Abstract
Ethanol induces the development of hepatic steatosis, increasingly recognized as causing vulnerability to subsequent liver injury. Ethanol has been shown to activate SREBP-1 (sterol regulatory element-binding protein) processing through the conventional cholesterol-sensitive pathway (1). The present study demonstrates that ethanol can also bring about SREBP-1 cleavage and activation through a novel pathway dependent on the endoplasmic reticulum-localized caspases-4 and -12. Evidence is presented that tumor necrosis factor can stimulate caspase-4 and -12 activation in ethanol-exposed cells, which cleaves SREBP-1 to a transcriptionally active form to induce the synthesis of lipogenic enzymes and triglycerides. Moreover, the caspase-4 and -12-dependent activation of SREBP-1 is insensitive to the normal negative feedback exerted by cholesterol and is mediated by the translocation of the scaffolding protein, TRAF-2, to the endoplasmic reticulum.
Hepatic steatosis is a frequent complication of excessive ethanol consumption. The etiology is due to the dual effects of decreasing fatty acid β-oxidation and increasing fatty acid synthesis (2-4). Steatosis was once thought to be a benign condition, but recent reevaluation has uncovered that hepatic steatosis can predispose the liver to more progressive and serious forms of liver injury such as alcoholic steatohepatitis and cirrhosis.
The etiology of the steatosis caused by ethanol was thought to typically involve an increase in the concentration of NADH caused by the metabolism of ethanol. Although this may be an important contributory factor, additional effects of ethanol abet in the surplus lipid accumulation. For instance, ethanol exposure activates sterol response element-binding proteins (SREBP)2 that elevate the levels and activities of lipogenic enzymes such as acetyl-CoA carboxylase (ACC) and fatty acid synthase (3).
Three isoforms of SREBP exist, SREBP-1a, -1c, and 2. SREBP-1a and 1c are transcribed from a common gene but utilize differing transcription start sites that result in alternate forms of exon 1 (5, 6). SREBP-1a regulates fatty acid and cholesterol synthesis, whereas SREBP-1c and SREBP-2 are involved mainly in regulating fatty acid and cholesterol synthesis, respectively. The SREBPs reside in the endoplasmic reticulum. SREBP is organized into an amino-terminal domain that contains the basic helix-loop-helix leucine zipper motif for binding to sterol response elements (SRE) in the promoter regions of SREBP responsive genes. For SREBP to perform its function as a transcription factor, the amino-terminal domain must be released from the intermembrane portion of the protein and translocate to the nucleus. This is accomplished by the proteolytic cleavage of the amino-terminal domain from the carboxylterminal domain by the sequential action of two proteases: site-1 protease (S1P) and site-2 protease (S2P) located in the Golgi apparatus. SREBP must first be escorted from its site in the ER to the Golgi by SREBP cleavage activating protein (SCAP). SCAP possesses a cholesterol sensing domain. When cholesterol levels are low, SCAP escorts SREBP from the ER to the Golgi where S1P initially cleaves the precursor form of SREBP in the luminal loop, producing a membrane-bound intermediate. S2P then cleaves the intermediate to release the transcription factor domain from the membrane. When the cholesterol level of the cell rises, SCAP undergoes a conformational change that causes it to bind to the ER resident proteins, Insig-1 or -2, which hinders the incorporation of the SCAP-SREBP complex into ER transport vesicles. In this instance, SREBP cannot be transported to the Golgi to undergo activation by S1P and S2P. In this way the activation of SREBP is suppressed when cholesterol levels are elevated.
Recent studies have uncovered that ethanol exposure can provoke the endoplasmic reticulum stress response (7). A number of adverse conditions are known to induce ER stress and provoke the unfolded protein response (8, 9). Ethanol brings about a myriad of alterations in cell function, any of which separately or in tandem may help promote ER stress. For instance, an increase in the levels of homocysteine brought about by ethanol has been shown to induce ER stress (10, 11). The homocysteine-induced response is associated with activation of SREBPs. The ER stress response entails the mobilization and synthesis of a number of proteins such as CHOP and activation of caspases-4 and -12 (10, 12). Indeed, ethanol exposure has been shown to bring about the ER-localized activation of caspase-12 in the liver of micropigs (13).
The present report demonstrates that sublethal concentrations of TNF can bring about the activation of caspases-4 and -12 in ethanol-exposed cells that in turn is capable of cleaving and activating SREBP-1. Moreover, the caspase-4 and -12-dependent cleavage and activation of SREBP-1 in ethanol-exposed cells is insensitive to the normal negative feedback of cholesterol, thus resulting in disengagement in the activation of SREBP-1 from the intracellular levels of cholesterol and causing an inappropriate induction of lipogenic enzymes in the face of cholesterol excess.
EXPERIMENTAL PROCEDURES
Cell Culture and Treatments—HepG2 cells and McA-RH 7777 cells were maintained in Dulbecco's modified Eagle's medium containing 25 mm glucose, 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin under an atmosphere of 95% air, 5% CO2 at 37 °C. Cells were subcultured 1:5 once a week. The cells were treated with 25 mm ethanol for 48 h. Where indicated, cells were first exposed to 25 mm ethanol for 24 h and then treated with TNF at a concentration of 1 ng/ml for an additional 24 h in the presence of 25 mm ethanol. Where indicated in the figure legends, cultures were supplemented with a combination of cholesterol and 25-hydroxycholesterol at concentrations of 10 and 1 μg/ml, respectively. The culture medium was replaced every 24 h with fresh medium containing ethanol. To prevent evaporation of ethanol, the flasks or plates were placed in a plastic dessicator in the incubator containing a mixture of water and ethanol. The level of ethanol in the culture medium was monitored spectrophotometrically by an alcohol dehydrogenase assay. The level of NADH was measured by an increase in absorbance at 340 nm. After 24 h of ethanol exposure, the cells (control or ethanol-treated) were trypsinized, counted in a hemocytometer, and plated into 6-well 9.3-cm2 plates at 1.0 × 106 cells for determination of caspase-4 and -12 activities. The cells were allowed to attach and spread for 24 h. When present, ethanol was added back to the cells during this time. On the day of the experiment, the cells were washed and placed in Dulbecco's modified Eagle's medium in the presence of 25 mm ethanol. Where indicated, TNF was dissolved in phosphate-buffered saline and added to the wells in a 0.2% volume to give a final concentration of 1 ng/ml (22 units/ml) and incubated for an additional 24 h (total ethanol exposure of 48 h).
Detection of Caspase-4 and -12 Activity and Triglyceride Assay—The caspase-4 and -12 assays are based on the ability of the active enzyme to cleave the fluorophore AFC from enzyme substrates LEVD-AFC and ATAD-AFC, respectively. Cell extracts were prepared and diluted 1:1 with 2× reaction buffer (10 mm Tris, pH 7.4, 1 mm dithiothreitol, 2 mm EDTA, 0.1% CHAPS, 1 mm phenylmethylsulfonyl fluoride, 10 μg/ml pepstatin, 10 μg/ml leupeptin). Caspase-4 or -12 substrate was added to a final concentration of 50 μm, and the reaction was incubated for 1 h at 37 °C. Samples were then transferred to a 96-well plate, and fluorescence measurements were made in a 96-well plate reader at 400 nm excitation and 505 nm emission. For measurement of triglyceride levels, cells where harvested and the level of triglycerides determined enzymatically with absorbance of the reaction measured spectrophotometrically at 540 nm (Sigma).
RNA Interference—The Dharmacon SMART selection and SMART pooling technologies are utilized for RNA interference studies. The SMART selection uses an algorithm composed of 33 criteria and parameters that attempt to limit nonfunctional small interfering RNAs (siRNAs). SMART pooling combines four SMART-selected siRNA duplexes in a single pool to increase the probability of reducing mRNA levels by at least 75%. The siRNAs targeting SREBP-1, caspases-4, -12, or -3, and tumor necrosis factor-associated factor 2 (TRAF-2) were delivered by a lipid-based method supplied from a commercial vendor (Gene Therapy Systems) at a final siRNA concentration of 100 nm. After formation of the siRNA-liposome complexes, the mixture was added to the McA-RH 7777 or HepG2 cells in serum-free medium for 4 h. Afterward, the medium was aspirated, and complete medium was added back with or without the addition of 25 mm ethanol for 48 h.
Isolation of Cytosolic and Microsomal Fractions and Western Blotting—Cells were plated in 25-cm2 flasks at 5.0 × 106 cells/flask. After treatments, the cells were harvested by trypsinization followed by centrifugation at 600 × g for 10 min at 4 °C. The cell pellets were washed once in phosphate-buffered saline and then resuspended in 3 volumes of isolation buffer (20 mm HEPES, pH 7.4, 10 mm KCl, 1.5 mm MgCl2, 1 mm sodium/EDTA, 1 mm dithiothreitol, 10 mm phenylmethylsulfonyl fluoride, 1.0 μg/ml leupeptin, and 1.0 μg/ml aprotinin in 250 mm sucrose). After being chilled on ice for 3 min, the cells were disrupted by 40 strokes of a glass homogenizer. The homogenate was centrifuged twice at 2,500 × g at 4 °C to remove unbroken cells and nuclei. The mitochondria were then pelleted by centrifugation at 12,000 × g at 4 °C for 30 min. The supernatant was re-centrifuged at 200,000 × g (Beckman Coulter Optima TLX Ultracentrifuge) for 1 h for the preparation of microsomes. The resulting supernatant contains the soluble cytosolic fraction and the microsomal pellet represents the ER and Golgi. Various fractions, as indicated in the figure legends (25 μg of protein) were separated on 12% SDS-polyacrylamide gels and electroblotted onto nitrocellulose membranes. Full-length and cleaved SREBP-1 were detected with a polyclonal antibody to SREBP-1 at a dilution of 1:500 (Affinity Bioreagents). TRAF-2 was detected with a rabbit polyclonal antibody at a dilution of 1:1000 (Cell Signaling Technologies). Acetyl-CoA carboxylase was detected with a rabbit monoclonal antibody at a dilution of 1:1000 (Cell Signaling Technologies). Caspase-4 was detected with a mouse monoclonal antibody at a dilution of 1:1,000 (4B9; MBL International). Caspases-3 and -9 were detected with rabbit polyclonal antibodies that recognize the activated cleavage products at a dilution of 1:500 (Calbiochem). In each case, the relevant protein was visualized by staining with the appropriate secondary horseradish peroxidase-labeled antibody (1:10,000) and was detected by enhanced chemiluminescence.
SRE Binding and Reporter Assay—After 24 h of ethanol exposure in the presence or absence of cholesterol supplementation, cells were transiently transfected with a native luciferase reporter construct or a luciferase reporter driven by an SRE promoter containing multiple SRE elements upstream of a minimal TA promoter derived from the TATA box of the herpes simplex virus-thymidine kinase promoter (Panomics). The transfected cells were incubated for a further 24 h in 25 mm ethanol in the absence or presence of cholesterol supplementation and/or TNF treatment. The cells were then lysed (25 mm Tris phosphate, pH 7.8, 2 mm dithiothreitol, 2 mm 1,2-diaminocyclohexane-N,N,N′,N′-tetraacetic acid, 10% glycerol, 1% Triton X-100) and luciferase activity measured with a luciferase activity assay kit (Stratagene) utilizing a single tube luminometer (Berthold). Luciferase activity measured in cells lysates was normalized to β-galactosidase expression and is expressed relative to values obtained using the empty luciferase reporter vector. For the SRE binding assay, nuclei were isolated from McA-RH 7777 or HepG2 cells exposed to the conditions indicated. Cleaved and active SREBP-1 binds specifically to a double-stranded DNA sequence containing the SREBP response element immobilized to the wells of a 96-well plate. The SREBP-1 was detected by the addition of a SREBP-1 antibody. Following washes, a secondary antibody conjugated to horseradish peroxidase was added and a colorimetric signal was detected at 450 nm on a plate reader.
Generation of Recombinant SREBP-1 and in Vitro Cleavage Assay—Knock-down and rescue experiments were performed on HepG2 and McA-RH 7777 cells. First, endogenous SREBP-1 expression was repressed by siRNA transfection targeting SREBP-1 as described above. SREBP-1 expression was then restored by transfection with a pcDNA3.1 plasmid containing a cDNA for the expression of the caspase site mutants SREBP-1(D460A) for the human HepG2 cells and SREBP-1(D451A) for the mouse McA-RH 7777 cells, respectively. Note that the human and mouse proteins differ slightly in their location of the caspase cleavage site and the S2P protease cleavage site. Arginines 527 and 524 for the human and mouse proteins, respectively, are crucial for S2P cleavage. Therefore mutants SREBP-1(R527A) and SREBP-1(R524A) were generated and expressed in HepG2 cells and McA-RH 7777 cells, respectively. Purified SREBP-1 containing a histidine tag was generated by in vitro transcription and translation followed by purification on a nickel resin. The purified SREBP-1 was incubated with 5 units of recombinant caspase-3, -4, or -2 (EMD Biosciences) in caspase reaction buffer (50 mm HEPES, pH 7.2, 50 mm NaCl, 0.1% CHAPS, 10 mm EDTA, 5% glycerol, and 10 mm dithiothreitol).
RESULTS
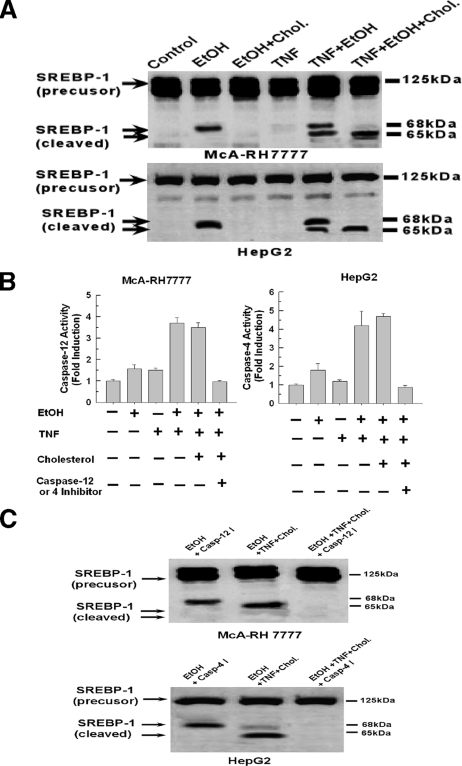
TNFα Provokes a Cholesterol-independent Activation of SREBP-1 in Ethanol-exposed Cells—Exposure of the rat hepatoma cell line McA-RH 7777 to ethanol has been shown to activate SREBP-1 (1). Indeed, as shown in Fig. 1A, top panel, exposure of McA-RH 7777 cells to 25 mm ethanol for 48 h results in the appearance of the cleaved form of SREBP-1 (second lane). The antibody used detects both SREBP-1a and -1c isoforms. Similarly, exposure of the human cell line HepG2 to 25 mm ethanol also resulted in cleavage of SREBP-1 (Fig. 1A, bottom panel, second lane). As expected and demonstrated previously, the cleavage of SREBP-1 elicited by ethanol exposure was prevented by the addition of exogenous cholesterol in both instances (Fig. 1A, third lane). We have previously shown that ethanol exposure sensitizes hepatocytes to TNF-induced cytotoxicity at a dose of 10 ng/ml. Importantly, in the present study, a 10-fold lower concentration of TNF (1 ng/ml) is utilized that did not bring about any significant cell death in the presence or absence of ethanol. The addition of TNF at a concentration of 1 ng/ml had no effect on SREBP-1 cleavage in control cells (Fig. 1A, fourth lane). However, in ethanol-exposed cells, TNF rendered the cleavage of SREBP-1 insensitive to inhibition by cholesterol. As shown in Fig. 1A, fifth lane, the addition of 1 ng/ml TNF to ethanol-exposed cells resulted in the appearance of two cleavage fragments of SREBP-1. Moreover, in contrast to the ability of cholesterol supplementation to suppress SREBP-1 cleavage in ethanol-exposed cells, it is demonstrated in Fig. 1, sixth lane, that the addition of sublethal doses of TNF made cleavage of SREBP-1 in ethanol-exposed cells insensitive to inhibition by cholesterol. Importantly, in this instance cholesterol supplementation suppressed the appearance of the 68-kDa fragment while having no effect on the 65-kDa fragment. It thus appears that the cholesterol-insensitive cleavage of SREBP-1 results in a product that migrates at 65 kDa compared with 68 kDa for the cholesterol-sensitive pathway, indicating differing sites of cleavage.
FIGURE 1.
Cleavage of SREBP-1 in ethanol-exposed cells treated with TNF is insensitive to inhibition by cholesterol but is dependent on activation of caspase-4 or -12. A, McA-RH 7777 and HepG2 cells were exposed to 25 mm ethanol for 48 h in the presence or absence of cholesterol and 25-hydroxycholesterol at concentrations of 10 and 1 μg/ml, respectively. Where indicated, TNF (1 ng/ml) was added to the cell culture medium after 24 h of ethanol exposure and the cells were then incubated for a further 24 h. The cells were then harvested and assessed for cleavage of SREBP-1 by Western blotting as described under “Experimental Procedures.” B, McA-RH 7777 or HepG2 cells were plated into 6-well 9.3-cm2 plates at 1.0 × 106 cells. Cells were exposed to 25 mm ethanol in the presence or absence of cholesterol and 25-hydroxycholesterol at concentrations of 10 and 1 μg/ml, respectively. TNF (1 ng/ml) was added to the cell culture medium after 24 h of ethanol exposure and the cells incubated for a further 24 h. Where indicated, 10 μm caspase-4 or -12 inhibitors, LEVD-FMK or ATAD-FMK, respectively, were added in tandem with the TNF. Caspase-4 or -12 activity was measured as detailed under “Experimental Procedures.” C, first lanes, McA-RH 7777 or HepG2 cells were exposed to 25 mm ethanol for 48 h in the presence of 10 μm of the caspase-4 or -12 inhibitors. In the second lane, the cells were first exposed to 25 mm ethanol in the presence of cholesterol and 25-hydroxycholesterol at concentrations of 10 and 1 μg/ml, respectively. TNF (1 ng/ml) was added to the medium after 24 h of ethanol exposure and the cells incubated for a further 24 h, whereas, in the third lane, 10 μm caspase-4 or -12 inhibitors were added in tandem with TNF.
TNFα Activates Caspase-4 or -12 in Ethanol-exposed Cells That Mediates the Cholesterol Insensitive Cleavage of SREBP-1—Caspases-4 and -12 are resident in the ER of human and mouse cells, respectively, and are activated by agents that incite the ER stress response such as ethanol (14-16). As shown in Fig. 1B, exposure of McA-RH 7777 or HepG2 cells to 25 mm ethanol for 48 h did cause a slight increase in the activity of caspases-4 or -12. Similarly, treatment of control cells with 1 ng/ml TNF caused a minor increase in caspase-4 or -12 activity. However, when ethanol-exposed cells were treated with TNFα (1 ng/ml), there was a robust increase in caspase-4 and -12 activities, reaching 3.5-4-fold above control levels. Moreover, the TNF-induced activation of caspases-4 and -12 was not prevented by the addition of cholesterol, but was inhibited by the presence of the selective caspase-4 or -12 inhibitors, LEVD-FMK and ATAD-FMK, respectively.
Caspase-3 has been shown to cleave and activate SREBP-1, however, it is unknown if caspases-4 or -12 share this property (17-19). Therefore we wanted to determine whether the ER-localized caspase-4 and -12 could be responsible for the cholesterol-insensitive cleavage of SREBP-1 in ethanol-exposed cells treated with TNF. As shown in Fig.1C, first lane, addition of the caspase-4 or -12 inhibitors did not prevent cleavage of SREBP-1 in ethanol-exposed cells, indicating this is mediated by the conventional SCAP-mediated pathway. As shown in Fig. 1C, second lane, cholesterol was unable to prevent cleavage of SREBP-1 in ethanol-exposed cells in the presence of TNF (Fig. 1B, second lane). However, preincubation with caspase-4 or -12 inhibitors prevented the cholesterol-insensitive cleavage of SREBP-1 induced by TNF in ethanol-exposed cells (Fig. 1B, third lane).
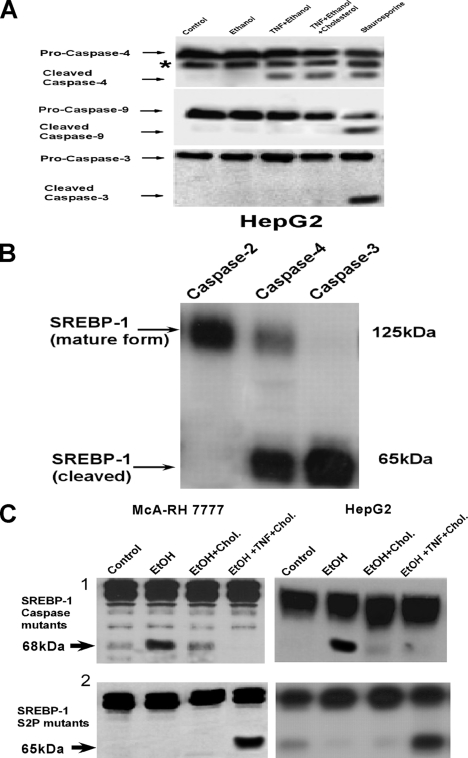
The activation of ER-localized caspases was selective in ethanol-exposed cells treated with TNF (1 ng/ml). As shown in Fig. 2A (third and fourth lanes), caspase-4 was cleaved to an active form in ethanol-exposed cells treated with TNF in the absence or presence of cholesterol. By contrast caspases-9 and -3 were not cleaved in ethanol-exposed cells treated with TNF. However, treatment with staurosporine caused the cleavage of caspases-4, -9, and -3 (Fig. 2A, fifth lane). Importantly, staurosporine resulted in significant cell killing, whereas TNF at 1 ng/ml did not cause a loss of cell viability in the ethanol-exposed cells. Next, the ability of caspase-4 to directly cleave SREBP-1 was determined. As shown in Fig. 2B, recombinant caspase-4 was able to cleave SREBP-1 in cell extracts as was caspase-3. By contrast, caspase-2 did not result in significant SREBP-1 cleavage.
FIGURE 2.
SREBP-1 can be cleaved in vitro by caspases-4 and -12 with mutation of the caspase cleavage site preventing the cholesterol-insensitive processing of SREBP-1 in ethanol-exposed cells treated with TNF. A, HepG2 cells were plated into 6-well 9.3-cm2 plates at 1.0 × 106 cells. Cells were exposed to 25 mm ethanol in the presence or absence of cholesterol and 25-hydroxycholesterol at concentrations of 10 and 1 μg/ml, respectively. TNF (1 ng/ml) was added to the cell culture medium after 24 h of ethanol exposure and the cells incubated for a further 24 h. For the fifth lane, cells were treated with 100 nm staurosporine for 24 h. The cells were then harvested and the cleavage of caspases-4, -9, and -3 were determined by Western blotting. B, purified SREBP-1 containing a histidine tag was generated by in vitro transcription and translation followed by purification on a nickel resin. The purified SREBP-1 was incubated with 5 units of either recombinant caspases-2, -3, or -4(EMD Biosciences) in reaction buffer, as indicated under “Experimental Procedures.” Cleavage of SREBP-1 was detected by Western blotting. C, McA-RH7777 or HepG2 cells were transfected with siRNA targeting SREBP-1. After 48 h, the cells were transfected with an expression plasmid (pcDNA3.1) containing a cDNA for the caspase site mutants; SREBP-1(D460A) and SREBP-1(D474A) for the HepG2 and McA-RH 7777 cells, respectively, or the S2P site mutants; SREBP-1(R527A) and SREBP-1(R524A) for the HepG2 and McA-RH7777 cells, respectively, which were all insensitive to siRNA suppression though the introduction of a “silent” mutation in the open reading frame of the gene where it is targeted by the siRNA. The cells were incubated for 24 h, then harvested, re-plated, and exposed to 25 mm ethanol for 24 h in the absence or presence of cholesterol. Where indicated, TNF was then added and the cells incubated for an additional 24 h. The cells were then harvested and the cleavage of SREBP-1 determined by Western blotting.
SREBP-1 possesses a consensus caspase cleavage site at aspartate 460 in the human protein or aspartate 474 in the mouse, a site very close to arginine 527 or 524 cleavage sites utilized by S2P in the human and mouse SREBP-1, respectively. Knock-down and rescue experiments were then performed wherein native SREBP-1 expression was first suppressed with siRNA followed by transfection with a plasmid construct that induced the expression of SREBP-1 where the caspase cleavage site at aspartate 460 or 474 in the human and mouse SREBP-1, respectively, were mutated to an alanine residue (SREBP(D460A) or -(D474A)). As shown in Fig. 2C, top panels, second lanes, ethanol exposure was able to induce the cleavage of SREBP-1 caspase site mutants. This is consistent with the asparagine 527 or 524 cleavage sites utilized by S2P in the human or mouse proteins, respectively, remaining intact in SBERP(D460A) or SREBP(D474A). In addition, the SREBP(D460A) and SREBP(D474A) cleavage induced by ethanol was prevented by supplementation with cholesterol (Fig. 2C, top panels, third lane). However, in contrast to native SREBP-1, TNF was unable to initiate cleavage of the caspase consensus site mutants, SREBP(D460A) or SREBP(D474A), in ethanol-exposed cells in the presence of cholesterol supplementation (Fig. 2C, top panels, fourth lane). By distinction, rescue of SREBP-1 expression with mutants where the S2P site of cleavage at arginine 527 or 524 in the human or mouse proteins, respectively, is mutated to alanine (SREBP(R527A) or -(R524A)) confirms that caspases-4 and -12 utilize different sites from S2P. As shown in Fig. 2C, bottom panels, second lane, ethanol exposure alone was unable to initiate cleavage of the S2P site mutants, SREBP(R527A) or SREBP(R524A). However, the S2P site mutants, SREBP(R527A) and SREBP(R524A), remained sensitive to cleavage induced by TNF in ethanol-exposed cells in the presence of cholesterol supplementation. As demonstrated in Fig. 2C, bottom panels, fourth lane, SREBP(R527A) or SREBP(R524A) were cleaved in ethanol-exposed cells treated with TNF in the presence of cholesterol supplementation. Together these data indicate that the cleavage sites of SREBP-1 utilized in ethanol-exposed cells are distinct depending on the absence or presence of TNF and cholesterol supplementation.
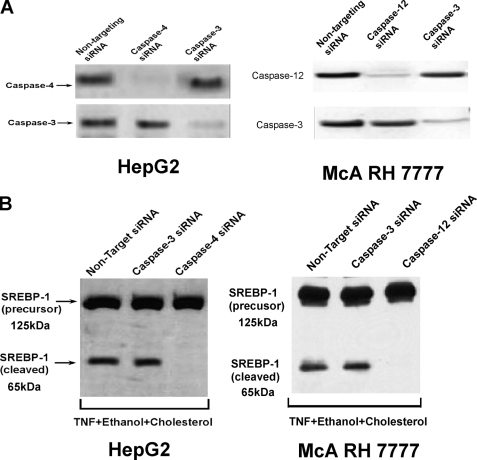
Suppression of caspase-4 and -12 expression was carried out utilizing siRNA. As shown in Fig. 3A, non-targeting siRNA had no effect on the expression of caspases-4 or -12 in HepG2 and McA-RH 7777 cells, respectively. By contrast siRNA targeting caspase-4 in HepG2 cells down-regulated the expression of caspase-4, whereas having no effect on the level of caspase-3. By contrast, siRNA targeting caspase-3 decreased the expression of caspase-3 significantly, whereas having no effect on caspase-4 levels. Similar results were obtained utilizing siRNA targeting caspase-12 in McA-RH 7777 cells (Fig. 3A, right panel). As can be seen in Fig. 3B, left panel, knock-down of caspase-4 prevented the cholesterol-insensitive cleavage of SREBP-1 in ethanol-exposed cells treated with TNF. However, knock-down of caspase-3 had no effect on SREBP-1 cleavage. Identical results were obtained with caspase-12 suppression in McA-RH 7777 cells (Fig. 3C, right panel).
FIGURE 3.
Suppression of caspase-4 but not caspase-3 by siRNA prevents the cholesterol-insensitive cleavage of SREBP-1 in ethanol-exposed cells treated with TNF. A, McA-RH 7777 or HepG2 cells were transfected with siRNAs targeting caspase-4, -12, or -3 or a non-targeting control. After 48 h, the cells were harvested and the levels of caspase-4, -12, or -3 were determined by Western blotting. B, McA-RH 7777 or HepG2 cells were transfected with siRNAs targeting caspase-4, -12, or -3 or a non-targeting control. After 24 h, the transfected cells were then incubated for a further 24 h in 25 mm ethanol with TNF (1 ng/ml) in the presence of cholesterol supplementation. The cells were then harvested and assessed for cleavage of SREBP-1 by Western blotting as described under “Experimental Procedures.”
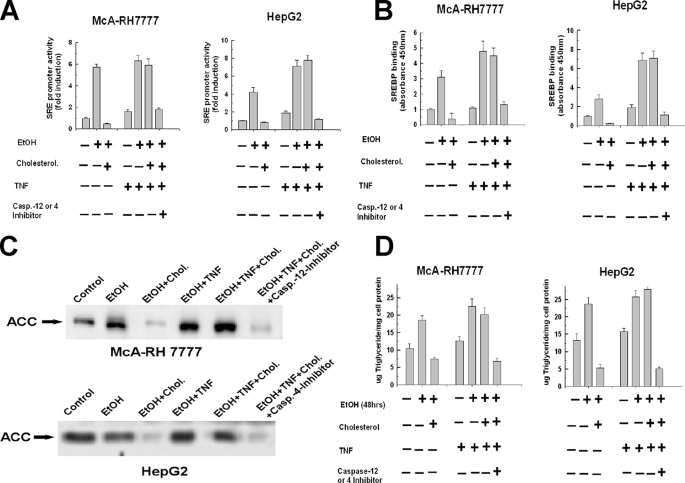
Caspase-cleaved SREBP-1 Is Functional—We next wanted to determine whether the cleavage product of SREBP-1 produced by caspase-4 or -12 activities in ethanol-exposed cells treated with TNF are transcriptionally active and can bind the SRE promoter. McA-RH 7777 and HepG2 cells were transfected with a luciferase reporter vector that contains a sterol response element in the promoter. As shown in Fig. 4A and reported previously, exposure of the cells to 25 mm ethanol for 48 h markedly induced luciferase reporter activation that was inhibited by cholesterol supplementation (1). TNF alone produced little activation of the luciferase reporter. However, the addition of TNF to ethanol-exposed cells rendered the increase in luciferase reporter activation insensitive to suppression by cholesterol supplementation. Importantly, the cholesterol-insensitive activation of the luciferase reporter by TNF in ethanol-exposed cells was repressed by the caspase-4 inhibitor in HepG2 cells and the caspase-12 inhibitor in the McA-RH 7777 cells (Fig. 4A).
FIGURE 4.
Caspase-4 and -12 cleaved SREBP-1 is functional. A, McA-RH 7777 or HepG2 cells were plated into 6-well 9.3-cm2 plates at 1.0 × 106 cells and were transiently transfected with a native luciferase reporter construct or a luciferase reporter driven by an SRE promoter. After 24 h, the transfected cells were then incubated for a further 24 h in 25 mm ethanol in the absence or presence of cholesterol supplementation. Where indicated, TNF (1 ng/ml) was added alone or in tandem with the caspase-4 or -12 inhibitors (10 μm) and incubated for another 24 h. The cells were then harvested and luciferase activity measured and normalized as described under “Experimental Procedures.” B, cells were treated as described in A and nuclei were then isolated from cells and the binding of SREBP-1 to the SRE consensus sequence was detected by enzyme-linked immunosorbent assay with absorbance measured at 450 nm. C, McA-RH 7777 or HepG2 cells were exposed to 25 mm ethanol for 48 h in the presence or absence of cholesterol and 25-hydroxycholesterol at concentrations of 10 and 1 μg/ml, respectively. Where indicated, TNF (1 ng/ml) was added to the cell culture medium after 24 h of ethanol exposure, alone or in tandem with 10 μm of the caspase-4 or -12 inhibitors. The cells were then harvested and assessed for the levels of acetyl-CoA carboxylase by Western blot. D, McA-RH 7777 or HepG2 cells were exposed to 25 mm ethanol for 48 h in the presence or absence of cholesterol and 25-hydroxycholesterol at concentrations of 10 and 1 μg/ml, respectively. Where indicated, TNF (1 ng/ml) was added to the cell culture medium after 24 h of ethanol exposure, alone or in tandem with 10 μm of the caspase-4 or -12 inhibitors. The cells were then harvested and assessed for triglyceride levels enzymatically as described under “Experimental Procedures.”
Similarly, an increase in the binding of SREBP-1 to the SRE was detected in ethanol-exposed cells treated with TNF, in the presence or absence of cholesterol. As shown in Fig. 4B, there was a 3-fold increase of SREBP-1 binding to the SRE in ethanol-exposed cells that was prevented by cholesterol. TNF alone produced no increase in SREBP-1 binding. However, in ethanol-exposed cells, TNF at 1 ng/ml caused a 5-fold stimulation of SREBP-1 binding to the SRE. This was not altered by the presence of cholesterol but was prevented by caspase-4 and -12 inhibitors is HepG2 and McA-RH 7777 cells, respectively (Fig. 4B).
Expression of the lipogenic protein, ACC, is induced by activation of SREBP-1. As shown in Fig. 4C, second and third lanes, exposure to 25 mm ethanol for 48 h increased the expression of ACC and this was repressed by cholesterol supplementation. By contrast, cholesterol was unable to prevent the induction of ACC in ethanol-exposed cells treated with TNF (Fig. 4C, fourth and fifth lanes). However, inhibition of caspase-4 or -12 activity by incubation with the respective caspase inhibitors prevented the cholesterol-insensitive induction of ACC expression in ethanol-exposed cells treated with TNF (Fig. 4C, sixth lane).
Triglyceride synthesis induced by exposure of the cells to ethanol was also differentially sensitive to cholesterol inhibition in the absence and presence of TNF. As shown in Fig. 4D, exposure to 25 mm ethanol for 48 h provoked a 50-55% increase of triglyceride levels in McA-RH 7777 cells and HepG2 cells. Cholesterol supplementation prevented the accumulation of triglycerides in the cells exposed to ethanol for 48 h and even decreased triglyceride levels below that of controls. Treatment with TNF (1 ng/ml) alone provoked a modest accumulation of triglycerides. By contrast, ethanol-exposed cells treated with TNF demonstrated a robust 60% elevation of triglycerides (Fig. 4D). However, in distinction to ethanol-exposure alone, the triglyceride accumulation induced in ethanol-exposed cells in the presence of TNF was insensitive to inhibition by cholesterol but was prevented by the caspase-4 and -12 inhibitors (Fig. 4D). Thus these data demonstrate that SREBP-1 cleaved by caspases-4 and -12 in ethanol-exposed cells treated with TNF is functional and can bind to and activate SRE containing promoters, stimulating the expression of lipogenic proteins such as ACC with a resultant accumulation of triglycerides.
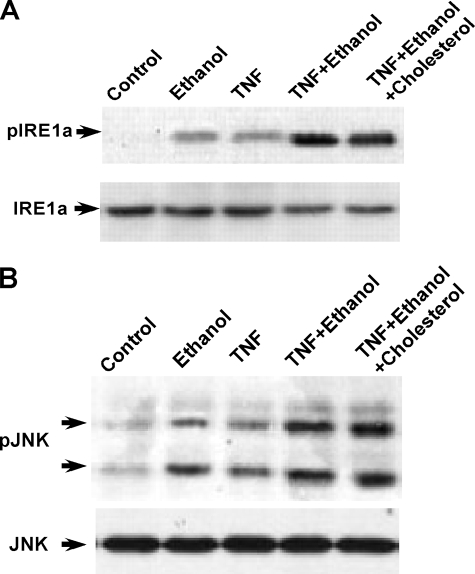
Ethanol Induces an ER Stress Response That Modifies TNF Signaling—Inositol-requiring protein 1α (IRE-1α) dimerizes and undergoes autophosphorylation upon induction of ER stress. As can been seen in Fig. 5A, ethanol exposure induced the phosphorylation of IRE-1α in HepG2 cells. Likewise, TNF (1 ng/ml) in control cells produced modest IRE-1α phosphorylation. However, ethanol-exposed cells treated with TNF (1 ng/ml) exhibited a synergistic stimulation of IRE-1α phosphorylation (Fig. 5A, fourth lane). The TNF augmented phosphorylation of IRE-1α in ethanol-exposed cells was not modified by cholesterol (fifth lane). Similar results were obtained with McA-RH 7777 cells.
FIGURE 5.
TNF synergistically stimulates indications of ER stress in ethanol-exposed cells. A and B, HepG2 cells were exposed to 25 mm ethanol in the presence or absence of cholesterol and 25-hydroxycholesterol at concentrations of 10 and 1 μg/ml, respectively. TNF (1 ng/ml) was added to the cell culture medium after 24 h of ethanol exposure and the cells incubated for a further 24 h. The cells were then harvested and the level of phosphorylated and total IRE-1α or JNK was determined by Western blotting.
JNK has been shown to be activated at the ER through mediation of TRAF-2. Because TNF can activate TRAF-2, we determined the degree of JNK activation under the present conditions. As shown in Fig. 5B, ethanol exposure alone produced a slight increase of active phosphorylated JNK. Likewise, TNF treatment of control cells did cause some degree of JNK activation. By contrast, treatment of ethanol-exposed cells with TNF resulted in a synergistic stimulation of JNK activity that was not modified by cholesterol supplementation. Such results indicate that TNF enhances signaling at the ER in cells exposed to ethanol. Similar results were observed in McA-RH 7777 cells.
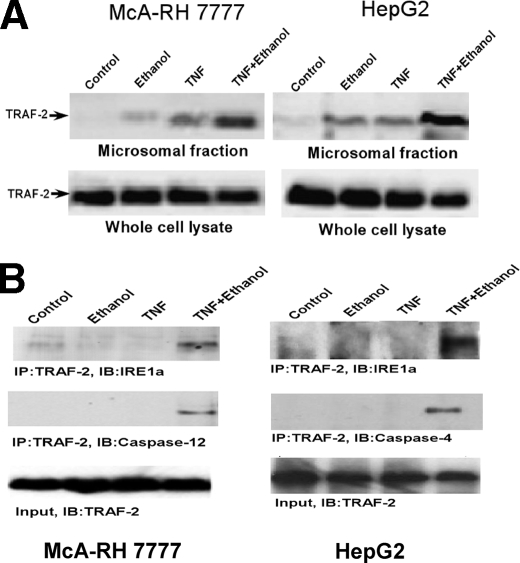
The TNF-induced Activation of Caspase-4 and -12 in Ethanol-exposed Cells Is Dependent on TRAF-2 Expression—IRE-1 is phosphorylated and oligomerizes during ER stress and forms a complex with TRAF-2 to activate caspase-12 or JNK (20). TRAF-2 also becomes activated by engagement of the TNF receptor by TNF (21). Therefore we wanted to explore the possibility that the augmentation of caspase-4 and -12 activation by TNF in ethanol-exposed cells is dependent on TRAF-2. TRAF-2 is a cytosolic protein that translocates to its site of action upon stimulation (22). Therefore microsomes that represent the ER fraction were isolated from HepG2 and McA-RH 7777 cells and assessed for their level of TRAF-2. As shown in Fig. 6A, first lane, control cells had little TRAF-2 binding to the ER. Cells exposed to ethanol or TNF alone exhibited a modest elevation of TRAF-2 in the ER fraction (Fig. 6A, second and third lanes). By contrast, in cells first exposed to ethanol for 24 h and then treated with TNF, there was a dramatic increase in the levels of TRAF-2 at the ER (Fig. 6A, fourth lane). Importantly, there was no increase or decrease of TRAF-2 in whole cell lysates under any of the conditions examined, indicating that the increased levels of TRAF-2 in the ER fraction of ethanol-exposed cells treated with TNF cannot be accounted for by a generalized elevation of TRAF-2 expression.
FIGURE 6.
TRAF-2 translocates to the ER in ethanol-exposed cells treated with TNF and forms a complex with IRE-1α and caspase-4 or -12. A, McA-RH 7777 or HepG2 cells were either untreated (first lane), exposed to 25 mm ethanol for 48 h (second lane) or treated with TNF (1 ng/ml) alone for 24 h (third lane). Alternatively, cells were first exposed to 25 mm ethanol for 24 h and then treated with TNF (1 ng/ml) in the presence of 25 mm ethanol for a further 24 h (fourth lane). The cells were then harvested and the microsomal fraction isolated as described under “Experimental Procedures.” The level of TRAF-2 in the microsomal fraction and whole cell lysates was assessed by Western blotting. B, McA-RH 7777 or HepG2 cells were either untreated (first lane), exposed to 25 mm ethanol for 48 h (second lane) or treated with TNF (1 ng/ml) alone for 24 h (third lane). Alternatively, cells were first exposed to 25 mm ethanol for 24 h and then treated with TNF (1 ng/ml) in the presence of 25 mm ethanol for a further 24 h (fourth lane). Association of TRAF-2 with IRE-1α and caspase-4 or -12 was determined by immunoprecipitation (IP) assay with TRAF-2 antibody followed by Western blotting (IB) with anti-IRE-1α or anti-caspase-4 or -12 antibody.
TRAF-2 has been shown to form a complex with IRE-1α. Therefore, immunoprecipitation experiments were conducted to determine whether TRAF-2, which is localized to the ER in ethanol-exposed cells treated with TNF, associates with IRE-1α. As shown in Fig. 6B (fourth lane), IRE-1α co-immunoprecipitated with TRAF-2 only in ethanol-exposed cells treated with TNF in both McA-RH 7777 and HepG2 cells. Importantly, caspase-4 in HepG2 cells and caspase-12 in McA-RH 7777 cells also co-immunoprecipitated with TRAF-2 in ethanol-exposed cells treated with TNF (Fig. 6B, second panel). Such data indicate the possibility that a complex forms between TRAF-2, IRE-1α, and caspase-4 or -12 in ethanol-exposed cells when they are treated with TNF.
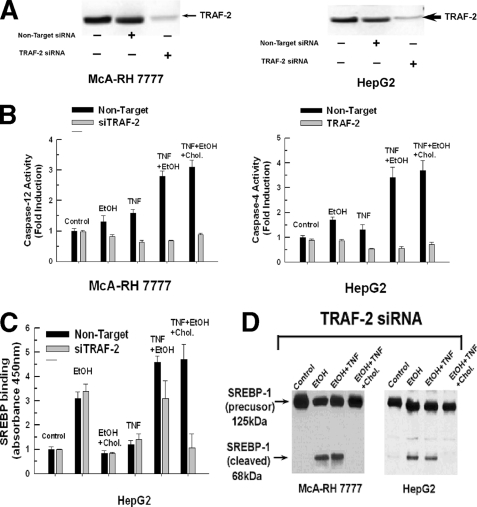
We next wanted to determine whether TRAF-2 expression was necessary for the activation of caspases-4 or -12 seen in ethanol-exposed cells treated with TNF. As shown in Fig. 7A, TRAF-2 expression was suppressed in McA-RH 7777 cells and HepG2 cells with siRNAs targeting TRAF-2, whereas the non-targeting control had no effect. Therefore TRAF-2 expression was suppressed in control and ethanol-exposed cells and its effect on caspase-4 and -12 activity was determined. As shown in Fig. 7B, exposure to TNF or ethanol alone caused only a slight increase in caspase-4 or -12 activity that was prevented in cells transfected with the siRNA targeting TRAF-2 but not the non-targeting control. By contrast, treatment of ethanol-exposed cells with TNF in the presence or absence of cholesterol resulted in a robust 3-fold stimulation of caspase-4 or -12 activity. Importantly, suppression of TRAF-2 expression prevented the activation of caspases-4 and -12 in ethanol-exposed cells treated with TNF, with the non-targeting control having no effect.
FIGURE 7.
The cholesterol-insensitive processing of SREBP-1 by caspase-4 or -12 in ethanol-exposed cells treated with TNF is prevented by suppressing TRAF-2 expression. A, McA-RH 7777 or HepG2 cells were transfected with siRNA targeting TRAF-2 or a non-targeting control. After 48 h, the cells were harvested and the levels of TRAF-2 expression determined. B, McA-RH 7777 or HepG2 cells were untreated, treated with TNF (1 ng/ml) alone for 24 h, or exposed to 25 mm ethanol for 48 h. Alternatively, cells were first exposed to 25 mm ethanol for 24 h in the presence or absence of cholesterol and then treated with TNF (1 ng/ml) for a further 24 h. The cells were then harvested and assayed for caspase-4 or -12 activity as described under “Experimental Procedures.” C, McA-RH 7777 or HepG2 cells were transfected with siRNA targeting TRAF-2. After 48 h, the cells were either left untreated or exposed to 25 mm ethanol for 24 h in the presence or absence of cholesterol (Chol.). Where indicated, the cells were then treated with TNF (1 ng/ml) for an additional 24 h. The binding of SREBP-1 to the sterol response consensus DNA sequence was determined by enzyme-linked immunosorbent assay and measured at 450 nm. D, McA-RH 7777 or HepG2 cells were transfected with siRNA targeting TRAF-2. After 48 h, the cells were either left untreated or exposed to 25 mm ethanol for 24 h in the presence or absence of cholesterol. Where indicated, the cells were then treated with TNF (1 ng/ml) for an additional 24 h in the presence of 25 mm ethanol with or without cholesterol supplementation. The cells were then harvested and the cleavage of SREBP-1 assessed by Western blotting.
Inhibition of caspase-4 and -12 activity afforded by suppression of TRAF-2 levels prevented the TNF-induced activation of SREBP-1 in ethanol-exposed cells treated with TNF. As shown in Fig. 7C, left panel, siRNA suppression of TRAF-2 expression prevented the increase in binding of SREBP-1 to the SRE seen in ethanol-exposed cells treated with TNF in the presence of cholesterol. Importantly, suppression of TRAF-2 did not prevent the activation of SREBP-1 induced by ethanol exposure alone and only partially lowered SREBP-1 binding in ethanol-exposed cells when treated with TNF in the absence of cholesterol. These results indicate that TRAF-2 is not required for the conventional cholesterol-sensitive pathway of SREBP-1 activation mediated by S1P and S2P proteases but is required for SREBP-1 cleavage and activation by caspase-4 or -12. This is further demonstrated in Fig. 7D. The inhibition of caspase-4 or -12 activation in ethanol-exposed cells treated with TNF afforded by suppression of TRAF-2 levels was accompanied by prevention of SREBP-1 cleavage. As expected, in cells exposed to ethanol alone or subsequently treated with TNF in the absence of cholesterol, SREBP-1 was cleaved (Fig. 7D, second and third lanes), indicating that TRAF-2 expression is not necessary for the conventional processing of SREBP-1. Note that suppression of TRAF-2 expression in ethanol-exposed cells treated with TNF in the absence of cholesterol prevented the appearance of the 65-kDa cleavage product of SREBP-1 that is caspase dependent, as noted in Fig. 1A, fifth lane. In this instance due to the inhibition of caspase-mediated cleavage brought about by suppressing TRAF-2 levels, only the sterol-sensitive cleavage product is generated (Fig. 7D, third lane). However, in the presence of cholesterol supplementation and TRAF-2 suppression, neither sterol-regulated or caspase-dependent cleavage occurs (Fig. 7D, fourth lane). Non-targeting siRNAs had no effect on SREBP-1 processing in either instance (result not shown).
DISCUSSION
The present study demonstrates that TNFα can trigger the cleavage and activation of SREBP-1 in cells exposed to ethanol. Moreover, the induction of SREBP-1 cleavage and activation in ethanol-exposed cells treated with TNF is insensitive to inhibition by cholesterol feedback and relies on a TRAF-2-dependent activation of ER resident caspases-4 or -12, rather than the conventional processing mediated by the S1P and S2P proteases. Inhibition of caspase-4 or -12 prevented the cholesterol-insensitive processing of SREBP-1, along with induction of the SRE promoter, binding of SREBP-1 to the SRE, expression of a lipogenic enzyme, and accumulation of triglycerides.
An initial manifestation of excessive alcohol consumption is the development of hepatic steatosis. Ethanol metabolism results in an increase in the levels of NADH, which hinders fatty acid oxidation. This was the mechanism thought to account for the development of alcoholic steatosis. However, recent studies have uncovered that the transcription factor SREBP-1 actuates a battery of lipogenic enzymes in ethanol-exposed cells (3, 4, 23). These observations were confirmed in vivo, where feeding mice an ethanol containing diet resulted in an increase in the mature form of SREBP-1 (24). As has been reported and seen in the present study, the cleavage and activation of SREBP-1 in ethanol-exposed cells is sensitive to the negative feedback regulation exerted by cholesterol, indicating that in this instance, SREBP-1 processing is mediated by S1P and S2P.
In addition to its effects on lipogenesis, ethanol exposure can also activate the endoplasmic reticulum stress response (7). The ER stress response is engaged under conditions where an increased malfolding of proteins occurs. Ethanol exposure is conductive to such an environment. This may be provoked in part by an elevation in the levels of homocysteine and acetal-dehyde brought about by ethanol. Homocysteine-induced endoplasmic reticulum stress activates SREBPs resulting in a dysregulation of cholesterol and triglyceride synthesis (10). The elevation of homocysteine caused by ethanol is thought to be mediated by a derangement in methionine metabolism induced by ethanol exposure. Indeed, chronic ethanol feeding to micopigs revealed a correlation between liver homocysteine levels and the accumulation of ER stress markers such as GRP78 and increase in the synthesis of lipogenic enzymes such as acetyl-CoA carboxylase. Importantly, the ER stress response induced by ethanol was accompanied by activation of caspase-12 (13).
Caspases-4 and -12 are expressed in human and rodents, respectively, and reside on the exterior of the endoplasmic reticulum (25). Caspase-12 is activated in a pathway involving the IRE-1α (20). IRE-1α is a ER stress transducer that oligomerizes when malfolded proteins accumulate (26, 27). The activation of caspase-12 by IRE-1α oligomerization is bridged by TRAF-2. TRAF family proteins possess a TRAF domain, containing a coiled-coil motif (21). The TRAF domain mediates the self-association exhibited by TRAF proteins and their interactions with receptors and downstream signaling proteins such as IRE-1α. In addition to mediating the activation of caspase-12, the interaction of TRAF-2 with IRE-1α couples stimulation of JNK to ER stress (28, 29). However, it has been shown that TRAF-2 can also mediate an anti-apoptotic effect through the activation of NF κB at the ER by its interaction with IRE-1α (30). This is similar to the ability of TRAF-2 to activate NFκB by acting as a scaffolding protein for the so called death inducing signaling complex, which forms at the TNF receptor (21-31). In both instances, TRAF-2 that is initially localized in the cytosol translocates to its respective site of action upon stimulation. In the present study, we demonstrate a robust increase in the association of TRAF-2 with the ER when ethanol-exposed cells are treated with TNF, indicating a translocation of TRAF-2 to the ER. Moreover, suppression of TRAF-2 expression prevented the TNF-induced activation of caspases-4 and -12 in ethanol-exposed cells along with the cholesterol-insensitive cleavage and activation of SREBP-1. It is also demonstrated that TRAF-2, IRE-1α, and caspase-4 or -12 form a complex in ethanol-exposed cells treated with TNF. However, the precise signaling pathway leading from engagement of the TNF receptor to the translocation of TRAF-2 to the ER is presently unclear.
The present study demonstrates that caspase-4 and -12 are capable of cleaving SREBP-1 to generate an active transcription factor. Caspase-3 also possesses the ability to cleave SREBP-1 and -2, generating transcriptionally active fragments that translocate to the nucleus and activate sterol-responsive genes (17, 19). Indeed it has been demonstrated in human hepatocytes that TNF is capable of stimulating the maturation of SREBP-1 in a cholesterol-independent fashion (32). This was dependent on the action of neutral sphingomyelinase and was recapitulated by ceramide, a product of sphingomyelinase. Even though the enzyme mediating the cleavage of SREBP-1 in this study was not definitively determined, the dose of TNF utilized (10 ng/ml) and the known ability of ceramide to activate caspase-3, lead the authors to conjecture that caspase-3 was the responsible mediator of the SREBP-1 cleavage. SREBP-2 is also activated by ER stress but in a caspase-independent mechanism (33). In the present study, no caspase-dependent cleavage or activation of SREBP-2 was detected (results not shown). Because SREBP-2 also contains a caspase consensus cleavage site, the reason for this is unclear. It may be speculated that the caspase cleavage site is inaccessible in SREBP-2 or there is a selective interaction between the activation complex composed of TRAF-2 and caspase-4 or -12 with SREBP-1 that does not occur with SREBP-2.
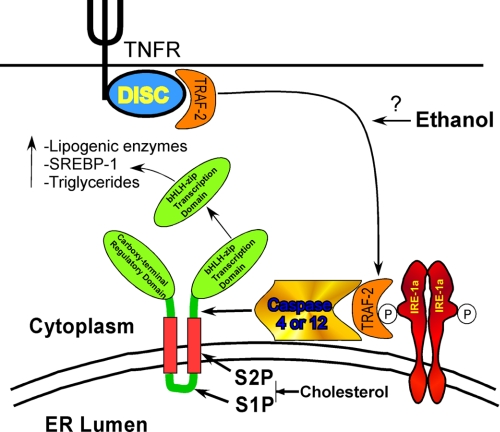
The pleiotrophic cytokine TNF stimulates hepatic lipogenesis mediated in part by an increase in the levels of the lipogenic enzymes, acetyl-CoA carboxylase and fatty acid synthase (34, 35). Additionally, TNF has recently been demonstrated to increase the expression of SREBP-1 (36). Indeed, in the present study, TNF alone provoked an increase of triglyceride levels. However, this was not accompanied by a detectable processing of SREBP-1. Ethanol is know to increase the plasma level of TNF (37-39). Through the ER stress promoted by ethanol, the lipogenic effects of TNF may be amplified by facilitating a cholesterol-insensitive mechanism of SREBP-1 activation. This would render ethanol-exposed cells insensitive to the normal negative feedback regulation exerted by cholesterol. This uncoupling of SREBP-1 activation by TNF in ethanol-exposed cells would greatly potentiate the synthesis of lipogenic enzymes and SREBP-1 itself, because the SREBP-1 gene contains a SRE in its promoter. This would thus create a self-reinforcing positive feedback, resulting in a runaway synthesis of lipids that would make the cell vulnerable to subsequent insults (Fig. 8).
FIGURE 8.
TNF provokes a cholesterol-insensitive processing of SREBP-1 in ethanol-exposed cells that is promoted by caspase-4 or -12 activation, resulting in the inappropriate induction of lipogenic enzymes and consequent accumulation of triglycerides. DISC, death inducing signaling complex.
This work was supported, in whole or in part, by National Institutes of Health Grants R01CA118356 and R01AA012897. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: SREBP, sterol response element-binding proteins; ACC, acetyl-CoA carboxylase; SRE, sterol response element; ER, endoplasmic reticulum; SCAP, SREBP cleavage activating protein; TNF, tumor necrosis factor; siRNA, small interfering RNA; CHAPS, 3-[(3-cholamidopropyl)dimethylammonio]-1-propanesulfonic acid; JNK, c-Jun NH2-terminal kinase; IRE-1, inositol-requiring protein 1; TRAF-2, tumor necrosis factor-associated factor 2.
References
- 1.You, M., Fischer, M., Deeg, M. A., and Crabb, D. W. (2002) J. Biol. Chem. 277 29342-29347 [DOI] [PubMed] [Google Scholar]
- 2.Lieber, C. S. (1997) Clin. Chim. Acta 257 59-84 [DOI] [PubMed] [Google Scholar]
- 3.Crabb, D. W., and Liangpunsakul, S. (2006) J. Gastroenterol. Hepatol. 21 Suppl. 3, S56-S60 [DOI] [PubMed] [Google Scholar]
- 4.Donohue, T. M., Jr. (2007) World J. Gastroenterol. 13 4974-4978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton, J. D., Goldstein, J. L., and Brown, M. S. (2002) Cold Spring Harbor Symp. Quant. Biol. 67 491-498 [DOI] [PubMed] [Google Scholar]
- 6.Rawson, R. B. (2003) Nat. Rev. Mol. Cell. Biol. 4 631-640 [DOI] [PubMed] [Google Scholar]
- 7.Kaplowitz, N., and Ji, C. (2006) J. Gastroenterol. Hepatol. 21 S7-S9 [DOI] [PubMed] [Google Scholar]
- 8.Schroder, M., and Kaufman, R. J. (2005) Annu. Rev. Biochem. 74 739-789 [DOI] [PubMed] [Google Scholar]
- 9.Schroder, M., and Kaufman, R. J. (2005) Mutat. Res. 569 29-63 [DOI] [PubMed] [Google Scholar]
- 10.Ji, C., and Kaplowitz, N. (2003) Gastroenterology 124 1488-1499 [DOI] [PubMed] [Google Scholar]
- 11.Ji, C., and Kaplowitz, N. (2004) World J. Gastroenterol. 10 1699-1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kalai, M., Lamkanfi, M., Denecker, G., Boogmans, M., Lippens, S., Meeus, A., Declercq, W., and Vandenabeele, P. (2003) J. Cell Biol. 162 457-467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esfandiari, F., Villanueva, J. A., Wong, D. H., French, S. W., and Halsted, C. H. (2005) Am. J. Physiol. 289 G54-G63 [DOI] [PubMed] [Google Scholar]
- 14.Pelletier, N., Casamayor-Palleja, M., De Luca, K., Mondiere, P., Saltel, F., Jurdic, P., Bella, C., Genestier, L., and Defrance, T. (2006) J. Immunol. 176 1340-1347 [DOI] [PubMed] [Google Scholar]
- 15.Kamada, S., Washida, M., Hasegawa, J., Kusano, H., Funahashi, Y., and Tsujimoto, Y. (1997) Oncogene 15 285-290 [DOI] [PubMed] [Google Scholar]
- 16.Lakshmanan, U., and Porter, A. G. (2007) J. Immunol. 179 8480-8490 [DOI] [PubMed] [Google Scholar]
- 17.Wang, X., Zelenski, N. G., Yang, J., Sakai, J., Brown, M. S., and Goldstein, J. L. (1996) EMBO J. 15 1012-1020 [PMC free article] [PubMed] [Google Scholar]
- 18.Wang, X., Pai, J. T., Wiedenfeld, E. A., Medina, J. C., Slaughter, C. A., Goldstein, J. L., and Brown, M. S. (1995) J. Biol. Chem. 270 18044-18050 [DOI] [PubMed] [Google Scholar]
- 19.Higgins, M. E., and Ioannou, Y. A. (2001) J. Lipid Res. 42 1939-1946 [PubMed] [Google Scholar]
- 20.Yoneda, T., Imaizumi, K., Oono, K., Yui, D., Gomi, F., Katayama, T., and Tohyama, M. (2001) J. Biol. Chem. 276 13935-13940 [DOI] [PubMed] [Google Scholar]
- 21.Bradley, J. R., and Pober, J. S. (2001) Oncogene 20 6482-6491 [DOI] [PubMed] [Google Scholar]
- 22.Arch, R. H., Gedrich, R. W., and Thompson, C. B. (2000) Biochem. Biophys. Res. Commun. 272 936-945 [DOI] [PubMed] [Google Scholar]
- 23.You, M., and Crabb, D. W. (2004) Alcohol 34 39-43 [DOI] [PubMed] [Google Scholar]
- 24.Ji, C., Chan, C., and Kaplowitz, N. (2006) J. Hepatol. 45 717-724 [DOI] [PubMed] [Google Scholar]
- 25.Lamkanfi, M., Kalai, M., and Vandenabeele, P. (2004) Cell Death Differ. 11 365-368 [DOI] [PubMed] [Google Scholar]
- 26.Hetz, C., Bernasconi, P., Fisher, J., Lee, A. H., Bassik, M. C., Antonsson, B., Brandt, G. S., Iwakoshi, N. N., Schinzel, A., Glimcher, L. H., and Korsmeyer, S. J. (2006) Science 312 572-576 [DOI] [PubMed] [Google Scholar]
- 27.Urano, F., Bertolotti, A., and Ron, D. (2000) J. Cell Sci. 113 3697-3702 [DOI] [PubMed] [Google Scholar]
- 28.Nishitoh, H., Saitoh, M., Mochida, Y., Takeda, K., Nakano, H., Rothe, M., Miyazono, K., and Ichijo, H. (1998) Mol. Cell 2 389-395 [DOI] [PubMed] [Google Scholar]
- 29.Urano, F., Wang, X., Bertolotti, A., Zhang, Y., Chung, P., Harding, H. P., and Ron, D. (2000) Science 287 664-666 [DOI] [PubMed] [Google Scholar]
- 30.Hu, P., Han, Z., Couvillon, A. D., Kaufman, R. J., and Exton, J. H. (2006) Mol. Cell. Biol. 26 3071-3084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hoek, J. B., and Pastorino, J. G. (2004) Semin. Liver Dis. 24 257-272 [DOI] [PubMed] [Google Scholar]
- 32.Lawler, J. F., Jr., Yin, M., Diehl, A. M., Roberts, E., and Chatterjee, S. (1998) J. Biol. Chem. 273 5053-5059 [DOI] [PubMed] [Google Scholar]
- 33.Colgan, S. M., Tang, D., Werstuck, G. H., and Austin, R. C. (2007) Int. J. Biochem. Cell Biol. 39 1843-1851 [DOI] [PubMed] [Google Scholar]
- 34.Feingold, K. R., and Grunfeld, C. (1987) J. Clin. Investig. 80 184-190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grunfeld, C., Verdier, J. A., Neese, R., Moser, A. H., and Feingold, K. R. (1988) J. Lipid Res. 29 1327-1335 [PubMed] [Google Scholar]
- 36.Endo, M., Masaki, T., Seike, M., and Yoshimatsu, H. (2007) Exp. Biol. Med. (Maywood) 232 614-621 [PubMed] [Google Scholar]
- 37.Enomoto, N., Yamashina, S., Kono, H., Schemmer, P., Rivera, C. A., Enomoto, A., Nishiura, T., Nishimura, T., Brenner, D. A., and Thurman, R. G. (1999) Hepatology 29 1680-1689 [DOI] [PubMed] [Google Scholar]
- 38.Pennington, H. L., Hall, P. M., Wilce, P. A., and Worrall, S. (1997) J. Gastroenterol. Hepatol. 12 305-313 [DOI] [PubMed] [Google Scholar]
- 39.Diehl, A. M. (1999) Alcohol Clin. Exp. Res. 23 1419-1424 [PubMed] [Google Scholar]