Abstract
Viral infection activates Toll-like receptor and RIG-I (retinoic acid-inducible gene I) signaling pathways, leading to phosphorylation of IRF3 (interferon regulatory factor 3) and IRF7 and stimulation of type I interferon (IFN) transcription, a process important for innate immunity. We show that upon vesicular stomatitis virus infection, IRF3 and IRF7 are modified not only by phosphorylation but by the small ubiquitin-related modifiers SUMO1, SUMO2, and SUMO3. SUMOylation of IRF3 and IRF7 was dependent on the activation of Toll-like receptor and RIG-I pathways but not on the IFN-stimulated pathway. However, SUMOylation of IRF3 and IRF7 was not dependent on their phosphorylation, and vice versa. We identified Lys152 of IRF3 and Lys406 of IRF7 to be their sole small ubiquitin-related modifier (SUMO) conjugation site. IRF3 and IRF7 mutants defective in SUMOylation led to higher levels of IFN mRNA induction after viral infection, relative to the wild type IRFs, indicating a negative role for SUMOylation in IFN transcription. Together, SUMO modification is an integral part of IRF3 and IRF7 activity that contributes to postactivation attenuation of IFN production.
Invading pathogens are recognized by Toll-like receptors (TLRs)3 and/or RNA helicases (i.e. RIG-I/MDA-5 (retinoic acid-inducible gene I/melanoma differentiation-associated gene 5)) (1-5). Binding of pathogen components to these molecules activates downstream signaling pathways, which results in the production of various proinflammatory cytokines, important for the establishment of innate and adaptive immunity (3). Among them, type I interferons (IFNs) play a major role in conferring antiviral and antimicrobial activities (6-8). Production of type I IFN depends on activation of IRF3 (interferon regulatory factor 3) and IRF7 (3, 9-11). IRF3 and IRF7 are phosphorylated by TBK-1 (TANK-binding kinase 1) and IKKε (IκB kinase ε), dimerized, translocated into the nucleus, and finally stimulate IFN gene transcription (3, 9-11).
Ubiquitin-like proteins (Ubls), including the small ubiquitin-related modifiers (SUMO) and ISG15 (interferon stimulated gene 15), among others, modify many proteins to regulate various biological processes (12-15). Ubls are conjugated to target proteins by an enzymatic cascade involving an activating enzyme (E1), a conjugating enzyme (E2), and a ligase (E3) (15-17). Ubl modification of signaling molecules and transcription factors has a large impact on gene expression (13, 14). Type I IFN induction involves ubiquitin and Ubl modifications of multiple signaling molecules. For example, RIG-I is modified by ubiquitin by at least two independent E3 ligases, TRIM25 and RNF125, to positively and negatively regulate type I IFN production, respectively (18-20). RIG-I is also modified by ISG15 (19, 21, 22). Furthermore, IRF7 is ubiquitinated by TRAF6, an event believed to be important for type I IFN transcription (23). IRF7 is reported to interact with the TNF receptor-associated adaptor protein RIP in the presence of an EBV oncoprotein, which enhances IRF7 ubiquitination and activation (24).
The SUMO proteins, ∼12 kDa in size, covalently attach to many proteins (13, 14, 25). In mammals, there are at least three SUMO isoforms (SUMO1, -2, and -3). SUMO2 and SUMO3 form a distinct subgroup known as SUMO2/3. They are very similar to each other in the amino acid sequence, differing in only 3 residues, but are different from SUMO1 with which they share only 50% amino acid identity (14). SUMO1 and SUMO2/3 appear to modify both common and different substrates, including a number of transcription factors (13, 14). Many SUMOylated proteins possess the consensus motif, ψKXE, where ψ is a hydrophobic residue, X is any residue, and K is the SUMO acceptor lysine (26). The unique SUMO E2 conjugating enzyme, Ubc9, recognizes the consensus motif and transfers SUMO to the acceptor lysine residue in the substrate (12). SUMOylation of transcription factors is generally associated with transcriptional repression, although there are some exceptions (13, 14).
Transcription factors of the IRF family regulate the entire type I IFN system from induction of IFNs to diverse IFN responses (9, 11, 27). Among IRF members, IRF1 is shown to be covalently conjugated to SUMO1, and this SUMOylation appears to be linked to transcriptional inhibition (28). Prompted by this report, we asked whether other members of the IRF family are also SUMOylated. In this paper, we show that, indeed, IRF3 and IRF7 are covalently conjugated to SUMO1, SUMO2, and SUMO3, and the SUMOylation of IRF3 and IRF7 was markedly increased following virus infection. Virus-induced SUMOylation of IRF3 and IRF7 was a consequence of TLR and RIG-I activation but not of IFN signaling. We also found that prevention of SUMOylation from IRF3 and IRF7 through the mutation of SUMOylation sites leads to increased IFNα4 and IFNβ mRNA expression following viral infection. Our findings support the view that virus-mediated IRF3 and IRF7 SUMOylation represents postactivation attenuation of IFN gene transcription.
EXPERIMENTAL PROCEDURES
Reagents—Mouse monoclonal antibodies against FLAG M2, α-tubulin, anti-FLAG-agarose beads, and protein A-agarose beads were purchased from Sigma. Mouse monoclonal antibody for the V5-tag and rabbit and mouse antibodies for the T7-tag were purchased from Invitrogen, Abcam (Cambridge, MA), and Novagen (Gibbstown, NJ), respectively. Antibodies against STAT1, STAT2, and PKR were from Santa Cruz Biotechnology, Inc. (Santa Cruz, CA), and those against SUMO1 and murine IRF3 were from Zymed Laboratories Inc. (South San Francisco, CA). Mouse monoclonal antibody against Ubc9 was from Transduction Laboratories (Lexington, KY), and recombinant human IFNβ was from Toray Industries, Inc. (Tokyo, Japan). Calf intestine alkaline phosphatase was obtained from New England Biolabs (Ipswich, MA).
Cell Culture and Virus Infection—Human embryonic kidney 293T cells and human 2fTGH, U3A, U4A, U5A, and U6A cells (gifts from Dr. G. Stark of Lerner Research Institute, Cleveland Clinic) were grown in Dulbecco's modified Eagle's medium containing 10% fetal bovine serum. Murine NIH3T3 cells were cultured in Dulbecco's modified Eagle's medium containing 10% calf serum. For virus inoculation, medium was removed from the culture dish, and cells were infected with vesicular stomatitis virus (VSV) or encephalomyocarditis virus (EMCV) at an MOI of 1 for 1 h. Then fresh medium was added to the cells, and the culture was allowed to continue for the indicated periods of time.
Constructs—cDNA fragments of IRF3, IRF7, VISA, TRIF, SUMO2, and SUMO3 were generated from total RNA prepared from NIH3T3 cells by RT-PCR and cloned into pcDNA3.1 with a FLAG tag at the C terminus or a V5 tag at the N terminus. To construct mutants for IRF3 and IRF7, appropriate substitutions were introduced into the pcDNA3.1-IRF7-FLAG and pcDNA3.1-IRF3-FLAG using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA). A plasmid expressing a T7-tagged SUMO1 (pCGT-T7-SUMO1) was a gift from Dr. H. Yokosawa of Hokkaido University. A T7-tagged SUMO2 expressing plasmid, pCGT-T7-SUMO2, was constructed by substitution of the SUMO1 open reading frame with SUMO2. For constructing an shRNA retroviral vector targeting Ubc9, an oligonucleotide fragment (5′-ggtccgagcacaagcgaagaa-3′) was inserted into pSUPER.retro (Oligoengine, Seattle, WA). Retroviral preparations were produced according to the manufacturer's instructions. As a control, a retroviral vector with a scrambled oligonucleotide fragment was prepared and tested in parallel.
Detection of SUMO-conjugated Proteins—To detect SUMO conjugation of IRF3 and IRF7, 293T cells (3 × 106) or 2fTGH cells (4 × 105) were transfected with a total of 3.3 or 2.5 μg of plasmid DNA using Lipofectamin 2000 and Lipofectamin LRX (Invitrogen), respectively. Twelve h later, cells were stimulated with VSV or IFNβ for indicated time periods. Cells were washed and lysed in Lysis buffer, containing 150 mm NaCl, 50 mm Tris-HCl (pH 7.5), 4 mm EDTA, 0.1% NaDOC, 1% Nonidet P-40, 0.1% SDS, complete protease inhibitor mixture (Roche Applied Science), and 20 mm N-ethylmaleimide (Sigma). Lysates were centrifuged, and supernatants were incubated with anti-FLAG-agarose overnight with gentle rotation at 4 °C. Immune complexes were washed 4 times with Lysis buffer, and separated on SDS-PAGE and subjected to immunobblot analysis. For detecting SUMOyated IRF3 and IRF7, NIH3T3 cells (5 × 107) expressing control or Ubc9 shRNA were infected by VSV at an MOI of 1 for the indicated periods of time. Whole cell extracts were incubated with 5 μg of anti-mouse IRF3 antibody and protein A-agarose overnight at 4 °C. Immune complexes were washed six times with Lysis buffer and analyzed by immunoblot using the indicated antibodies.
Phos-tag SDS-PAGE—293T cells were transiently transfected with pcDNA3.1-IRF3-FLAG together with a serial dilution of V5-VISA expression plasmid. Twenty-four h after transfection, cells were lysed in Lysis buffer without EDTA. Whole cell extracts were separated on SDS-PAGE with or without 75 mm phos-tag acrylamide AAL-107 (NARD institute, Hyogo, Japan) according to the manufacturer's suggestions. For dephosphorylation experiments, extracts were incubated with or without calf intestine alkaline phosphatase in calf intestine alkaline phosphatase buffer (50 mm Tris-HCl, pH 9.0, 10 mm MgCl2) at 37 °C for 30 min. Samples were diluted with lysis buffer and separated on SDS-PAGE with or without 75 mm phos-tag acrylamide.
Quantitative Reverse Transcription-PCR—NIH-3T3 cells (2 × 105) were transfected with 2.5 μg of indicated plasmids using Lipofectamine LTX. Twelve h after transfection, cells were infected with VSV or EMCV at an MOI of 1 for the indicated periods. Total RNA prepared with the Trizol reagent (Invitrogen) was reverse transcribed with the Transcriptor First Strand cDNA synthesis kit (Roche Applied Sciences). The amount of IFNβ, IFNα4, and hypoxanthine guanine phosphoribosyltransferase cDNA were measured by using Universal ProbeLibrary and LightCycler 480 (Roche Applied Science) according to the manufacturer's instructions. Primers for quantitative reverse transcription-PCR were designed by the ProbeFinder software (Roche Applied Science).
RESULTS
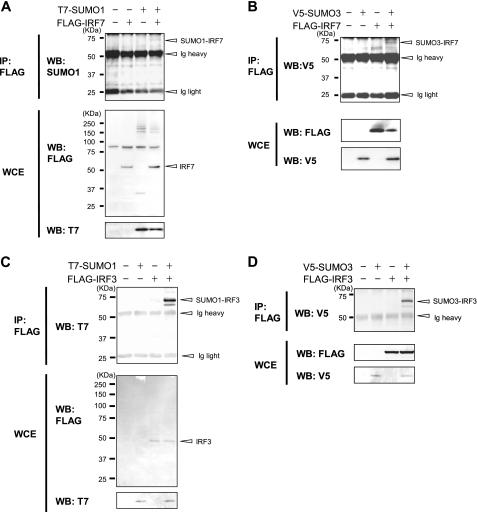
IRF3 and IRF7 Are Modified by SUMO1, -2, and -3—To investigate whether IRF7 has a potential to be conjugated to SUMO molecules, FLAG-tagged IRF7 was introduced into 293T cells together with T7-tagged SUMO1 (Fig. 1A). Whole cell extracts were immunoprecipitated with anti-FLAG antibody-bound agarose beads, and precipitates were tested for T7-SUMO by immunoblot analysis. A band of ∼70 kDa that reacted with antibody for SUMO1 was detected in samples transfected with FLAG-IRF7 and T7-SUMO1 (open arrowhead in Fig. 1A, top). This band was not detected in cells transfected with SUMO1 alone. Assuming the size of SUMO1 to be ∼12 kDa and that of IRF7 to be 55 kDa, this 70 kDa band was most likely to be a SUMOylated IRF7. Similarly, when FLAG-IRF7 was co-expressed with V5-tagged SUMO3, IRF7 precipitates showed a ∼70 kDa band with V5 SUMO3 reactivity (Fig. 1B, top). Likewise, FLAG-IRF3, when co-introduced with T7-SUMO1 or V5-SUMO3 precipitated a ∼70-kDa band with a respective SUMO reactivity (Fig. 1, C and D, top). Similarly, we observed SUMOylation of IRF7 and IRF3 by SUMO2 (Fig. S1A, lane 6; data not shown). These results indicate that both IRF3 and IRF7 can be modified by SUMO1, -2, and -3.
FIGURE 1.
SUMO conjugation of IRF7 and IRF3. 293T cells were transfected with plasmids for FLAG-IRF7 (A and B) or FLAG-IRF3 (C and D) along with T7-SUMO1 (A and C) or V5-SUMO3 (B and D). Whole cell extracts (WCE) were immunoprecipitated (IP) with anti-FLAG antibody-agarose beads, and SUMO-conjugated proteins were detected with anti-T7, anti-V5, or anti-SUMO1 antibody in Western blot (WB) (top). Expression of transfected proteins was verified by Western blot analysis of whole cell extracts using the indicated antibody (bottom).
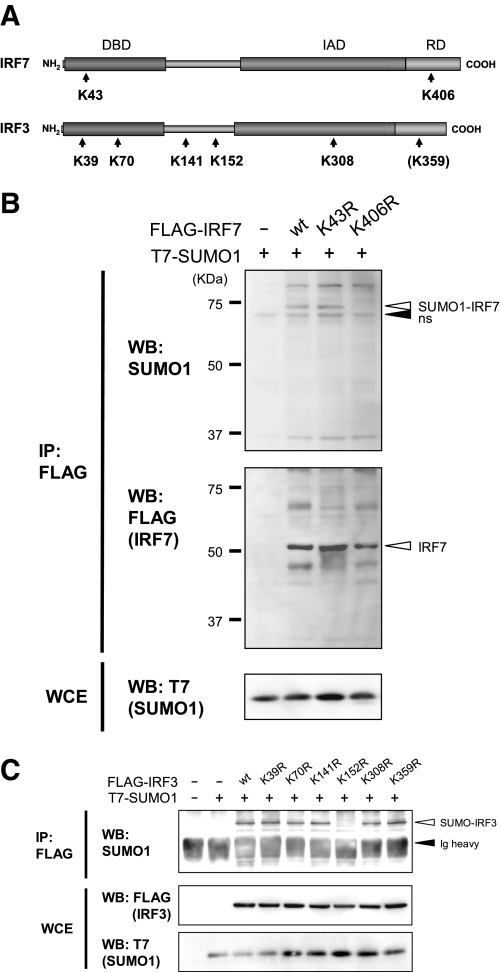
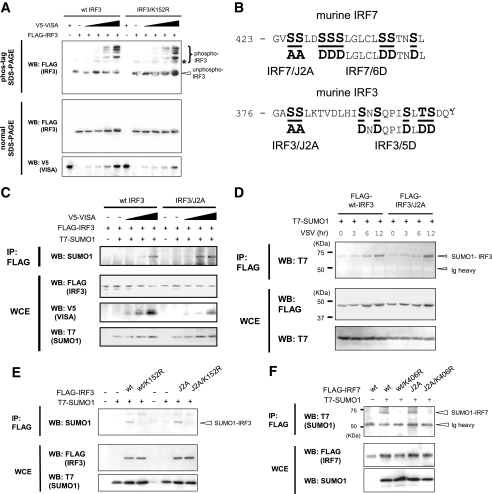
Identification of SUMO Acceptor Sites in IRF3 and IRF7—The SUMOplot prediction program (available on the World Wide Web) revealed two putative SUMOylation sites in IRF7 that carried the ψKXE motif and five in IRF3 (Fig. 2A). To test if these sites are functional SUMO conjugation sites, a Lys → Arg mutation was introduced into each of these sites in IRF3 and IRF7, and the mutants were tested for SUMO conjugation. As shown in Fig. 2B, the IRF7 K406R mutant failed to conjugate SUMO1 (top), whereas wild type (WT) IRF7 and the K43R mutant were clearly SUMOylated. IRF7 K406R was expressed at levels comparable with WT IRF7 as well as K43R, indicating that the lack of SUMOylation in this mutant was not due to a reduced protein expression. For IRF3, all mutants except K152R produced bands conjugated to SUMO1 (Fig. 2C). This was true for IRF3 K359R, in which the mutation was placed in the position equivalent to that of IRF7 K406R. This mutant was SUMOylated as well as WT IRF3. Similar results were obtained when IRF7 and IRF3 mutants were co-expressed with SUMO2 or SUMO3 (Fig. S1, A-C; data not shown). These results indicate that Lys406 of IRF7 and Lys152 of IRF3 serve as a conjugation site for SUMO1, -2, and -3.
FIGURE 2.
Identification of SUMO conjugation sites in IRF3 and IRF7. A, a schematic presentation of murine IRF7 and IRF3. The positions of lysine residues that conform to the putative SUMO conjugation sites are indicated by arrows. IRF3 and IRF7 mutants contained an Arg substitution at each of the Lys residues. Lys359 corresponds to Lys406 of IRF7 (in parenthesis). DBD, DNA binding domain; IAD, IRF association domain; RD, regulatory domain. B and C, 293T cells were transfected with mutant FLAG-IRF7 (B) or mutant FLAG-IRF3 (C) along with T7-SUMO1 for 48 h. Extracts were immunoprecipitated (IP) with anti-FLAG-agarose beads, and precipitates were tested by Western blotting (WB) with the indicated antibodies (top). ns, nonspecific band. Whole cell extracts were tested for expression of transfected proteins.
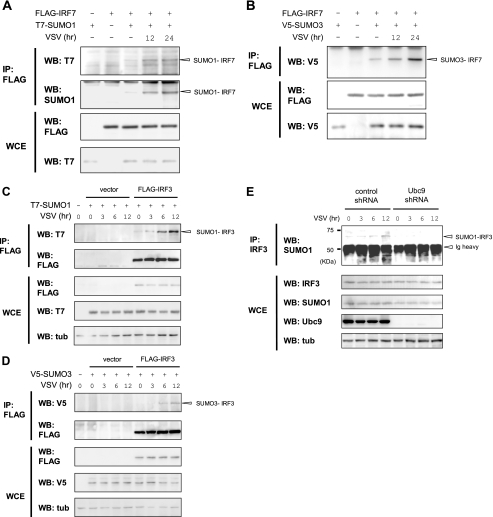
Virus Infection Increases SUMOylation of IRF3 and IRF7—Having found that IRF3 and IRF7 can conjugate SUMO1, -2, and -3, it was of interest to assess events that may trigger SUMOylation of IRF3 and IRF7. Given that IRF3 and IRF7 are activated upon virus infection, we tested whether their SUMOylation is stimulated by virus infection. In Fig. 3, A and B, 293T cells expressing FLAG-IRF7 and T7-SUMO1 or V5-SUMO3 were infected with VSV, and IRF7 SUMOylation was tested by immunoprecipitation and immunoblot analysis. Little SUMO conjugation was observed without viral infection. However, IRF7 showed a clear increase in SUMO1 and SUMO3 conjugation at 12 and 24 h after virus infection. Similarly, VSV infection led to SUMOylation of IRF3 both with SUMO1 and SUMO3 (Fig. 3, C and D). Interestingly, the levels of SUMOylation were the greatest at 12 h after viral infection for both IRF3 and IRF7; little SUMO conjugation was seen at 3 h for IRF3. Increased SUMOylation observed after viral infection was not due to changes in the expression levels for IRF3/IRF7 or SUMO1/3, since they remained constant for 12 h after viral infection (Fig. 3, A-D, bottom).
FIGURE 3.
Increased SUMOylation of IRF3 and IRF7 following virus infection. A-D, 293T cells were transfected with FLAG-IRF7 (A and B) or FLAG-IRF3 (C and D) with or without T7-SUMO1 (A and C) or V5-SUMO3 (B and D) for 12 h. Cells were infected with VSV at an MOI of 1 for the indicated periods. Extracts were immunoprecipitated (IP) with anti-FLAG-agarose beads and tested for SUMO conjugation by Western blotting (WB) with the indicated antibodies. E, NIH3T3 cells expressing control shRNA or Ubc9 shRNA were infected with VSV at an MOI of 1, and cells were allowed to proceed for the indicated periods. Extracts were immunoprecipitated with anti-IRF3 antibody and tested in Western blotting with anti-SUMO1 antibody (top). Expression of endogenous IRF3, SUMO1, Ubc9, and tubulin α (tub) was tested by Western blot analysis of whole cell extracts (WCE).
We felt it important to determine whether endogenous IRF3 and IRF7 are SUMOylated in a natural setting. To this end, NIH3T3 cells were infected with VSV, and IRF3 immunoprecipitates were tested for reactivity with SUMO1 (Fig. 3E, left in top panel). A very low level of SUMOylated IRF3 was observed before infection, and SUMOylated IRF3 was markedly increased at 6 and 12 h after VSV infection (see the ∼70 kDa band in the top panel). The level of SUMO conjugation was greater at 12 h than 6 h, consistent with the data in Fig. 3, A-D. These results demonstrate that VSV infection triggers SUMOylation of endogenous IRF3. To further substantiate the authenticity of the virus-induced IRF3 SUMOylation, we tested the effect of shRNA for Ubc9, a sole E2 enzyme for SUMO conjugation (12-14). Expression of the shRNA drastically reduced Ubc9 expression without affecting IRF3 expression (Fig. 3E, right in top panel). IRF3 SUMOylation was completely abrogated in these cells, indicating that this SUMOylation was dependent on Ubc9 and the endogenous SUMOylation cascade. It should be noted here that it was not possible to test SUMOylation of endogenous IRF7 or SUMO3 modifications due to their very low expression levels.
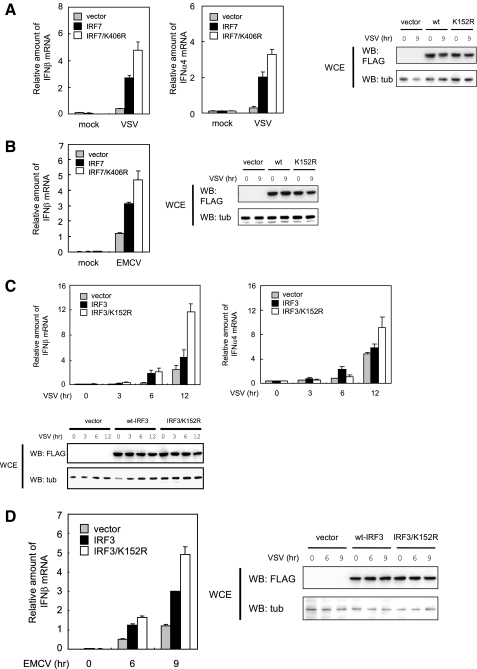
SUMOylation of IRF3 and IRF7 Represses Type I IFN Gene Expression—Virus-mediated SUMOylation observed above pointed to the possibility that SUMO modification regulates functional activity of IRF3 and IRF7. To test this possibility, we examined whether mutations in SUMOylation sites in IRF3 and IRF7 affect virus-induced type I IFN induction. NIH3T3 cells transfected with WT IRF7 or the SUMOylation-defective mutant IRF7 K406R were stimulated with VSV for 9 h. Then IFNβ and IFNα4 mRNA levels were measured by quantitative reverse transcription-PCR (Fig. 4A, top left and middle). Expression of WT IRF7 markedly increased both IFN mRNAs upon VSV infection. Importantly, overexpression of IRF7 K406R mutant led to a further increase in the expression of both IFNβ mRNA and IFNα4 mRNAs. Similarly, this mutant gave increased IFNβ mRNA following EMCV infection relative to WT IRF7 (Fig. 4B). Expression of WT IRF7 and the K406R mutant was comparable throughout 9 h of viral infection (Fig. 4A, right). Considering that transfection efficiency monitored by immunofluorescent staining of FLAG-IRF7 was consistently ∼50% (data not shown) and that the endogenous IRF7 participated in IFN induction, the increase in IFN mRNA expression detected by the Lys406 mutant probably represents an underestimate. Results with WT IRF3 and the K152R mutant are shown in Fig. 4, C and D. In agreement with the data for IRF7, the K152R mutant yielded substantially higher levels of IFNβ mRNA and IFNα4 mRNA compared with WT IRF3 upon VSV infection. Similarly, the mutant gave higher IFNβ induction upon infection with EMCV than WT IRF3. Interestingly, the enhanced type I IFN mRNA expression by the mutant was evident at 9-12 h after infection and was not seen at 6 h, which appeared in line with delayed IRF3 and IRF7 SUMOylation after virus infection (compare with data in Fig. 3). These results indicate that SUMOylation of IRF3 and IRF7 negatively regulate virus-induced type I IFN expression.
FIGURE 4.
Mutation of the SUMOylation site in IRF7 and IRF3 increases type I IFN production. NIH3T3 cells were transfected with WT IRF7 or IRF7/K406R (A and B) or with WT IRF3 or IRF3/K152R (C and D) for 12 h. For A and B, cells were infected with VSV or EMCV at an MOI of 1 for 9 h prior to harvest. For C and D, cells were harvested at varying periods after infection. The amounts of IFNβ or IFNα4 mRNAs were quantified by quantitative reverse transcription-PCR by normalizing with hypoxanthine guanine phosphoribosyltransferase mRNA. The values represent the average of three samples ± S.D. Comparable expression of WT IRF3/IRF7 and the mutants was verified by Western blotting (WB) of whole cell extracts (WCE) (right and bottom).
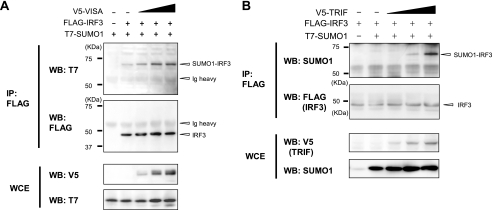
TLR and RIG-I/MDA-5 Signaling Mediates SUMOylation of IRF3 and IRF7—The above data indicate that virus induced SUMOylation of IRF3 and IRF7 leads to inhibition of type I IFN transcription. We next investigated signaling pathways that may mediate SUMOylation of IRF3 and IRF7. VISA (also called Cardif, MAVS, or IPS-1) is a signaling molecule involved in the RIG-I/MDA-5-activated pathway (3, 29), and exogenously expressed VISA can activate IRF3 and IRF7 (30-33). In Fig. 5A, co-transfection of V5-tagged VISA led to SUMOylation of IRF3 in a VISA dose-dependent manner. SUMOylation of IRF7 was also increased by co-expression of VISA (Fig. S2). TRIF is an adaptor molecule contributing within the TLR signaling pathway, and ectopic TRIF expression results in activation of IRF3 and IRF7 (34, 35). Similar to VISA, co-expression of V5-TRIF markedly increased SUMOylated IRF3 (Fig. 5B). These results indicate that IRF3 and IRF7 are SUMOylated as a result of TLR and RIG-I/MDA-5 signaling.
FIGURE 5.
Induction of IRF3 and IRF7 SUMOylation by VISA and TRIF. 293T cells were transfected with FLAG-IRF3 along with increasing amounts of V5-VISA (A) or V5-TRIF (B) plus a constant amount of T7-SUMO1 for 12 h. SUMOylated IRF3 was detected by immunoprecipitation (IP) with anti-FLAG-agarose, followed by Western blot analysis (WB) with the indicated antibodies. WCE, whole cell extract.
VISA-induced SUMOylation Does Not Affect Phosphorylation of IRF3—IRF3 and IRF7 are phosphorylated in response to TLR and RIG-I/MDA-5 signaling, marking their activation (3, 11). We sought to address whether phosphorylation and SUMOylation of IRF3 and IRF7 were internally coupled and take place in a mutually dependent manner. To this end, we first examined whether WT IRF3 and IRF3 K152R, the SUMOylation-defective mutant, could be phosphorylated upon VISA stimulation. To detect phosphorylated IRF3, the phos-tag SDS-PAGE was employed, in which phosphorylated proteins migrate more slowly than unphosphorylated counterparts (36). As shown in Fig. 6A (top), when co-expressed with VISA, both WT IRF3 and IRF3 K152R produced multiple slower migrating IRF3 bands, in addition to the unphosphorylated IRF3 band in a VISA dose-dependent manner. The band patterns produced by the WT IRF3 and IRF3 K152R in the phos-tag SDS-PAGE were very similar to each other. The appearance of the slowly migrating IRF3 bands detected after VISA co-expression was attributed to phosphorylation, since these bands were no longer detected following phosphatase (calf intestine alkaline phosphatase) treatment (Fig. S3A). A faint, slowly migrating band was found both in WT IRF3 and IRF3 K152R without VISA treatment, which was abolished after calf intestine alkaline phosphatase treatment, indicating a low level of constitutively activated IRF3. In the conventional SDS-PAGE (Fig. 6A, middle; data not shown), both WT IRF3 and IRF3 K152R migrated to a single position irrespective of VISA expression. These results show that VISA activation leads to IRF3 phosphorylation both in WT IRF3 and IRF3 Lys152, indicating that phosphorylation of IRF3 does not require SUMOylation.
FIGURE 6.
Independence of SUMOylation of IRF3 and IRF7 from the phosphorylation. A, 293T cells were transfected with WT FLAG-IRF3 or FLAG-IRF3/K152R along with increasing doses of V5-VISA for 24 h. Whole cell extracts (WCE) were tested in the phos-tag SDS-PAGE (top panel) or normal SDS-PAGE (lower panels) by Western blot analysis (WB). B, the serine/threonine cluster phosphorylation sites involved in the activation of type I IFN genes. Phosphorylated Ser and Thr are in boldface type and underlined. Alanine or aspartic acid substitutions were placed in the indicated residues to create IRF7/J2A, IRF7/6D, IRF3/J2A, and IRF3/5D. C, cells were transfected with WT IRF3 or IRF3/J2A along with T7-SUMO1 and increasing amounts of V5-VISA. SUMOylated IRF3 was detected by immunoprecipitation (IP) and Western blotting as in Fig. 1. D, cells transfected with WT IRF3 or IRF3/J2A along with T7-SUMO1 for 12 h were infected with VSV at an MOI of 1. Cells were harvested at the indicated period, and SUMOylated IRF3 was detected by immunoprecipitation (IP) and Western blot analysis as above. E and F, cells were transfected with WT IRF3, WT IRF7, or mutants with indicated substitutions along with T7-SUMO1. SUMOylated IRF3 or IRF7 was detected by immunoprecipitation and Western blot analysis as above.
IRF3 and IRF7 SUMOylation Does Not Depend on Phosphorylation—We then sought to examine whether SUMOylation of IRF3 and IRF7 is dependent on phosphorylation. To this end, mutants defective in phosphorylation were constructed. In Fig. 2, serines at amino acid positions 425 and 426 in IRF7 and those at 378 and 379 in IRF3 were replaced by alanine to generate IRF3/J2A and IRF7/J2A (Fig. 6B). These serine residues are phosphorylated after TLR/RIG-I activation, and their mutation blocks the subsequent activation events (37-40). We also generated constitutively active forms of IRF3 and IRF7 by replacing additional serine and threonine residues with aspartic acid (see 5D and 6D in Fig. 6B), as described (37-40). As expected, VISA treatment did not produce slowly migrating bands both in IRF3/J2A and IRF3/J2A/K152R in the phos-tag SDS-PAGE, confirming the inability of these mutants to be phosphorylated (supplemental Fig. S3, B and C). However, both WT IRF3 and IRF3/J2A were SUMO-conjugated in the presence of SUMO1, in a VISA dose-dependent manner (Fig. 6C). SUMOylation was not observed when WT IRF3 and IRF3/J2A contained the K152R mutation. Conjugation of SUMO1 and SUMO3 to the IRF3/J2A mutant was also observed after VSV infection in a manner similar to WT IRF3 (Figs. 6D and S3D), whereas the bands for the SUMOylated form of WT IRF3 and IRF3/J2A were eliminated by K152R mutation (Fig. 6E). Similarly for IRF7, IRF7/J2A was SUMOylated as well as the WT IRF7 in the presence of SUMO1, whereas WT IRF7/K406R and IRF7/J2A/K406R mutants were not (Fig. 6F). These results indicate that SUMOylation of IRF3 and IRF7 does not depend on their phosphorylation. Consistent with phosphorylation-independent SUMOylation of IRF3 and IRF7, the constitutively activated forms of IRF3 and IRF7 (IRF3/5D and IRF7/6D, respectively, in Fig. 6B) were SUMOylated in a similar manner as the wild type proteins (supplemental Fig. S4, A and B). These data indicate that SUMOylation of IRF3 and IRF7 takes place independently of their phosphorylation.
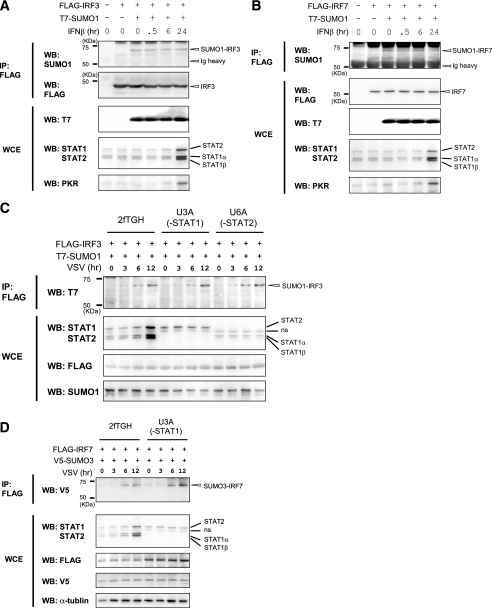
The Type I IFN-responsive JAK/STAT Pathway Does Not Contribute to IRF3 and IRF7 SUMOylation—SUMOylation of IRF3 and IRF7 occurs downstream of TLR and RIG-I/MDA-5 pathways (Fig. 5). Given that activation of these pathways leads to type I IFN production (3, 11), it was possible that type I IFN signaling causes SUMOylation of IRF3 and IRF7. In a similar vein, since IFNs stimulate expression of many genes through the activation of JAK/STAT pathway (41), an IFN-induced factor (s) may be responsible for SUMOylation of IRF3 and IRF7. To test these possibilities, we examined whether IFNβ treatment itself causes SUMOylation of IRF3 and IRF7. In Fig. 7, A and B, 293T cells were transfected with T7-SUMO1 and FLAG-IRF3 or FLAG-IRF7, and SUMOylation of IRF3 or IRF7 was tested at varying times after IFNβ treatment. Whereas IRF3 and IRF7 were both SUMOylated in the presence of SUMO1, IFNβ treatment up to 24 h did not increase SUMOylation of IRF3 and IRF7. After 24 h of IFNβ treatment, levels of IFN-stimulated genes, STAT1α, STAT1β, STAT2, and PKR, were increased, verifying that the IFN-signaling pathway was activated in these cells (Fig. 7, A and B, bottom). These data support the notion that induction of IFNs and activation of the JAK/STAT signaling pathway are not involved in the SUMOylation of IRF3 and IRF7. To substantiate this notion, we tested mutant cell lines defective in JAK/STAT pathway activation (42, 43). In Fig. 7C, the parental 2fTGH cells, the STAT1-deficient U3A cells, and the STAT2-deficient U6A cells were transfected with IRF3 and SUMO1 and tested for SUMOylation after VSV infection. IRF3 was SUMOylated in all three cells in a similar manner at 6 and 12 h following virus infection (top). Levels of STAT1 and STAT2 were increased only in 2fTGH cells, not in U3A or U6A cells (middle), confirming that JAK/STAT signaling was functional only in the parental cells and not in mutant cells. IRF3 was similarly SUMOylated in other JAK/STAT pathway mutants, such as U4A and U5A cells (data not shown). Likewise, IRF7 was SUMOylated in U3A cells when co-expressed with SUMO3, following VSV infection (Fig. 7D). These results indicate that SUMOylation of IRF3 or IRF7 occurs as a consequence of activation of TLR and RIG-I/MDA-5 pathways and does not depend on the IFN-activated JAK/STAT pathway.
FIGURE 7.
The lack of requirement of IFN signaling for SUMOylation of IRF7 and IRF3. A and B, 293T cells were transfected with FLAG-IRF3 (A) or FLAG-IRF7 (B) along with T7-SUMO1 for 12 h and treated with 1000 units/ml human IFNβ for the indicated periods. SUMOylated IRF3 and IRF7 were detected by immunoprecipitation (IP), followed by Western blot (WB) (top). Whole cell extracts (WCE) were analyzed for the IFN-stimulated expression of STAT1, STAT2, and PKR by Western blotting (bottom). C and D, the parental 2fTGH, STAT1-deficient U3A, and STAT2-deficient U6A cells were transfected with FLAG-IRF3 and FLAG-IRF7, along with T7-SUMO1 or V5-SUMO3, and infected with VSV at an MOI of 1 for the indicated periods. SUMOylated IRF3 and IRF7 were detected by immunoprecipitation, followed by Western blot analysis (top). Whole cell extracts (WCE) were analyzed for expression STAT1α, STAT1β, and STAT2 as above. ns, nonspecific band.
DISCUSSION
We report here that IRF3 and IRF7 are SUMOylated in response to virus infection, each through a single residue at Lys152 and Lys406, respectively. We identified the signaling pathways that trigger this SUMOylation, since SUMOylation of IRF3 and IRF7 was an event downstream of TLR and RIG-I pathway activation. TLR pathways are activated by a wide range of pathogen components, whereas RIG-I is activated by double-stranded RNA and single-stranded RNA with 5′-triphosphate (3, 44). Significantly, IFN activated JAK/STAT pathway was not involved in the SUMOylation of IRF3 and IRF7, indicating that this SUMOylation is coupled directly to the pathogen recognition events rather than indirectly to host cytokine responses. Thus, the virus-induced SUMOylation of IRF3 and IRF7 is likely to be a part of complex host innate responses against pathogens. Our finding that blocking SUMOylation enhanced type I IFN induction for both IRF3 and IRF7 points to the idea that SUMOylation plays a role in the discontinuation of IFN transcription after activation. Additional data indicating that pronounced SUMOylation of IRF3 and IRF7 was detected relatively late, 12 h after viral stimulation, also favor this idea. Proper down-regulation of inflammatory cytokine expression is necessary for the host to avoid excessive cytotoxicity and tissue injury after infection. There are multiple mechanisms that allow timely attenuation of inflammatory reactions. For example, cytokine signaling by JAK/STAT pathways is negatively regulated by the proteins of the SOCS (suppressors of cytokine signaling) family, that are responsible for feedback inhibition of cytokine induction (45). The PIAS family of SUMO E3 ligases also represses cytokine signaling by inhibiting the activity of factors belonging to the STAT and NFκB families (46-48). Ubiquitin-mediated destabilization of IRF3 and RIG-I also plays a role in down-regulation of innate immunity, although ubiquitination of RIG-I has an opposite role as well (20, 21, 49). Further, IFN-stimulated ISGylation of RIG-I inhibits activation of downstream target genes (18, 19). The multiplicity of negative feedback regulation indicates the presence of diverse mechanisms that collectively safeguard timely cessation of cytokine induction. SUMOylation of IRF3 and IRF7 may contribute to the overall feedback inhibition by introducing a distinct mechanism that acts at the level of IFN transcription (see below) (13, 14).
It is possible that SUMOylation of IRF3 and IRF7 occurs as a result of TLR/RIG-I-mediated activation of a SUMO E3 ligase (s). Conversely, this SUMOylation may be a result of inhibition of a SUMO-specific protease (s). Although signal-dependent activities of SUMO ligases or proteases have not been fully elucidated, it has been reported that PIAS1 is phosphorylated by TLR and other stress signals through the activation of IKKα (48). IKKα-mediated PIAS1 phosphorylation is apparently linked to its function, since it is required for transcriptional repression.
At present, however, the E3 ligase that mediates SUMOylation of IRF3 and IRF7 after virus stimulation has not been identified. Among the known SUMO E3 ligases, including the PIAS family proteins, RanBP2, and Pc2, PIAS proteins are shown to play a critical role in regulating innate immunity (46, 47, 50-52). They control IFN-responsive JAK/STAT pathways and NFκB-dependent transcription. In addition, IRF1, another member of the IRF family, is reported to be SUMOylated by PIAS3 (28). Thus, a ligase within the PIAS family may be a plausible candidate for the SUMOylation of IRF3 and IRF7 observed after viral infection.
SUMO-mediated regulation of IFN transcription may involve not only IRF3 and IRF7 but other transcription factors. For IFNβ, in addition to IRF3/IRF7, AP-1 and NFκB participate in transcription (53). c-Jun, components of the AP-1 transcription complex, are targets of modification by SUMO1-SUMO3, and this SUMOylation is associated with reduced transcription (54, 55). Furthermore, IκBα that binds to NFκB is also SUMOylated (56). SUMOylated IκBα blocks NFκB activation by preventing ubiquitination and degradation of IκBα. It may be of interest to test whether virus stimulation causes increased SUMOylation of c-Jun and IκBα along with that of IRF3 and IRF7.
Our study found that SUMOylation of IRF3 and IRF7 is an event independent of their phosphorylation. Similarly, IRF3 and IRF7 were phosphorylated irrespective of their capacity to be SUMOylated. These results support the view that SUMO modification of IRF3 and IRF7 is not restricted to the already activated IRF3 or IRF7 molecules, but it can occur in fresh, unactivated molecules as well. The broad availability of IRF3 and IRF7 for SUMOylation may suggest that this modification causes long lasting inhibition of IRF3 and IRF7 activity, which may help in reinforcing transcriptional repression required after activation. It is of note here that although phosphorylation is important for SUMOylation of some proteins, such as HSF1, HSF4b, and GATA-1 (57), the relationships between phosphorylation and SUMOylation are complex, and there are examples where these modifications antagonize with each other as reported for p53 and c-Fos (54, 58). It has been shown that ubiquitin and SUMO modifications have extensive cross-talk, and in some cases they are seen in a mutually exclusive manner (56). It has been shown that IRF7 is ubiquitinated through TRAF6 in response to TLR stimulation, and this modification is believed to be important for IFN transcription (23). At present, it is not clear whether SUMOylation of IRF7 regulates its ubiquitination or vice versa. On the other hand, it is reported that activated IRF3 interacts with PIN1 and is degraded by a protea-some-mediated mechanism, presumably by ubiquitination (49). Based on our observation that IRF3 is SUMOylated irrespective of its phosphorylation, IRF3 degradation does not seem to be directly linked to its SUMOylation.
Our finding that SUMOylation mutants increased expression of both IFNα and IFNβ following virus infection is consistent with a number of studies showing that SUMOylation of transcription factors results in repressed transcription (13, 14, 59). In some cases, SUMO-mediated transcriptional repression coincides with the recruitment of histone deacetylases (HDACs). It has been shown that SUMOylated peroxisome proliferator-activated receptor-γ recruits HDAC3 via co-repressor NCoR to repress NFκB target genes (60). Similarly, SUMOylated ELK1 binds to HDAC2 and reduces acetylation of chromatin, leading to repressed promoter activity (61). Further, the coactivator p300 is SUMOylated through the CRD1 domain and binds to HDAC6, correlating with transcriptional repression (62). Supporting a possible role of HDACs in SUMO-mediated repression, HDAC1 itself is modified by SUMO (63). By analogy with these examples, SUMO modification of IRF3 and IRF7 may recruit HDACs to IFN promoters in a timely manner, leading to changes in chromatin environment to cause stable transcriptional repression. Since the IFNβ promoter is shown to undergo increased histone acetylation upon viral stimulation (64), it is possible that SUMOylated IRF3 and IRF7 may help reverting an open chromatin configuration to a repressed chromatin by recruiting HDACs. SUMO-mediated transcriptional repression is also associated with changes in subnuclear localization of transcription factors, as reported for Sp3 (65) and SATB2 (66). SUMO-mediated factor relocalization and HDAC recruitment may not be mutually exclusive events, however. It would be of importance to study how SUMO-modified IRF3 and IRF7 change local chromatin architecture.
In conclusion, IRF3 and IRF7 are modified by SUMO following viral infection, and this modification restricts the extent and duration of virus-stimulated IFN transcription, thereby contributing to down-regulation of IFN production postactivation.
Supplementary Material
Acknowledgments
We thank Drs. Hideyoshi Yokosawa and Koji Nakagawa for the gift of T7-SUMO1-expressing plasmid and technical advice and Dr. George R. Stark for the gift of 2fTGH and mutant cells.
This work was supported, in whole or in part, by the National Institutes of Health Intramural Research Program, NICHD, and the Trans-National Institutes of Health Biodefense Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S4.
Footnotes
The abbreviations used are: TLR, Toll-like receptor; IRF, interferon regulatory factor; IFN, interferon; Ubl, ubiquitin-like protein; SUMO, small ubiquitin-related modifier; VSV, vesicular stomatitis virus; EMCV, encephalomyocarditis virus; MOI, multiplicity of infection; shRNA, short hairpin RNA; WT, wild type; JAK, Janus kinase; STAT, signal transducers and activators of transcription; HDAC, histone deacetylase.
References
- 1.Akira, S., Uematsu, S., and Takeuchi, O. (2006) Cell 124 783-801 [DOI] [PubMed] [Google Scholar]
- 2.Andrejeva, J., Childs, K. S., Young, D. F., Carlos, T. S., Stock, N., Good-bourn, S., and Randall, R. E. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 17264-17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kawai, T., and Akira, S. (2006) Nat. Immunol. 7 131-137 [DOI] [PubMed] [Google Scholar]
- 4.Takeda, K., and Akira, S. (2005) Int. Immunol. 17 1-14 [DOI] [PubMed] [Google Scholar]
- 5.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S., and Fujita, T. (2004) Nat. Immunol. 5 730-737 [DOI] [PubMed] [Google Scholar]
- 6.Durbin, J. E., Fernandez-Sesma, A., Lee, C. K., Rao, T. D., Fery, A. B., Moran, T. M., Vukmanovic, S., Garcia-Sastre, A., and Levy, D. E. (2000) J. Immunol. 164 4220-4228 [DOI] [PubMed] [Google Scholar]
- 7.Hengel, H., Koszinowski, U. H., and Conzelmann, K. K. (2005) Trends Immunol. 26 396-401 [DOI] [PubMed] [Google Scholar]
- 8.Levy, D. E., and Garcia-Sastre, A. (2001) Cytokine Growth Factor Rev. 12 143-156 [DOI] [PubMed] [Google Scholar]
- 9.Honda, K., and Taniguchi, T. (2006) Nat. Rev. Immunol. 6 644-658 [DOI] [PubMed] [Google Scholar]
- 10.Fitzgerald, K. A., McWhirter, S. M., Faia, K. L., Rowe, D. C., Katz, E., Golenbock, D. T., Cole, A. J., Liao, S. M., and Maniatis, T. (2003) Nat. Immunol. 4 491-496 [DOI] [PubMed] [Google Scholar]
- 11.Honda, K., Takaoka, A., and Taniguchi, T. (2006) Immunity 25 349-360 [DOI] [PubMed] [Google Scholar]
- 12.Gill, G. (2004) Gene Dev. 18 2046-2059 [DOI] [PubMed] [Google Scholar]
- 13.Gill, G. (2005) Curr. Opin. Genet. Dev. 15 536-541 [DOI] [PubMed] [Google Scholar]
- 14.Hay, R. T. (2005) Mol. Cell 18 1-12 [DOI] [PubMed] [Google Scholar]
- 15.Schwartz, D. C., and Hochstrasser, M. (2003) Trends Biochem. Sci. 28 321-328 [DOI] [PubMed] [Google Scholar]
- 16.Hodges, M., Tissot, C., and Freemont, P. S. (1998) Curr. Biol. 8 R749-R752 [DOI] [PubMed] [Google Scholar]
- 17.Huang, D. T., Walden, H., Duda, D., and Schulman, B. A. (2004) Oncogene 23 1958-1971 [DOI] [PubMed] [Google Scholar]
- 18.Arimoto, K., Takahashi, H., Hishiki, T., Konishi, H., Fujita, T., and Shimotohno, K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 7500-7505 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Arimoto, K., Konishi, H., and Shimotohno, K. (2008) Mol. Immunol. 45 1078-1084 [DOI] [PubMed] [Google Scholar]
- 20.Gack, M. U., Shin, Y. C., Joo, C. H., Urano, T., Liang, C., Sun, L., Takeuchi, O., Akira, S., Chen, Z., Inoue, S., and Jung, J. U. (2007) Nature 446 916-920 [DOI] [PubMed] [Google Scholar]
- 21.Kim, M. J., Hwang, S. Y., Imaizumi, T., and Yoo, J. Y. (2008) J. Virol. 82 1474-1483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhao, C., Denison, C., Huibregtse, J. M., Gygi, S., and Kreg, R. M. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 10200-10205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kawai, T., Sato, S., Ishii, K. J., Coban, C., Hemmi, H., Yamamoto, M., Terai, K., Matsuda, M., Inoue, J. I., Uematsu, S., Takeuchi, O., and Akira, S. (2004) Nat. Immunol. 5 1061-1068 [DOI] [PubMed] [Google Scholar]
- 24.Huye, L. E., Ning, S., Kelliher, M., and Pagano, J. S. (2007) Mol. Cell. Biol. 27 2910-2918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Geiss-Friedlander, R., and Melchior, F. (2007) Nat. Rev. Mol. Cell. Biol. 8 947-956 [DOI] [PubMed] [Google Scholar]
- 26.Rodriguez, M. S., Dargemont, C., and Hay, R. T. (2001) J. Biol. Chem. 276 12654-12659 [DOI] [PubMed] [Google Scholar]
- 27.Ozato, K., Tailor, P., and Kubota, T. (2007) J. Biol. Chem. 282 20065-20069 [DOI] [PubMed] [Google Scholar]
- 28.Nakagawa, K., and Yokosawa, H. (2002) FEBS Lett. 530 204-208 [DOI] [PubMed] [Google Scholar]
- 29.Qi, B., Huang, Y., Rowe, D., and Halliday, G. (2007) Int. J. Biochem. Cell Biol. 39 287-291 [DOI] [PubMed] [Google Scholar]
- 30.Kawai, T., Sato, S., Coban, C., Kumar, H., Kato, H., Ishii, K. J., Takeuchi, O., and Akira, S. (2005) Nat. Immunol. 6 981-988 [DOI] [PubMed] [Google Scholar]
- 31.Meylan, E., Curran, J., Hofmann, K., Moradpour, D., Binder, M., Barten-schlager, R., and Tschopp, J. (2005) Nature 437 1167-1172 [DOI] [PubMed] [Google Scholar]
- 32.Seth, R. B., Sun, L., Ea, C. K., and Chen, Z. J. (2005) Cell 122 669-682 [DOI] [PubMed] [Google Scholar]
- 33.Xu, L. G., Wang, Y. Y., Han, K. J., Li, L. Y., Zhai, Z., and Shu, H. B. (2005) Mol. Cell 19 727-740 [DOI] [PubMed] [Google Scholar]
- 34.Oshiumi, H., Matsumoto, M., Funami, K., Akazawa, T., and Seya, T. (2003) Nat. Immunol. 4 161-167 [DOI] [PubMed] [Google Scholar]
- 35.Yamamoto, M., Sato, S., Mori, K., Hoshino, K., Takeuchi, O., Takeda, K., and Akira, S. (2002) J. Immunol. 169 6668-6672 [DOI] [PubMed] [Google Scholar]
- 36.Kinoshita, E., Kinoshita-Kikuta, E., Takiyama, K., and Koike, T. (2005) Mol. Cell. Proteomics 5 749-757 [DOI] [PubMed] [Google Scholar]
- 37.Caillaud, A., Hovanessian, A. G., Levy, D. E., and Marie, I. J. (2005) J. Biol. Chem. 280 17671-17677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lin, R., Heylbroeck, C., Pitha, P. M., and Hiscott, J. (1998) Mol. Cell. Biol. 18 2986-2996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mori, M., Yoneyama, M., Ito, T., Takahashi, K., Inagaki, F., and Fujita, T. (2004) J. Biol. Chem. 279 9698-9702 [DOI] [PubMed] [Google Scholar]
- 40.Servant, M., Grandvaux, N., tenOever, B. R., Duguay, D., Lin, R., and Hiscott, J. (2003) J. Biol. Chem. 278 9441-9447 [DOI] [PubMed] [Google Scholar]
- 41.Darnell, J. E., Jr., Kerr, I. M., and Stark, G. R. (1994) Science 264 1415-1421 [DOI] [PubMed] [Google Scholar]
- 42.Leung, S., Oureshi, S. A., Kerr, I. M., Darnell, J. E., Jr., and Stark, G. R. (1995) Mol. Cell. Biol. 15 1312-1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muller, M., Lacton, C., Briscoe, J., Schindler, C., Importa, T., Darnell, J. E., Jr., Stark, G. R., and Kerr, I. M. (1993) EMBO J. 12 4221-4228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cui, S., Eisenacher, K., Kirchhofer, A., Brzozka, K., Lammens, A., Lammens, K., Fujita, T., Conzelmann, K. K., Krug, A., and Hopfner, K. P. (2008) Mol. Cell 29 169-179 [DOI] [PubMed] [Google Scholar]
- 45.O'Shea, J. J., and Murray, P. J. (2008) Immunity 28 477-487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lui, B., Mink, S., Wong, K. A., Stein, N., Getman, C., Dempsey, P. W., Wu, H., and Shuai, K. (2004) Nat. Immunol. 5 891-898 [DOI] [PubMed] [Google Scholar]
- 47.Lui, B., Yang, R., Wong, K. A., Getman, C., Stein, N., Teitell, M. A., Cheng, G., Wu, H., and Shuai, K. (2005) Mol. Cell. Biol. 25 1113-1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lui, B., Yang, Y., Chenishof, V., Loo, R. R. O., Jang, H., Tahk, S., Yang, R., Mink, S., Shultz, D., Bellon, C. J., Loo, J. A., and Shuai, K. (2007) Cell 129 903-914 [DOI] [PubMed] [Google Scholar]
- 49.Saitoh, T., Tun-Kyi, A., Ryo, A., Yamamoto, M., Finn, G., Fujita, T., Akira, S., Yamamoto, N., and Yamaoka, S. (2006) Nat. Immunol. 7 598-605 [DOI] [PubMed] [Google Scholar]
- 50.Melchior, F., Schergaut, F., and Pichler, A. (2003) Trends Biochem. Sci. 28 612-618 [DOI] [PubMed] [Google Scholar]
- 51.Roth, W., Sustmann, C., Kieslinger, M., Gilmozzi, A., Irmer, D., Kermmer, E., Turck, C., and Grosschedl, R. (2004) J. Immunol. 173 6189-6199 [DOI] [PubMed] [Google Scholar]
- 52.Tahk, S., Lui, B., Chernishof, V., Wong, K. A., Wu, H., and Shuai, K. (2007) Proc. Natl. Acad. Sci. U. S. A. 104 11643-11648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Thanos, D., and Maniatis, T. (1995) Cell 83 1091-1100 [DOI] [PubMed] [Google Scholar]
- 54.Bossis, G., Malnou, C. E., Farras, R., Andermarcher, E., Hipskind, R., Rodriguez, M., Schmidt, D., Muller, S., Jariel-Encontre, I., and Piechaczyk, M. (2005) