Abstract
The 55-kDa TNFR1 (type I tumor necrosis factor receptor) can be released to the extracellular space by two mechanisms, the proteolytic cleavage and shedding of soluble receptor ectodomains and the release of full-length receptors within exosome-like vesicles. We have shown that the brefeldin A-inhibited guanine nucleotide exchange protein BIG2 associates with TNFR1 and selectively modulates the release of TNFR1 exosome-like vesicles via an ARF1- and ARF3-dependent mechanism. Here, we assessed the role of BIG2 A kinase-anchoring protein (AKAP) domains in the regulation of TNFR1 exosome-like vesicle release from human vascular endothelial cells. We show that 8-bromo-cyclic AMP induced the release of full-length, 55-kDa TNFR1 within exosome-like vesicles via a protein kinase A (PKA)-dependent mechanism. Using RNA interference to decrease specifically the levels of individual PKA regulatory subunits, we demonstrate that RIIβ modulates both the constitutive and cAMP-induced release of TNFR1 exosome-like vesicles. Consistent with its AKAP function, BIG2 was required for the cAMP-induced PKA-dependent release of TNFR1 exosome-like vesicles via a mechanism that involved the binding of RIIβ to BIG2 AKAP domains B and C. We conclude that both the constitutive and cAMP-induced release of TNFR1 exosome-like vesicles occur via PKA-dependent pathways that are regulated by the anchoring of RIIβ to BIG2 via AKAP domains B and C. Thus, BIG2 regulates TNFR1 exosome-like vesicle release by two distinct mechanisms, as a guanine nucleotide exchange protein that activates class I ADP-ribosylation factors and as an AKAP for RIIβ that localizes PKA signaling within cellular TNFR1 trafficking pathways.
Tumor necrosis factor signals via two receptors, the type I, 55-kDa TNFR1 (TNFRSF1A, CD120a) and the type II, 75-kDa TNFR2 (TNFRSF1B, CD120b), to mediate inflammation, apoptosis, and innate immune responses (1–3). TNFR1, which is considered the major receptor for TNFR1 signaling and contains death domains in its intracytoplasmic tail, can be released to the extracellular space, where it binds tumor necrosis factor and modulates its bioactivity (3, 4). Two distinct mechanisms regulate the release of TNFR1 to the extracellular space, proteolytic cleavage of TNFR1 ectodomains and the release of TNFR1 exosome-like vesicles. Proteolytic cleavage of the TNFR1 ectodomain, which occurs primarily in the spacer region between Asn-172 and Val-173, with a minor site between Lys-174 and Gly-175, results in the shedding of soluble receptors from the cell surface (5–11). Tumor necrosis factor-α-converting enzyme (ADAM17) was identified as a TNFR1 sheddase based upon the finding that tumor necrosis factor-α-converting enzyme-deficient cells have lower ratios of shed to cell surface TNFR1 than tumor necrosis factor-α-converting enzyme-reconstituted cells (12). Similarly, depletion of tumor necrosis factor-α-converting enzyme protein by RNA interference was recently reported to decrease significantly the quantity of TNFR1 released into culture medium from airway epithelial cells in response to Staphylococcus aureus protein A, which activates tumor necrosis factor-α-converting enzyme through epidermal growth factor receptor-dependent signaling (13).
Full-length TNFR1 can also be released from cells to the extracellular space within the membranes of exosome-like vesicles (14). Exosomes are membrane-enclosed vesicles, which are typically 50–100 nm in diameter, that correspond to the internal vesicles of endolysosome-related multivesicular bodies and are released from cells via exocytic fusion with the plasma membrane (15–21). Exosomes also typically have a density of 1.13–1.21 g/ml on sucrose gradients and contain lipid raft microdomains (17–19, 22–24). Human vascular endothelial cells (HUVEC)2 in vitro constitutively release TNFR1 exosome-like vesicles that are 20–50 nm in diameter, sediment to a density of 1.1 g/ml, and are also capable of binding tumor necrosis factor (14). HUVEC-derived TNFR1 exosome-like vesicles do not contain lipid raft microdomains (14). Therefore, these TNFR1 exosome-like vesicles appear to be distinct from typical exosomes based upon their smaller size, lower density, and absence of lipid raft microdomains.
Both the proteolytic shedding of TNFR1 ectodomains and the release of TNFR1 exosome-like vesicles appear to be regulated by pathways that mediate the translocation of intracytoplasmic TNFR1 vesicles. For example, stimuli, such as histamine, induced the redistribution of TNFR1 from intracellular storage pools in the Golgi apparatus to the cell surface, where they can be proteolytically cleaved and shed (13, 25). We previously described the calcium-dependent formation of a complex composed of ARTS-1 (aminopeptidase regulator of tumor necrosis factor receptor shedding), a type II integral membrane aminopeptidase, and NUCB2 (nucleobindin 2), a putative DNA- and calcium-binding protein that associated with cytoplasmic TNFR1 before its commitment to pathways resulting in either constitutive release of TNFR1 exosome-like vesicles or inducible proteolytic cleavage of TNFR1 ectodomains (26, 27).
Many vesicular trafficking pathways are controlled by 20-kDa ADP-ribosylation factors, which are inactive in the GDP-bound state and are activated when the GDP is replaced by a GTP, a process catalyzed by guanine nucleotide exchange proteins (GEPs) (28). ARF-GEPs, some of which can be inhibited by brefeldin A, contain ∼200-amino acid Sec7 domains that accelerate the exchange of ARF-GDP for GTP (29, 30). Recently, we reported that human BIG2 (brefeldin A-inhibited guanine nucleotide exchange protein 2), a brefeldin A-inhibited ARF-GEP, regulates the constitutive release of TNFR1 exosome-like vesicles in an ARF1- and ARF3-dependent fashion but did not affect the inducible proteolytic cleavage of TNFR1 ectodomains (31). The functional interaction between TNFR1 and BIG2 that mediates constitutive release of TNFR1 exosome-like vesicles is downstream from the ARTS-1-NUCB2 complex (31). BIG2, in addition to its role as an ARF-GEP, contains three A kinase-anchoring protein (AKAP) domains that may coordinate cAMP and ARF regulatory functions (32). We hypothesized that BIG2 might regulate the release of TNFR1 exosome-like vesicles via its AKAP as well as its Sec7 domains. Here, we report that cAMP-dependent protein kinase A (PKA) signaling induced TNFR1 exosome-like vesicle release via anchoring of PKA regulatory subunit RIIβ to AKAP domains B and C of BIG2. Thus, BIG2 can modulate the release of TNFR1 exosome-like vesicles via two distinct mechanisms, the activation of class I ARFs and cAMP-dependent PKA signaling.
MATERIALS AND METHODS
Cells and Reagents—HUVEC (passages 3 and 8) and EGM-2 medium were purchased from Cambrex BioScience (Walkersville, MD). Cell-permeable myristoylated protein kinase A inhibitor 14–22 amide (Myr-PKI) and cell-permeable ERK activation inhibitor peptide I (ERKI) were from Calbiochem. 8-Br-cAMP, 8-Br-cGMP, and H-89 were from Sigma.
Antibodies—Murine IgG2b monoclonal (H5) antibody that reacted with TNFR1 was from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), as were antibodies against β-tubulin (D10) and PKA regulatory subunits RIα (N15), RIβ (C19), RIIα (M20), and RIIβ (E20). Chicken polyclonal anti-NUCB2 antibodies were generated against a glutathione S-transferase fusion protein with sequence corresponding to amino acids 326–420 of the NUCB2 C-terminal leucine zipper domain (Sigma) (27). Rabbit polyclonal antibodies against ARTS-1 and BIG2 were used as previously described (26, 31). The murine monoclonal penta-His antibody was from Qiagen (Valencia, CA).
RNA Interference—Individual siGENOME RNA duplexes for BIG2; siGENOME SMARTpool RNA duplexes for the PKA regulatory subunits RIα, RIβ, RIIα, and RIIβ; and siCONTROL nontargeting siRNA 1 were purchased from Dharmacon (Lafayette, CO). HUVEC were transfected with 100 nm siRNA using DharmaFECT 1 transfection reagent (Dharmacon) for 3 days prior to performance of experiments.
BIG2 Expression Plasmids—A pCMV6-XL4 mammalian expression plasmid encoding full-length human BIG2 was purchased from Origene Technologies (Rockville, MD). The BIG2 cDNA clone contained seven nucleotide substitutions that differed from the ARFGEF2 reference sequence (NCBI accession number NM_006420). Among these putative polymorphisms, two represented silent mutations (C3663T and C4131T), whereas five were nonsynonymous mutations (A619G, G620A, A2884G, A3145G, and A5287G) that resulted in four amino acid replacements. As previously described, the five nonsynonymous mutations were corrected by site-directed mutagenesis using the QuikChange multisite-directed mutagenesis kit (Stratagene) (31). An amino-terminal His6 tag was inserted in the BIG2 plasmid using the QuikChange multisite-directed mutagenesis kit (stratagene) and primer 5′-TTGTAATACGACTCACTATAGGGGGGCCGCATGGGTCATCACCATCACCATCACGAATTCGCCCTTATGCAGGAGAGCCAGACC-3′). Complete sequence of the His-BIG2 plasmid was confirmed by DNA sequencing.
BIG2 AKAP domain point mutations were introduced using the QuikChange XL site-directed mutagenesis kit (Stratagene) and the following primers: AKAP domain B (His-BIG2(V289W)), 5′-GGAGCCCAGGAGGTGTGGAAGGACATCTTGG-3′; AKAP domain C (His-BIG2(V534W)), 5′-GCTAACATTTTTGAGCGCCTTTGAAATGATTTATCCAAAATTGC-3′. Both primers were utilized to construct an expression plasmid with mutations in domains B and C (His-BIG2(V289W/V534W)). All mutations were confirmed by sequencing. HUVEC, grown in 6-well plates that contained 2 ml of medium, were transfected with plasmids using FuGENE 6, according to the manufacturer's instructions (Roche Applied Science), as previously described (31). Briefly, 3 μl of Fugene 6 were mixed with 1 μg of plasmid DNA in 100 μl of serum-free medium and added to cells for 14 h. Culture medium was changed, and experiments were performed 2 days later.
Immunoblotting—HUVEC were lysed in buffer containing 1% Triton X-100, 1% n-octyl β-d-glucopyranoside, 50 mm Tris, pH 7.5, and 120 mm NaCl (Sigma), supplemented with Complete™ protease inhibitor (Roche Applied Science). For immunoblots of HUVEC conditioned medium, cells were grown in medium that contained FBS depleted of exosomes by centrifugation at 175,000 × g at 4 °C for 16 h. Conditioned medium was cleared of cells and debris by sequential centrifugation at 200 × g for 10 min, 500 × g for 10 min, 1,200 × g for 20 min, and 10,000 × g for 30 min before immunoblotting of cellular proteins (50 μg/lane) or medium (26 μl/lane). Quantification was performed using NIH Image software (version 1.63) for densitometry (27).
Immunofluorescence Confocal Laser-scanning Microscopy—HUVEC grown on collagen I-coated slides (BD Biosciences, San Jose, CA) were fixed in 4% paraformaldehyde for 10 min, washed three times with PBS, permeabilized with 0.1% saponin in PBS (5 min), washed three times with PBS, and blocked with PBS containing 10% donkey and 10% goat serum for 1 h. Cells were incubated overnight at 4 °C with the following primary antibodies: murine monoclonal anti-TNFR1 (H5) antibody (2 μg/ml), rabbit polyclonal anti-BIG2 antibody (1:500), or goat polyclonal anti-RIIβ (E20) antibody (1:200), diluted in PBS containing 1% donkey and 1% goat serum. After washing three times in PBS containing 0.1% bovine serum albumin, slides were incubated with species-specific secondary antibodies conjugated to Alexa Fluor® 488 or Alexa Fluor® 568, (Invitrogen) at a 1:200 dilution. Slides were mounted with Vectashield mounting medium that contained 4′,6-diamidino-2-phenylindole (Vector Laboratories, Burlingame, CA) and imaged using a Leica SP laser-scanning confocal microscope (Leica, Heidelberg, Germany).
Immunoprecipitation—HUVEC were lysed in buffer containing 0.1% Triton X-100, 50 mm Tris, pH 7.5, and 120 mm NaCl (Sigma) supplemented with Complete™ protease inhibitor (Roche Applied Science). Proteins from HUVEC lysates (400 μg) were incubated for 2 h with 5 μg of mouse anti-penta-His antibodies immobilized on 200 μl of protein A/G beads (Pierce) that had been blocked with 1% ovalbumin in PBS. Beads were washed six times with cold lysis buffer, and immunoblots were performed as previously described (27). Proteins from HUVEC supernatants after immunoprecipitation were precipitated with 10% trichloroacetic acid (Sigma) for immunoblotting.
Quantification of Extracellular TNFR1 by ELISA—HUVEC were transfected with siRNA for 3 days and then incubated for 24 h in fresh, exosome-depleted medium. Medium cleared of cells and debris as described for immunoblotting was analyzed for TNFR1 using a Quantikine sandwich ELISA kit with a sensitivity of 7.8 pg/ml (R&D Systems, Minneapolis, MN).
Statistical Analyses—Data were analyzed by a paired Student's t test with a Boneferroni correction for multiple comparisons. A p value of ≤0.05 was considered significant.
