified in mammalian QTL studies. Our cross-disciplinary genetic study implicates a member of the laminin gene family as a novel candidate gene affecting variation in body composition traits in natural populations. These observations motivate future studies in independent human populations to verify the effects of this gene.
Methods
Drosophila deficiency and mutant complementation mapping
Drosophila stocks
Deficiency stocks used for the deficiency complementation mapping were obtained from the Bloomington Drosophila Stock Center http://www.flybase.org. All deficiencies used in our study are from the Exelixis and DrosDel collections that have been generated in co-isogenic w1118 backgrounds [9,10].
Mutant stocks were obtained from the Bloomington Drosophila Stock Center and from Trudy Mackay at NC State. Except for LanABG02469, all the mutations are DrosDel and Exelixis P and piggyBac insertions in the w1118co-isogenic background. LanABG02469 is a hypomorphic mutation generated by the insertion in the w1118;Canton S strain of a P-element that is located 339 bp upstream the coding region of the LanA gene. Flies were maintained in vials containing 10 ml of standard cornmeal, agar, sugar, and yeast medium at 25°C.
Experimental design and phenotypic measurements
We conducted the QC tests with deficiencies and mutations using ORE and 2b, the parental lines used to establish the mapping population for the recombination mapping study [3]. In our experiments, we crossed virgin females from ORE and 2b to males from each deficiency stock and to males from the w1118 strain. These crosses produce four possible genotypes: ORE/Deficiency, ORE/w1118 and 2b/Deficiency, 2b/w1118. We measured four replicate trait values of each genotypic class for each sex using the same experimental design described in [3], with the only exception being the way TAG content was measured. Briefly, we kept each genotypic class of flies in four replicate vials, each containing a group of 10 single-sexed individuals. After 4–5 days, we anesthetized each group of flies and measured live weight to 0.01 mg accuracy with an analytical balance. We then homogenized the flies using the protocol described in. We assayed TAG content spectrophotometrically using a commercially available kit (Sigma-Triglyceride Assay Kit) following the manufacturer's suggested protocol. To account for difference in body weight and total protein content, we used the live weight and total protein content as covariates in the analysis of the data. We measured total proteins for each homogenate using a standard Lowry protein assay.
We conducted the QC test with mutant stocks using the same experimental design described for deficiencies, with the only exception being that nine replicate trait values of each genotypic class were measured for each sex.
Statistical analyses
All quantitative complementation tests to deficiencies were carried out simultaneously. A quantitative failure of ORE and 2b QTL alleles to complement a deficiency was inferred if the difference in the mean trait value between the ORE and 2b alleles over the deficiency was significantly greater than the difference in the mean trait value of the ORE and 2b alleles over w1118 [35]. In a three-way factorial analysis of covariance (ANCOVA), these differences are indicated by a line-by-genotype (L × G) or line-by-genotype-by-sex (L × G × S) interaction terms according to the model: y = μ + L + G + S + LW + PRO + L × G + L × S + L × G × S + E, in which μ is the overall mean, L is the fixed main effect of line (ORE or 2b), S is the fixed main effect of sex, G is the fixed main effect of genotype (Def or w1118), LW and PRO are the covariates live body weight and total protein content, and E is the error term. A significant L × G × S interaction term is indicative of a sex-specific failure to complement. The deficiencies that showed a significant failure to complement were confirmed by re-testing nine replicate trait values of each genotypic class for each sex. Bonferroni corrections were performed to control for the effect of multiple comparisons.
The data from QC tests with mutations were analyzed for each sex separately using the two-way factorial model of ANCOVA: y = μ + L + G + LW + PRO + L × G + E, in which μ is the overall mean, L is the fixed main effect of line (ORE or 2b), G is the fixed main effect of genotype (Def or w1118), LW and PRO are the covariates live body weight and total protein content, and E is the error term. We inferred a quantitative failure of ORE and 2b QTL alleles to complement the mutant allele if the L × G interaction term was significant. In addition, as no significant difference in TAG content was observed between the parental lines ORE and 2b[3], we also considered a significant L term as a failure to complement, if the difference between the parental strains was significant in the mutant background but not in the w1118 chromosome background [3]. Bonferroni corrections were performed to control for the effect of multiple comparisons.
The statistical analyses were carried out using the SAS GLM procedures (Version 9.0; SAS Institute, 2002, Cary, NC, USA).
Human study
Subjects
A total of 228 European-American (n = 101) and African-American (n = 127) women were evaluated for the human association study. Subjects were participants of two ongoing longitudinal studies on the role of metabolism in the etiology of obesity conducted in AA and EA pre-menopausal women at the University of Alabama at Birmingham. Prior to testing, subjects were maintained in a weight-maintenance state for 4 weeks. During the final 2 weeks, meals were provided through the General Clinical Research Center at UAB to ensure weight stability of less than 1% variation and to maintain daily macronutrient intake in the range of 20–23% fat, 16–23% protein, and 55–64% carbohydrate. Subjects were then admitted as inpatients to the GCRC for 4 days, during the follicular phase of the menstrual cycle. All metabolic testing took place during this inpatient period. At the time of testing, subjects were sedentary (no previous history of exercise training), had a BMI range between 24 – 30 kg/m2, were nonsmokers, and were not taking any medication known to alter body composition (including hormones). Race was determined by self-reported African-American or Caucasian ancestry in both parents and grandparents.
The study protocol was approved by the Institutional Review Board for human studies at the University of Alabama at Birmingham. A written informed consent was obtained from all study participants before enrolling in the study.
Anthropometrical and serum lipid measurements
Height and body weight were measured in light indoor clothes and without shoes. Blood samples were withdrawn after 12-h overnight fast. Analyses for serum lipids were performed in the Core Laboratory of the General Clinical Research Center and the Clinical Nutrition Research Center (CNRU) at UAB. Total cholesterol, HDL-C, and TAGs were measured with the Ektachem DT II System. With this system, HDL-C is measured after precipitation of low-density lipoprotein cholesterol (LDL-C) and very-low-density lipoprotein cholesterol with dextran sulfate and magnesium chloride. Control sera of low and high substrate concentration are analyzed with each group of samples, and values for these controls must fall within accepted ranges before samples are analyzed. The DT II is calibrated every six months with reagents supplied by the manufacturer. LDL-C was estimated using the Friedewald formula [36].
Body composition
Body composition [TFM and LTM] was measured by dual energy X-ray absorptiometry using either a Lunar DPX-L densitometer (LUNAR Radiation Corp., Madison, WI) or a LUNAR Prodigy densitometer in the Department of Nutrition Sciences at UAB. Body composition assessed by these instruments generally differs by a coefficient of variation of 4% or less [37]. Subjects were scanned in light clothing while lying flat on their backs with arms at their sides.
Genotyping
To test for associations between genetic variants in human LAMA5 and phenotypic traits, we selected three of the human LAMA5 SNPs identified by the International HapMap project. Using HapMap data release #21a, we estimated that these three SNPs captured 95.6% of common variation (Minor Allele Frequency >0.05, n = 23) at an r2 > 0.8 across LAMA5 gene. The genotypes of these polymorphisms were determined by Pyrosequencing technology [38] at the CNRU Genetics Core at UAB.
To account for the confounding effects of population stratification, we used estimates of genetic admixture as a covariate in statistical models. The genetic admixture estimates were obtained from the genotyping of ancestry informative markers (AIMs) across the human genome. These AIMs are informative for parental ancestry, defined as those long-separated populations that intermixed during historical periods to produce new admixed populations. Genotyping for the measures of genetic admixture was performed at Prevention Genetics http://www.preventiongenetics.com using the McSNP method and agarose gel electrophoresis, as previously described [39]. Molecular techniques for the allelic identification and methodology for genetic admixture application have been described elsewhere[40,41]. Approximately 100 ancestry informative markers were utilized for the study. Information regarding marker sequences, experimental details, and parental population allele frequencies has been submitted to dbSNP http://www.ncbi.nlm.nih.gov/SNP/ under the handle PSU-ANTH.
Data Analyses
We assessed Hardy-Weinberg equilibrium, estimated haplotype frequencies, and r2 linkage disequilibrium coefficients by methods implemented in Arlequin program 3.01 [42]. Allele and genotype frequency comparisons between EA and AA samples were performed by the χ2 test. To test the effect of each genotyped SNP on trait variation, we performed genotypic associations for dominant, additive, and recessive models using linear regression analysis. Age and genetic admixture were used as covariates in all the analyses. As strong correlations have been shown between fat mass and lipid profile [43], lipoprotein levels were additionally adjusted for TFM and TAGs. TAG levels were also adjusted for TFM. Dummy variables were assigned to code the three genotypes in each model. In the additive model, we used 0, 1 and 2 to code for individuals homozygous for the major allele, heterozygous, and homozygous for the minor allele, respectively. In the dominant and recessive models, we used 0 to code for individuals homozygous for the major and minor alleles, respectively, and 1 to code for individuals carrying at least one copy of the other allele. Pair-wise haplotype-based association analyses were also performed. For all regression models, studentized residuals were evaluated for normality and logarithmic transformations of the dependent variable was performed to improve normality. When normality of the residuals was not obtained after transformations, the observations that were above and below three standard deviations were removed from the analyses. To test for significant differences among means according to genotype, data from the final regression model was analyzed by analysis of variances and mean differences assessed by post-hoc Duncan tests at P < 0.05. To control for the effect of multiple comparisons, we performed permutation tests (1000 simulations) to generate empirical P values under the null hypotheses of no association between genotypes and traits [19]. All the analyses were performed using SAS (Version 9.0; SAS Institute, 2002, Cary, NC, USA).
Authors' contributions
MDL conceived the study, participated in its design and coordination, carried out the Drosophila data analysis, and wrote the manuscript. MMC carried out the Drosophila complementation tests. KC carried out the human statistical analyses. JRF participated in the design and coordination of the study and the human statistical analyses. KHL carried out the human genotyping. JRF, BAG, and GRH contributed to design and acquisition of human data. BAG and JRF revised critically the manuscript. All authors read and approved the final manuscript.
Supplementary Material
Summary of quantitative complementation tests with mutants of positional candidate genes in D. melanogaster. The table contains a list of all the positional candidate genes and the corresponding mutant alleles analyzed by quantitative complementation tests. In the table are also reported the cytological positions of the candidate genes and the P values for Line and Line × Genotype effects of two-way factorial ANOVAs (see text for further explanation).
Acknowledgments
Acknowledgements
We thank Jeff Leips and two anonymous reviewers for comments on earlier versions of the manuscript. This work was supported by NIH grants R01HL80812 (MDL), R01DK49779 (GRH), and R01DK51694 (BAG), General Clinical research Center grant M01RR00032, CNRU grant P30DK56336. Stouffer's Lean Cuisine entrees were provided by the Nestlé Food Co., Solon OH and Smart Ones entrees were provided by H.J. Heinz Foods, Pittsburg, PA.
Contributor Information
Maria De Luca, Email: mdeluca2@uab.edu.
Michelle Moses Chambers, Email: mmoses@uab.edu.
Krista Casazza, Email: kristac@uab.edu.
Kerry H Lok, Email: kerrylok@uab.edu.
Gary R Hunter, Email: ghunter@uab.edu.
Barbara A Gower, Email: bgower@uab.edu.
José R Fernández, Email: jose@uab.edu.
References
- Klein S, Burke LE, Bray GA, Blair S, Allison DB, Pi-Sunyer X, Hong YL, Eckel RH. Clinical implications of obesity with specific focus on cardiovascular disease - A statement for professionals from the American Heart Association Council on Nutrition, Physical Activity, and Metabolism - Endorsed by the American College of Cardiology Foundation. Circulation. 2004;110:2952–2967. doi: 10.1161/01.CIR.0000145546.97738.1E. [DOI] [PubMed] [Google Scholar]
- Ruden DM, De Luca M, Garfinkel MD, Bynum KL, Lu X. Drosophila nutrigenomics can provide clues to human gene-nutrient interactions. Annu Rev Nutr. 2005;25:499–522. doi: 10.1146/annurev.nutr.25.050304.092708. [DOI] [PubMed] [Google Scholar]
- De Luca M, Yi N, Allison DB, Leips J, Ruden DM. Mapping quantitative trait loci affecting variation in Drosophila triacylglycerol storage. Obes Res. 2005;13:1596–1605. doi: 10.1038/oby.2005.196. [DOI] [PubMed] [Google Scholar]
- Mackay TF, Anholt RR. Of Flies and Man: Drosophila as a Model for Human Complex Traits. Annu Rev Genomics Hum Genet. 2006;7:339–367. doi: 10.1146/annurev.genom.7.080505.115758. [DOI] [PubMed] [Google Scholar]
- Mott R. Finding the molecular basis of complex genetic variation in humans and mice. Philos Trans R Soc Lond B Biol Sci. 2006;361:393–401. doi: 10.1098/rstb.2005.1798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackay TF. The genetic architecture of quantitative traits: lessons from Drosophila. Curr Opin Genet Dev. 2004;14:253–257. doi: 10.1016/j.gde.2004.04.003. [DOI] [PubMed] [Google Scholar]
- De Luca M, Leips J. Mapping genetic polymorphisms affecting natural variation in Drosophila longevity. Methods Mol Biol. 2007;371:307–320. doi: 10.1007/978-1-59745-361-5_22. [DOI] [PubMed] [Google Scholar]
- Yalcin B, Willis-Owen SAG, Fullerton J, Meesaq A, Deacon RM, Rawlins JNP, Copley RR, Morris AP, Flint J, Mott R. Genetic dissection of a behavioral quantitative trait locus shows that Rgs2 modulates anxiety in mice. Nat Genet. 2004;36:1197–1202. doi: 10.1038/ng1450. [DOI] [PubMed] [Google Scholar]
- Ryder E, Ashburner M, Bautista-Llacer R, Drummond J, Webster J, Johnson G, Morley T, Chan YS, Blows F, Coulson D, Reuter G, Baisch H, Apelt C, Kauk A, Rudolph T, Kube M, Klimm M, Nickel C, Szidonya J, Maróy P, Pal M, Rasmuson-Lestander A, Ekström K, Stocker H, Hugentobler C, Hafen E, Gubb D, Pflugfelder G, Dorner C, Marhold J, Serras F, Corominas M, Punset A, Roote J, Russell S. The DrosDel Deletion Collection: A Drosophila Genomewide Chromosomal Deficiency Resource. Genetics. 2007;177:615. doi: 10.1534/genetics.107.076216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks AL, Cook KR, Belvin M, Dompe NA, Fawcett R, Huppert K, Tan LR, Winter CG, Bogart KP, Deal JE, Deal-Herr ME, Grant D, Marcinko M, Miyazaki WY, Robertson S, Shaw KJ, Tabios M, Vysotskaia V, Zhao L, Andrade RS, Edgar KA, Howie E, Killpack K, Milash B, Norton A, Thao D, Whittaker K, Winner MA, Friedman L, Margolis J, Singer MA, Kopczynski C, Curtis D, Kaufman TC, Plowman GD, Duyk G, Francis-Lang HL. Systematic generation of high-resolution deletion coverage of the Drosophila melanogaster genome. Nat Genet. 2004;36:288–292. doi: 10.1038/ng1312. [DOI] [PubMed] [Google Scholar]
- Thibault ST, Singer MA, Miyazaki WY, Milash B, Dompe NA, Singh CM, Buchholz R, Demsky M, Fawcett R, Francis-Lang HL. A complementary transposon tool kit for Drosophila melanogaster using P and piggyBac. Nat Genet. 2004;36:283–287. doi: 10.1038/ng1314. [DOI] [PubMed] [Google Scholar]
- Drysdale RA, Crosby MA. FlyBase: genes and gene models. Nucleic Acids Res. 2005;33:D390–D395. doi: 10.1093/nar/gki046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulloo AG, Seydoux J, Jacquet J. Adaptive thermogenesis and uncoupling proteins: a reappraisal of their roles in fat metabolism and energy balance. Physiol Behav. 2004;83:587–602. doi: 10.1016/j.physbeh.2004.07.028. [DOI] [PubMed] [Google Scholar]
- Schonfeld-Warden NA, Warden CH. Physiological effects of variants in human uncoupling proteins: UCP2 influences body-mass index. Biochem Soc Trans. 2001;29:777–784. doi: 10.1042/0300-5127:0290777. [DOI] [PubMed] [Google Scholar]
- Colognato H, Yurchenco PD. Form and function: The laminin family of heterotrimers. Dev Dyn. 2000;218:213–234. doi: 10.1002/(SICI)1097-0177(200006)218:2<213::AID-DVDY1>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- Miner JH, Lewis RM, Sanes JR. Molecular-Cloning of A Novel Laminin Chain, Alpha-5, and Widespread Expression in Adult-Mouse Tissues. J Biol Chem. 1995;270:28523–28526. doi: 10.1074/jbc.270.48.28523. [DOI] [PubMed] [Google Scholar]
- Durkin ME, Loechel F, Mattei MG, Gilpin BJ, Albrechtsen R, Wewer UM. Tissue-specific expression of the human laminin a5-chain, and mapping of the gene to human chromosome 20q13. 2-13.3 and to distal mouse chromosome2 near the locus for the ragged (Ra) mutation. FEBS Letters. 1997;411:296–300. doi: 10.1016/s0014-5793(97)00686-8. [DOI] [PubMed] [Google Scholar]
- Rankinen T, Zuberi A, Chagnon YC, Weisnagel SJ, Argyropoulos G, Walts B, Perusse L, Bouchard C. The human obesity gene map: the 2005 update. Obesity (Silver Spring) 2006;14:529–644. doi: 10.1038/oby.2006.71. [DOI] [PubMed] [Google Scholar]
- Doerge RW, Churchill GA. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legare ME, Bartlett FS, Frankel WN. A major effect QTL determined by multiple genes in epileptic EL mice. Genome Res. 2000;10:42–48. [PMC free article] [PubMed] [Google Scholar]
- Wandstrat A, Wakeland E. The genetics of complex autoimmune diseases: non-MHC susceptibility genes. Nat Immunol. 2001;2:802–809. doi: 10.1038/ni0901-802. [DOI] [PubMed] [Google Scholar]
- Christensen K, Murray JC. What Genome-wide Association Studies Can Do for Medicine. N Engl J Med. 2007;356:1094. doi: 10.1056/NEJMp068126. [DOI] [PubMed] [Google Scholar]
- Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]
- Shannon MB, Patton BL, Harvey SJ, Miner JH. A Hypomorphic Mutation in the Mouse Laminin {alpha} 5 Gene Causes Polycystic Kidney Disease. J Am Soc Nephrol. 2006;17:1913–22. doi: 10.1681/ASN.2005121298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin PI, Vance JM, Pericak-Vance MA, Martin ER. No gene is an island: The flip-flop phenomenon. Am J Hum Genet. 2007;80:531–538. doi: 10.1086/512133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ungerer MC, Linder CR, Rieseberg LH. Effects of genetic background on response to selection in experimental populations of Arabidopsis thaliana. Genetics. 2003;163:277–286. doi: 10.1093/genetics/163.1.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leips J, Mackay TFC. Quantitative Trait Loci for Life Span in Drosophila melanogaster Interactions With Genetic Background and Larval Density. Genetics. 2000;155:1773–1788. doi: 10.1093/genetics/155.4.1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi N, Diament A, Chiu S, Kim K, Allison DB, Fisler JS, Warden CH. Characterization of Epistasis Influencing Complex Spontaneous Obesity in the BSB Model. Genetics. 2004;167:399–409. doi: 10.1534/genetics.167.1.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engel J. EGF-like domains in extracellular matrix proteins: localized signals for growth and differentiation? FEBS Lett. 1989;251:1–7. doi: 10.1016/0014-5793(89)81417-6. [DOI] [PubMed] [Google Scholar]
- Timpl R, Tisi D, Talts JF, Andac Z, Sasaki T, Hohenester E. Structure and function of laminin LG modules. Matrix Biol. 2000;19:309–317. doi: 10.1016/s0945-053x(00)00072-x. [DOI] [PubMed] [Google Scholar]
- Kolset SO, Salmivirta M. Cell surface heparan sulfate proteoglycans and lipoprotein metabolism. Cell Mol Life Sci. 1999;56:857–870. doi: 10.1007/s000180050031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilsie LC, Chanchani S, Navaratna D, Orlando RA. Cell surface heparan sulfate proteoglycans contribute to intracellular lipid accumulation in adipocytes. Lipids Health Dis. 2005;4 doi: 10.1186/1476-511X-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Divers J, Vaughan LK, Padilla MA, Fernandez JR, Allison DB, Redden DT. Correcting for measurement error in individual ancestry estimates in structured association tests. Genetics. 2007;176:1823–1833. doi: 10.1534/genetics.107.075408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanock SJ, Manolio T, Boehnke M, Boerwinkle E, Hunter DJ, Thomas G, Hirschhorn JN, Abecasis G, Altshuler D, Bailey-Wilson JE, Brooks LD, Cardon LR, Daly M, Donnelly P, Fraumeni JF, Jr., Freimer NB, Gerhard DS, Gunter C, Guttmacher AE, Guyer MS, Harris EL, Hoh J, Hoover R, Kong CA, Merikangas KR, Morton CC, Palmer LJ, Phimister EG, Rice JP, Roberts J, Rotimi C, Tucker MA, Vogan KJ, Wacholder S, Wijsman EM, Winn DM, Collins FS. Replicating genotype-phenotype associations. Nature. 2007;447:655–660. doi: 10.1038/447655a. [DOI] [PubMed] [Google Scholar]
- Pasyukova EG, Vieira C, Mackay TF. Deficiency mapping of quantitative trait loci affecting longevity in Drosophila melanogaster. Genetics. 2000;156:1129–1146. doi: 10.1093/genetics/156.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedewald WT, Levy RI, Fredrickson DS. Estimation of the concentration of low-density lipoprotein cholesterol in plasma, without use of the preparative ultracentrifuge. Clin Chem. 1972;18:499–502. [PubMed] [Google Scholar]
- Huffman DM, Landy NM, Potter TR, Nagy TR, Gower BA. Comparison of the LUNA DPX-L and Prodigy dual-energy X-ray absorptiometers for assesing total and regional body composition. Int J Body Composition Res. 2005;3:25–30. [PMC free article] [PubMed] [Google Scholar]
- Ronaghi M, Uhlen M, Nyren P. A sequencing method based on real-time pyrophosphate. Science. 1998;281:363, 365. doi: 10.1126/science.281.5375.363. [DOI] [PubMed] [Google Scholar]
- Akey JM, Sosnoski D, Parra E, Dios S, Hiester K, Su B, Bonilla C, Jin L, Shriver MD. Melting curve analysis of SNPs (McSNP (R)): A gel-free and inexpensive approach for SNP genotyping. Biotechniques. 2001;30:358–367. doi: 10.2144/01302tt05. [DOI] [PubMed] [Google Scholar]
- Parra EJ, Kittles RA, Argyropoulos G, Pfaff CL, Hiester K, Bonilla C, Sylvester N, Parrish-Gause D, Garvey WT, Jin L, McKeigue PM, Kamboh MI, Ferrell RE, Pollitzer WS, Shriver MD. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am J Phys Anthropol. 2001;114:18–29. doi: 10.1002/1096-8644(200101)114:1<18::AID-AJPA1002>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Shriver MD, Parra EJ, Dios S, Bonilla C, Norton H, Jovel C, Pfaff C, Jones C, Massac A, Cameron N, Baron A, Jackson T, Argyropoulos G, Jin L, Hoggart CJ, McKeigue PM, Kittles RA. Skin pigmentation, biogeographical ancestry and admixture mapping. Hum Genet. 2003;112:387–399. doi: 10.1007/s00439-002-0896-y. [DOI] [PubMed] [Google Scholar]
- Excoffier L, Laval G, Schneider S. Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online. 2005;1:47–50. [PMC free article] [PubMed] [Google Scholar]
- Choi JW, Choe HW, Pai SH. Serum lipid concentrations correlate more strongly with total body fat than with body mass index in obese humans. Clin Chim Acta. 2003;329:83–87. doi: 10.1016/s0009-8981(03)00018-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Summary of quantitative complementation tests with mutants of positional candidate genes in D. melanogaster. The table contains a list of all the positional candidate genes and the corresponding mutant alleles analyzed by quantitative complementation tests. In the table are also reported the cytological positions of the candidate genes and the P values for Line and Line × Genotype effects of two-way factorial ANOVAs (see text for further explanation).