RESULTS
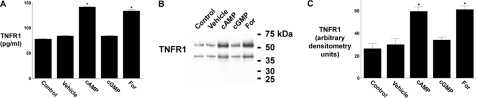
cAMP-induced Release of TNFR1 Exosome-like Vesicles from HUVEC—The role of cAMP and cGMP signaling pathways in the release of TNFR1 exosome-like vesicles from HUVEC was assessed using the cyclic nucleotide analogues, 8-Br-cAMP and 8-Br-cGMP, and forskolin, an activator of adenylyl cyclase. As quantified by ELISA, TNFR1 released into the medium from cells treated with 8-Br-cAMP or forskolin was increased by 68.4 and 59.2%, respectively, as compared with cells treated with vehicle, whereas 8-Br-cGMP treatment had no effect (Fig. 1A). Western blots of medium confirmed that 8-Br-cAMP and forskolin increased the release of the 55-kDa TNFR1 exosome-like vesicles by 99 and 104%, respectively (Fig. 1, B and C). A ∼40-kDa TNFR1 band, as previously described, was similarly increased (27, 31).
FIGURE 1.
Effect of cAMP and forskolin on the release of TNFR1 exosome-like vesicles. HUVEC were incubated without or with 1 mm 8-Br-cAMP (cAMP), 1 mm 8-Br-cGMP (cGMP), or 50 μm forskolin (For) for 24 h. A, TNFR1 concentration in medium quantified by ELISA. *, significant difference (n = 6) between cells treated with vehicle (DMSO) and those treated with 8-Br-cAMP (p < 10-12) or forskolin (p < 10-7). B, Western blot of TNFR1 in medium from one of five experiments that demonstrated similar results. C, densitometry was performed on the Western blots from B, and the quantity of the 55-kDa TNFR1 that was released into conditioned medium is presented as arbitrary densitometry units. *, significant difference (n = 5) between cells treated with vehicle and those treated with 8-Br-cAMP (p < 0.006) or forskolin (p < 0.003).
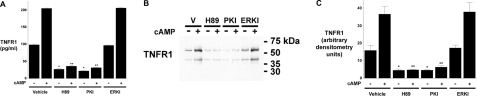
The cAMP-induced Release of TNFR1 Exosome-like Vesicles is PKA-dependent—Because cAMP can signal via PKA-dependent and PKA-independent pathways, we investigated whether the cAMP-induced increase in TNFR1 exosome-like vesicle release required PKA (33–35). As quantified by ELISA (Fig. 2A), the amount of TNFR1 released constitutively or in response to 8-Br-cAMP stimulation was significantly reduced when cells were treated with H-89, a relatively specific inhibitor of PKA. Since H-89 can also inhibit MSK1 (mitogen- and stress-activated protein kinase-1), additional experiments were performed using the cell-permeable Myr-PKI, which contains a PKA substrate consensus sequence with the serine replaced by alanine (36). Other cells were treated with cell-permeable ERKI to block the ERK signaling pathway that activates MSK1 (36, 37). As shown in Fig. 2A, the quantity of TNFR1 released, as measured by ELISA, was decreased by Myr-PKI but not ERKI treatment, which is consistent with the involvement of a cAMP-induced PKA-dependent signaling pathway. Western blots of medium confirmed that H-89 and Myr-PKI, but not ERKI, decreased both the constitutive and cAMP-induced release of full-length 55-kDa TNFR1 in exosome-like vesicles (Fig. 2, B and C).
FIGURE 2.
Constitutive and cAMP-mediated TNFR1 exosome-like vesicle release are PKA-dependent. HUVEC were incubated for 24 h without or with 1 mm 8-Br-cAMP (cAMP) and 10 μm H-89, 50 μm Myr-PKI, or 50 μm ERKI. A, TNFR1 concentration in medium quantified by ELISA. *, significant reduction (n = 6) in constitutive release of TNFR1 between cells treated with vehicle (DMSO) and those treated with H-89 (p < 10-9) or Myr-PKI (p < 10-9). **, significant reduction (n = 6) in cAMP-mediated release of TNFR1 from cells treated with vehicle (DMSO) and those treated with H-89 (p < 10-14) or Myr-PKI (p < 10-14). B, Western blot of TNFR1 in medium from one of five experiments that demonstrated similar results. C, densitometry was performed on the Western blots from B, and the quantity of the 55-kDa TNFR1 that was released into conditioned medium is presented as arbitrary densitometry units. *, significant reduction (n = 5) in the constitutive release of TNFR1 into culture medium from cells treated with vehicle (DMSO) and those treated with H-89 (p < 0.015) or Myr-PKI (p < 0.015). **, significant reduction (n = 6) in cAMP-mediated release of TNFR1 from cells treated with vehicle (DMSO) and those treated with H-89 (p < 10-3) or Myr-PKI (p < 0.004).
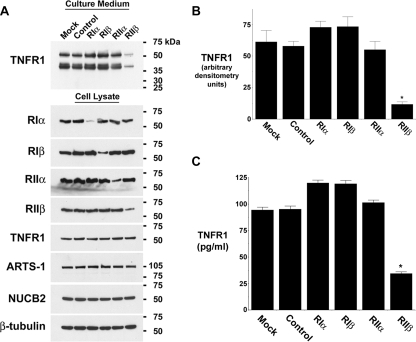
PKA Regulatory Subunit RIIβ Modulates cAMP-induced Release of TNFR1 Exosome-like Vesicles—Having demonstrated that both the constitutive and cAMP-induced release of TNFR1 exosome-like vesicles are mediated via a PKA-dependent pathway, we assessed the role of individual PKA regulatory subunits in this process. RNA interference was utilized to decrease specifically levels of mRNA (data not shown) and protein for each of the PKA regulatory subunits RIα, RIβ, RIIα, and RIIβ, without altering the quantities of TNFR1, ARTS-1, NUCB2 or β-tubulin protein in cell lysates (Fig. 3A). Western blots showed that medium from cells transfected with siRNA targeting RIIβ contained 69% less 55-kDa TNFR1 than did medium from cells transfected with nontargeting siRNA (Fig. 3, A and B). As quantified by ELISA, TNFR1 constitutively released into the medium from cells transfected with siRNA targeting RIIβ was 63% less than that from cells transfected with control nontargeting siRNA (Fig. 3C). In contrast, the siRNA-mediated depletion of RIα, RIβ, or RIIα did not attenuate the release of TNFR1 exosome-like vesicles. The specificity of the RIIβ siRNA experiments, which utilized a pool of four siRNA duplexes, was confirmed by additional experiments demonstrating that two individual siRNA duplexes targeting RIIβ decreased both RIIβ protein levels and TNFR1 release as quantified by ELISA (data not shown).
FIGURE 3.
The constitutive release of TNFR1 exosome-like vesicles is regulated by PKA RIIβ. HUVEC were transfected with vehicle (Mock); 100 nm control, nontargeting siRNA (Control); or 100 nm siRNA targeting RIα, RIβ, RIIα, or RIIβ for 3 days prior to the addition of fresh, exosome-depleted medium for 24 h. A, representative Western blot, from one of three experiments, showing TNFR1 in medium and cell lysates and showing RIα, RIβ, RIIα, RIIβ, ARTS-1, NUCB2, and β-tubulin in cell lysates. B, densitometry was performed on the Western blots from A, and the quantity of the 55-kDa TNFR1 that was constitutively released into conditioned medium is presented as arbitrary densitometry units. *, a significant decrease in the quantity of the 55-kDa TNFR1 present in conditioned medium from cells transfected with siRNA targeting RIIβ as compared with those transfected with control, nontargeting siRNA (p < 0.002, n = 3). C, TNFR1 in medium was quantified by ELISA. *, significant difference from cells transfected with siRNA targeting RIIβ as compared with those transfected with control, nontargeting siRNA (p < 10-7, n = 6).
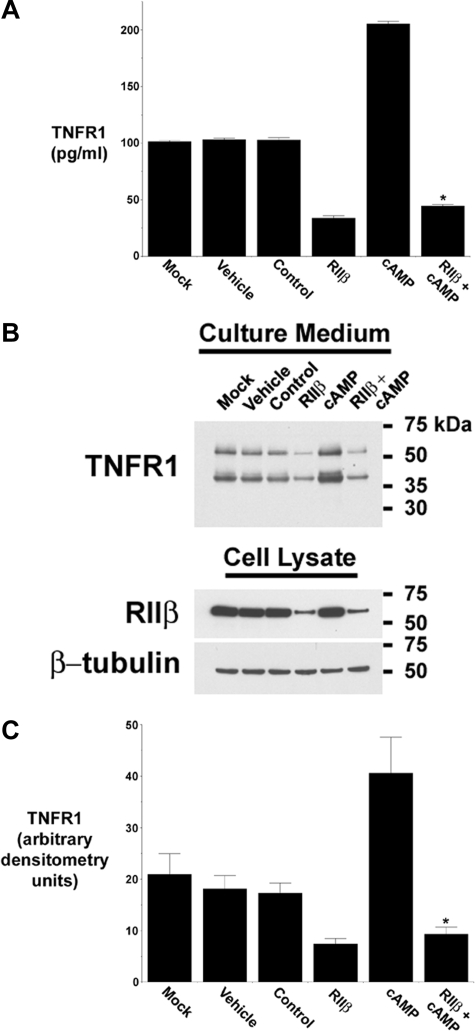
Additional experiments were performed to assess the role of RIIβ in the cAMP-induced release of TNFR1 exosome-like vesicles. As shown in Fig. 4A, the siRNA-mediated depletion of RIIβ inhibited the cAMP-induced release of TNFR1 into the medium, as quantified by ELISA. Western blots of medium confirmed that the ability of cAMP to induce the release of full-length 55-kDa TNFR1 was inhibited when RIIβ protein was reduced by RNA interference (Fig. 4, B and C). Taken together, these data demonstrate that PKA RIIβ was required for both the constitutive and cAMP-induced release of TNFR1 exosome-like vesicles.
FIGURE 4.
cAMP-mediated TNFR1 exosome-like vesicle release is regulated by PKA RIIβ. HUVEC were transfected with vehicle (Mock), DMSO (Vehicle); 100 nm control, nontargeting siRNA (Control); or 100 nm siRNA targeting RIIβ, without or with 1 mm cAMP for 3 days prior to the addition of fresh, exosome-depleted medium for 24 h. A, TNFR1 concentration in medium was quantified by ELISA. *, significant reduction in the quantity of TNFR1 present in medium from cells treated with cAMP and siRNA targeting RIIβ as compared with those treated with cAMP alone (p < 10-9, n = 6). B, representative Western blot, from one of five experiments, showing TNFR1 in medium and cell lysates, and RIIβ and β-tubulin in cell lysates. C, densitometry was performed on the Western blots from B, and the quantity of the 55-kDa TNFR1 that was constitutively released into conditioned medium is presented as arbitrary densitometry units. *, significant decrease in the quantity of the 55-kDa TNFR1 present in conditioned medium from those transfected with siRNA targeting RIIβ as compared with cells treated with cAMP alone (p < 0.003, n = 5).
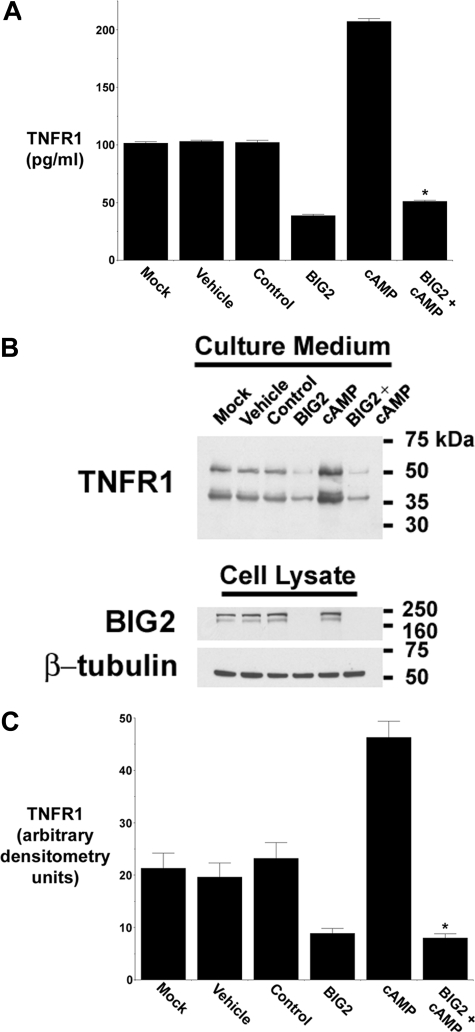
The PKA-induced Release of TNFR1 Exosome-like Vesicles Requires the Anchoring of RIIβ to BIG2 AKAP Domains B and C—We had previously reported that BIG2 co-immunoprecipitated with TNFR1 and regulated the constitutive release of TNFR1 exosome-like vesicles from HUVEC (31). To demonstrate that BIG2 participates in the cAMP-dependent PKA signaling pathway that mediates TNFR1 exosome-like vesicle release, HUVEC were transfected with siRNA targeting BIG2, which we had previously shown to specifically deplete BIG2 mRNA and protein and thereby attenuate the release of TNFR1 exosome-like vesicles (31). As quantified by ELISA (Fig. 5A) and Western blotting (Fig. 5, B and C), the cAMP-induced increases in TNFR1 exosome-like vesicle release were inhibited in cells in which BIG2 expression had been attenuated by siRNA, demonstrating that BIG2 participates in the cAMP-mediated PKA signaling pathway that regulates TNFR1 exosome-like vesicle release.
FIGURE 5.
cAMP-mediated TNFR1 exosome-like vesicle release requires BIG2. HUVEC were transfected with vehicle (Mock); DMSO (Vehicle), 100 nm control, nontargeting siRNA (Control); or 100 nm siRNA targeting BIG2, without or with 1 mm cAMP for 3 days prior to the addition of fresh, exosome-depleted medium for 24 h. A, TNFR1 concentration in medium was quantified by ELISA. *, significant reduction in the quantity of TNFR1 present in medium from cells treated with cAMP and siRNA targeting BIG2 as compared with those treated with cAMP alone (p < 10-13, n = 6). B, representative Western blot, from one of three experiments, showing TNFR1 in medium and cell lysates and showing BIG2 and β-tubulin in cell lysates. C, densitometry was performed on the Western blots from B, and the quantity of the 55-kDa TNFR1 that was released into conditioned medium is presented as arbitrary densitometry units. *, significant decrease in the quantity of the 55-kDa TNFR1 present in conditioned medium from cells transfected with siRNA targeting BIG2 as compared with those treated with cAMP alone (p = 0.006, n = 3).
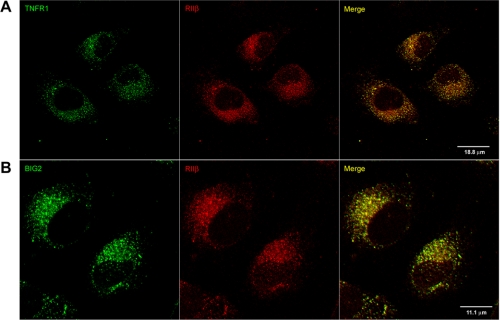
Confocal immunofluorescence microscopy was performed to confirm that RIIβ and TNFR1 reside in overlapping intracellular compartments in HUVEC. As shown in Fig. 6A, RIIβ and TNFR1 showed partial co-localization in diffusely distributed cytoplasmic vesicles. Similarly, RIIβ and BIG2 were partially co-localized in diffusely distributed cytoplasmic vesicles (Fig. 6B). All of these findings were consistent with the conclusion that BIG2 functions as an AKAP for RIIβ, which spatially localizes a subset of PKA catalytic activity to TNFR1 cytoplasmic vesicles.
FIGURE 6.
Constitutive association between PKA RIIβ and TNFR1 in HUVEC. HUVEC were grown on collagen I-coated slides and reacted with antibodies against TNFR1 and RIIβ (A) or BIG2 and RIIβ (B) and secondary antibodies conjugated with Alexa Fluor 488 (green) or Alexa Fluor 568 (red), respectively, before immunofluorescence confocal laser-scanning microscopy.
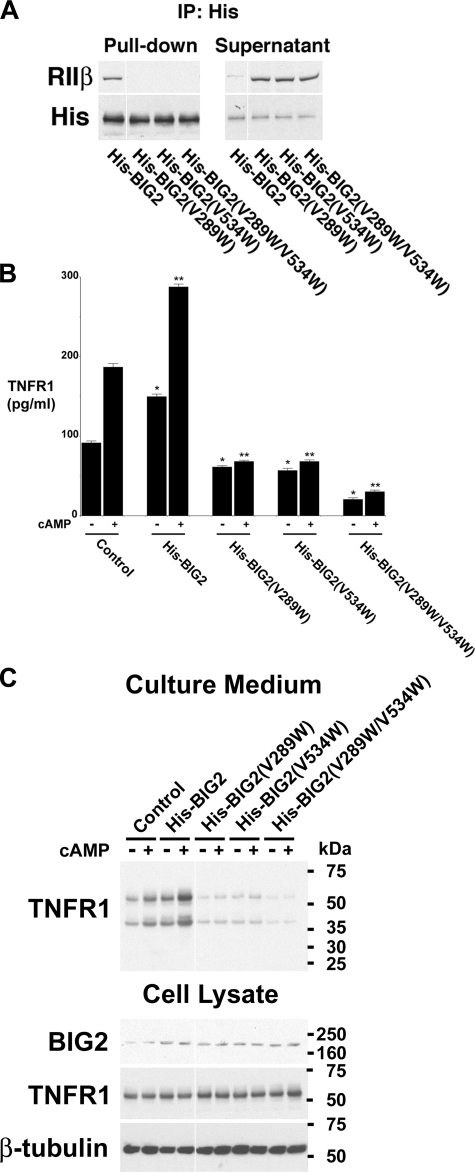
To confirm that BIG2 anchoring of RIIβ is required for cAMP-induced TNFR1 exosome-like vesicle release, histidine-tagged BIG2 expression constructs were generated that contained mutations in key conserved amino acids that mediate binding of RII subunits to AKAP domains B and C (32). The AKAP domain B and C mutants contained single amino acid replacements at positions 289 (V289W) and 534 (V534W), respectively. A double mutant contained single amino acid substitutions in both domains B and C (V289W and V534W). Immunoprecipitations of proteins from HUVEC lysates using an anti-His antibody confirmed that the His-BIG2 AKAP domain mutants failed to bind RIIβ (Fig. 7A). Quantification by ELISA of TNFR1 released into the medium from cells overexpressing wild-type His-BIG2 showed significantly greater constitutive and 8-Br-cAMP-induced TNFR1 release than that from cells transfected with empty (control) vector (Fig. 7B). The quantity of TNFR1 released into culture medium, constitutively or following 8-Br-cAMP stimulation, was significantly reduced from cells transfected with either the AKAP domain B (His-BIG2(V289W)), domain C (His-BIG2(V534W), or domain B/C (His-BIG2(V289W/V534W) mutants, as compared with cells transfected with empty (control) vector, which is consistent with a dominant negative effect. Furthermore, the amount of TNFR1 released into culture medium from cells overexpressing the domain B/C mutant (His-BIG2(V289W/V534W) was significantly less than that from cells overexpressing either the B or C mutants, which suggests that the B and C AKAP domains of BIG2 were functioning in an additive fashion to mediate both constitutive and cAMP-induced TNFR1 exosome-like vesicle release from HUVEC. Western blot analyses of culture medium and cell lysates confirmed the ELISA results (Fig. 7C).
FIGURE 7.
Constitutive and cAMP-mediated TNFR1 exosome-like vesicle release requires the anchoring of RIIβ to BIG2 AKAP domains B and C. HUVEC were transfected with plasmids encoding either His-tagged wild-type BIG2 (His-BIG2) or BIG2 mutants that contained single amino acid substitutions in AKAP domain B (His-BIG2(V289W)), domain C (His-BIG2(V534W)), or domains B and C (His-BIG2(V289W/V534W)) or empty plasmid (Control), for 2 days before the addition of fresh, exosome-depleted medium for 24 h, without or with 1 mm 8-Br-cAMP (cAMP). A, proteins immunoprecipitated (IP) with the anti-His antibody (Pull-down) or remaining in the supernatant were immunoblotted with antibodies against RIIβ or the His tag. This blot is representative of three individual experiments. B, TNFR1 concentration in medium was quantified by ELISA. *, significant increase (His-BIG2, p < 10-3) or decrease (His-BIG2(V289W), p < 10-3; His-BIG2(V534W), p < 0.002; His-BIG2(V289W/V534W), p < 10-3) in the quantity of TNFR1 constitutively released into the medium as compared with control cells transfected with the empty plasmid (n = 6). **, significant increase (His-BIG2, p < 0.012) or decrease (His-BIG2(V289W), p < 10-4; His-BIG2(V534W), p < 10-4; His-BIG2(V289W/V534W), p < 10-4) in the quantity of TNFR1 released into the medium following stimulation with 1 mm 8-Br-cAMP for 24 h as compared with control cells transfected with the empty plasmid (n = 6). The quantity of constitutive and 8-Br-cAMP-mediated TNFR1 release from cells transfected with the domain B/C double mutant (His-BIG2(V289W/V534W)) was significantly decreased as compared with those transfected with His-BIG2(V289W) (p < 10-3) or His-BIG2(V534W) (p < 10-3), respectively (n = 6). C, representative Western blot, from one of three experiments, showing TNFR1 in medium and cell lysates, and BIG2 and β-tubulin in cell lysates. Images were grouped from adjacent parts of the same gel in A and C.
DISCUSSION
Pathways that regulate the trafficking of intracytoplasmic TNFR1 vesicles appear to play a key role in modulating the release of TNFR1 exosome-like vesicles from cells. For example, calcium-dependent complexes containing ARTS-1 and NUCB2 associate with intracytoplasmic TNFR1 prior to the commitment of TNFR1 to pathways leading to either the release of TNFR1 exosome-like vesicles or the inducible cleavage of TNFR1 ectodomains (14, 26, 27). Consistent with the critical role of vesicular trafficking in these processes, the ARF-GEP BIG2 was shown to regulate selectively the constitutive release of TNFR1 exosome-like vesicles via the activation of class I ARFs 1 and 3, which mediate this process in an additive and nonredundant fashion (31). Furthermore, TNFR1 exosome-like vesicle release involves the brefeldin A-sensitive association between BIG2 and TNFR1. Thus, BIG2 functions as a key regulator of TNFR1 exosome-like vesicle release (31).
In addition to its role as an activator of ARF, BIG2 can function as an AKAP (32). PKA, a primary intracellular receptor for cAMP, is critical for cAMP regulation of diverse vital functions. The broad substrate specificity of PKA requires accurate limitation of its action temporally and spatially. That is achieved via proteins with AKAP domains that bind the R subunit dimer to establish kinase localization (38–41). These scaffolding proteins have also sites for interaction with substrates, additional enzymes, and regulatory proteins that assemble to constitute macromolecular machines for integration of signaling and metabolic or mechanical functions. PKA is a tetramer with two catalytic subunits that are maintained in an inactive state via association with a regulatory subunit dimer (39). Binding of cAMP by the regulatory subunits releases active catalytic units to phosphorylate PKA targets. Type I PKA contains RIα or -β subunits, and type II has RIIα or -β subunits (39, 41). PKA is anchored by an AKAP amphipathic helix of 14–18 amino acids that binds the N-terminal dimerization and docking (D/D) domains of PKA regulatory subunits (40, 42, 43). Type II PKAs are generally associated with specific cellular organelles and are less sensitive to cAMP signaling, whereas type I PKAs are predominantly cytosolic but also can be localized and are more sensitive to cAMP (41, 44). Sequence differences between RI and RII contribute to the specificity of PKA signaling by influencing the affinity of PKA binding to AKAPs, which typically bind RII subunits with high affinity, whereas dual specificity AKAPs bind both RI and RII subunits (43). Several RI-specific AKAPs have also been identified (39). Furthermore, RI binding to AKAPs is thought to be more dynamic due to weaker binding affinities and faster off-rates, whereas RII binding is more static (41, 43, 45).
Consistent with its role as an AKAP, BIG2 moved from the cytosol to Golgi in HepG2 cells treated with 8-Br-cAMP or forskolin (32). Since BIG2 was associated with a fraction of cytoplasmic TNFR1, we investigated whether the BIG2 AKAP domains regulate TNFR1 exosome-like vesicle release from HUVEC by targeting PKA catalytic activity to these sites (31). We found that both constitutive and cAMP-induced TNFR1 exosome-like vesicle release were regulated in a PKA-dependent fashion by a mechanism that involves the association of RIIβ with BIG2. BIG2 contains three AKAP domains (32). Yeast two-hybrid experiments have identified that domain A (BIG2 residues 27–48) interacted with RIα and RIβ, domain B (residues 284–301) interacted with RIIα and RIIβ, and domain C (residues 517–538) interacted with RIα, RIIα, and RIIβ (32). Consistent with these findings, we found that expression of BIG2 constructs containing point mutations in AKAP domain B or C abrogated RIIβ binding and TNFR1 exosome-like vesicle release.
Molecular determinants of RII binding were elucidated by analysis of the crystal structure of the D/D domain of RII in complex with an RII-selective AKAP peptide (AKAP-IS) (40, 46). AKAP-IS folds into an amphipathic α-helix that binds in a preformed shallow groove on the surface of the RII dimer D/D domains. Using another model AKAP peptide (SuperAKAP-IS) that is highly selective for RII, it was shown that RII-AKAP interactions are particularly sensitive to mutations of Super-AKAP-IS residues Ala9, Val13, and Ala16, which are buried deeply in the RII-AKAP interface and cannot accommodate amino acids larger than Ala or Val (40, 46). Similarly, the crystal structure of the helical motif from a 22-amino acid peptide corresponding to the AKAP domain of D-AKAP2, a dual-specificity AKAP, bound to a RIIα D/D domain, revealed that a stable, hydrophobic docking grove is formed by the helical interface of the two RIIα protomers (43). Alanine scanning revealed that the most critical residues for RIIα binding were Ile8, Ala9, Ile12, and Val13, with Val13 the crucial determinant of specificity for RII binding (43). Based upon these structural requirements, we generated BIG2 AKAP domain B and C mutants that failed to bind RIIβ. The domain B mutant contained a V289W substitution, which corresponded to Ala9, whereas the domain C mutant contained a V534W substitution, which corresponded to Val13 of the AKAP-IS and D-AKAP2 peptides, respectively (40, 43). This is consistent with the D-AKAP2 crystal structure, which revealed that tryptophan is too bulky to fit in the hydrophobic groove of RIIα and disrupts docking by causing steric interference with surface amino acids (43). Overexpression of His-BIG2(V289W) or His-BIG2(V534W), with an inactivating mutation in one or the other of its AKAP domains that interact with RIIβ, attenuated constitutive and cAMP-induced TNFR1 exosome-like vesicle release. This finding is consistent with the conclusion that both constitutive and cAMP-induced TNFR1 exosome-like vesicle release are dependent upon the ability of BIG2 to function as an AKAP for RIIβ, which localizes the spatiotemporal action of PKA in this pathway (39).
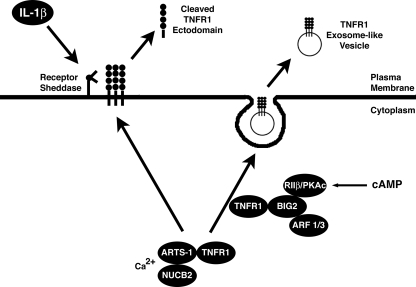
Thus, we have identified a novel mechanism by which BIG2 regulates the release of TNFR1 exosome-like vesicles. BIG2 had earlier been reported to associate with and mediate TNFR1 exosome-like vesicle release through the activation of class I ARFs (31). We now show that BIG2 localizes PKA signaling to an intracellular TNFR1 trafficking pathway by anchoring of RIIβ via its AKAP domains B and C, which act in an additive fashion (Fig. 8). Thus, the anchoring of PKA to BIG2, with resulting spatiotemporal localization of cAMP action, is required for both the constitutive and cAMP-induced release of TNFR1 exosome-like vesicles.
FIGURE 8.
Proposed model by which cAMP-dependent PKA signaling induces TNFR1 exosome-like vesicle release via anchoring of PKA regulatory subunit RIIβ to BIG2. The calcium-dependent intracellular NUCB2-ARTS-1 complex associates with TNFR1 before divergence of the pathways that lead either to the inducible proteolytic cleavage of TNFR1 ectodomains, shown here in response to IL-1β stimulation, or the constitutive release of TNFR1 exosome-like vesicles. The association of TNFR1 with BIG2 occurs after its interaction with ARTS-1 and NUCB2 and is related selectively to the extracellular release of TNFR1 exosome-like vesicles. BIG2, via AKAP domains B and C, spatiotemporally anchors constitutive and cAMP-mediated PKA catalytic activity (PKAc) via regulatory subunit RIIβ and thereby regulates the release of TNFR1 exosome-like vesicles to the extracellular compartment. Consistent with the ability of BIG2 to activate class I ARFs, the constitutive release of TNFR1 exosome-like vesicles also requires the nonredundant actions of both ARF1 and ARF3 (31).
Acknowledgments
We acknowledge Drs. Christian A. Combs and Daniela Malide, of the Light Microscopy Core Facility, NHLBI, for valuable expertise regarding the analysis of the confocal fluorescence microscopy experiments.
This work was supported, in whole or in part, by the Division of Intramural Research, NHLBI, National Institutes of Health. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: HUVEC, human umbilical vein endothelial cell(s); ARF, ADP-ribosylation factor; GEP, guanine nucleotide exchange protein; AKAP, A kinase-anchoring protein; PKA, protein kinase A; siRNA, small interfering RNA duplexes; Myr-PKI, myristoylated protein kinase A inhibitor 14–22 amide; ERK, extracellular signal-regulated kinase; ERKI, ERK activation inhibitor peptide I; 8-Br-cAMP, 8-bromo-cyclic AMP; 8-Br-cGMP, 8-bromo-cyclic GMP; ELISA, enzyme-linked immunosorbent assay.
References
- 1.Chen, G., and Goeddel, D. V. (2002) Science 296 1634-1635 [DOI] [PubMed] [Google Scholar]
- 2.Locksley, R. M., Killeen, N., and Lenardo, M. J. (2001) Cell 104 487-501 [DOI] [PubMed] [Google Scholar]
- 3.Wajant, H., Pfizenmaier, K., and Scheurich, P. (2003) Cell Death Differ. 10 45-65 [DOI] [PubMed] [Google Scholar]
- 4.Wallach, D., Varfolomeev, E. E., Malinin, N. L., Goltsev, Y. V., Kovalenko, A. V., and Boldin, M. P. (1999) Annu. Rev. Immunol. 17 331-367 [DOI] [PubMed] [Google Scholar]
- 5.Engelmann, H., Aderka, D., Rubinstein, M., Rotman, D., and Wallach, D. (1989) J. Biol. Chem. 264 11974-11980 [PubMed] [Google Scholar]
- 6.Nophar, Y., Kemper, O., Brakebusch, C., Engelmann, H., Zwang, R., Aderka, D., Holtmann, H., and Wallach, D. (1990) EMBO J. 9 3269-3278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Olsson, I., Lantz, M., Nilsson, E., Peetre, C., Thysell, H., Grubb, A., and Adolf, G. (1989) Eur. J. Haematol. 42 270-275 [DOI] [PubMed] [Google Scholar]
- 8.Schall, T. J., Lewis, M., Koller, K. J., Lee, A., Rice, G. C., Wong, G. H. W., Gatanaga, T., Granger, G. A., Lentz, R., Raab, H., Kohr, W. J., and Goeddel, D. V. (1990) Cell 61 361-370 [DOI] [PubMed] [Google Scholar]
- 9.Seckinger, P., Isaaz, S., and Dayer, J. M. (1989) J. Biol. Chem. 264 11966-11973 [PubMed] [Google Scholar]
- 10.Brakebusch, C., Varfolomeev, E. E., Batkin, M., and Wallach, D. (1994) J. Biol. Chem. 269 32488-32496 [PubMed] [Google Scholar]
- 11.Wallach, D., Aderka, D., Engelmann, H., Nophar, Y., Kemper, O., Holtmann, H., Brakebusch, C., Villa, S., Gondi, F. G., Bucciarelli, U., and Brakebusch, C. (1991) in Tumor Necrosis Factor: Structure-Function Relationship and Clinical Application (Osawa, T., and Bonavida, B., eds) Karger, Basel, Switzerland
- 12.Reddy, P., Slack, J. L., Davis, R., Cerretti, D. P., Kozlosky, C. J., Blanton, R. A., Shows, D., Peschon, J. J., and Black, R. A. (2000) J. Biol. Chem. 275 14608-14614 [DOI] [PubMed] [Google Scholar]
- 13.Gomez, M. I., Seaghdha, M. O., and Prince, A. S. (2007) EMBO J. 26 701-709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hawari, F. I., Rouhani, F. N., Cui, X., Yu, Z. X., Buckley, C., Kaler, M., and Levine, S. J. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 1297-1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Denzer, K., Kleijmeer, M. J., Heijnen, H. F., Stoorvogel, W., and Geuze, H. J. (2000) J. Cell Sci. 113 3365-3374 [DOI] [PubMed] [Google Scholar]
- 16.Gould, S. J., Booth, A. M., and Hildreth, J. E. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 10592-10597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Raposo, G., Nijman, H. W., Stoorvogel, W., Leijendekker, R., Harding, C. V., Melief, C. J. M., and Geuze, H. J. (1998) J. Exp. Med. 183 1161-1172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Théry, C., Zitvogel, L., and Amigorena, S. (2002) Nat. Rev. Immunol. 2 569-579 [DOI] [PubMed] [Google Scholar]
- 19.Chaput, N., Flament, C., Viaud, S., Taieb, J., Roux, S., Spatz, A., Andre, F., LePecq, J. B., Boussac, M., Garin, J., Amigorena, S., Thery, C., and Zitvogel, L. (2006) J. Leukocyte Biol. 80 471-478 [DOI] [PubMed] [Google Scholar]
- 20.Fevrier, B., and Raposo, G. (2004) Curr. Opin. Cell Biol. 16 415-421 [DOI] [PubMed] [Google Scholar]
- 21.Fevrier, B., Vilette, D., Laude, H., and Raposo, G. (2005) Traffic 6 10-17 [DOI] [PubMed] [Google Scholar]
- 22.Calzolari, A., Raggi, C., Deaglio, S., Sposi, N. M., Stafsnes, M., Fecchi, K., Parolini, I., Malavasi, F., Peschle, C., Sargiacomo, M., and Testa, U. (2006) J. Cell Sci. 119 4486-4498 [DOI] [PubMed] [Google Scholar]
- 23.de Gassart, A., Geminard, C., Fevrier, B., Raposo, G., and Vidal, M. (2003) Blood 102 4336-4344 [DOI] [PubMed] [Google Scholar]
- 24.Wubbolts, R., Leckie, R. S., Veenhuizen, P. T., Schwarzmann, G., Mobius, W., Hoernschemeyer, J., Slot, J. W., Geuze, H. J., and Stoorvogel, W. (2003) J. Biol. Chem. 278 10963-10972 [DOI] [PubMed] [Google Scholar]
- 25.Wang, J., Al-Lamki, R. S., Zhang, H., Kirkiles-Smith, N., Gaeta, M. L., Thiru, S., Pober, J. S., and Bradley, J. R. (2003) J. Biol. Chem. 278 21751-21760 [DOI] [PubMed] [Google Scholar]
- 26.Cui, X., Hawari, F., Alsaaty, S., Lawrence, M., Combs, C. A., Geng, W., Rouhani, F. .N., Miskinis, D., and Levine, S. J. (2002) J. Clin. Invest. 110 515-526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Islam, A., Adamik, B., Hawari, F. I., Ma, G., Rouhani, F. N., Zhang, J., and Levine, S. J. (2006) J. Biol. Chem. 281 6860-6873 [DOI] [PubMed] [Google Scholar]
- 28.Moss, J., and Vaughan, M. (1998) J. Biol. Chem. 273 21431-21434 [DOI] [PubMed] [Google Scholar]
- 29.Lippincott-Schwartz, J., Yuan, L. C., Bonifacino, J. S., and Klausner, R. D. (1989) Cell 56 801-813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Misumi, Y., Misumi, Y., Miki, K., Takatsuki, A., Tamura, G., and Ikehara, Y. (1986) J. Biol. Chem. 261 11398-11403 [PubMed] [Google Scholar]
- 31.Islam, A., Shen, X., Hiroi, T., Moss, J., Vaughan, M., and Levine, S. J. (2007) J. Biol. Chem. 282 9591-9599 [DOI] [PubMed] [Google Scholar]
- 32.Li, H., Adamik, R., Pacheco-Rodriguez, G., Moss, J., and Vaughan, M. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 1627-1632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Seino, S., and Shibasaki, T. (2005) Physiol. Rev. 85 1303-1342 [DOI] [PubMed] [Google Scholar]
- 34.de Rooij, J., Zwartkruis, F. J., Verheijen, M. H., Cool, R. H., Nijman, S. M., Wittinghofer, A., and Bos, J. L. (1998) Nature 396 474-477 [DOI] [PubMed] [Google Scholar]
- 35.Kawasaki, H., Springett, G. M., Mochizuki, N., Toki, S., Nakaya, M., Matsuda, M., Housman, D. E., and Graybiel, A. M. (1998) Science 282 2275-2279 [DOI] [PubMed] [Google Scholar]
- 36.Guan, H., Hou, S., and Ricciardi, R. P. (2005) J. Biol. Chem. 280 9957-9962 [DOI] [PubMed] [Google Scholar]
- 37.Delghandi, M. P., Johannessen, M., and Moens, U. (2005) Cell. Signal. 17 1343-1351 [DOI] [PubMed] [Google Scholar]
- 38.Walsh, D. A., Perkins, J. P., and Krebs, E. G. (1968) J. Biol. Chem. 243 3763-3765 [PubMed] [Google Scholar]
- 39.Wong, W., and Scott, J. D. (2004) Nat. Rev. Mol. Cell. Biol. 5 959-970 [DOI] [PubMed] [Google Scholar]
- 40.Gold, M. G., Lygren, B., Dokurno, P., Hoshi, N., McConnachie, G., Tasken, K., Carlson, C. R., Scott, J. D., and Barford, D. (2006) Mol. Cell 24 383-395 [DOI] [PubMed] [Google Scholar]
- 41.Burns-Hamuro, L. L., Ma, Y., Kammerer, S., Reineke, U., Self, C., Cook, C., Olson, G. L., Cantor, C. R., Braun, A., and Taylor, S. S. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4072-4077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carr, D. W., Stofko-Hahn, R. E., Fraser, I. D., Bishop, S. M., Acott, T. S., Brennan, R. G., and Scott, J. D. (1991) J. Biol. Chem. 266 14188-14192 [PubMed] [Google Scholar]
- 43.Kinderman, F. S., Kim, C., von Daake, S., Ma, Y., Pham, B. Q., Spraggon, G., Xuong, N. H., Jennings, P. A., and Taylor, S. S. (2006) Mol. Cell 24 397-408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Taylor, S. S., Buechler, J. A., and Yonemoto, W. (1990) Annu. Rev. Biochem. 59 971-1005 [DOI] [PubMed] [Google Scholar]
- 45.Herberg, F. W., Maleszka, A., Eide, T., Vossebein, L., and Tasken, K. (2000) J. Mol. Biol. 298 329-339 [DOI] [PubMed] [Google Scholar]
- 46.Alto, N. M., Soderling, S. H., Hoshi, N., Langeberg, L. K., Fayos, R., Jennings, P. A., and Scott, J. D. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 4445-4450 [DOI] [PMC free article] [PubMed] [Google Scholar]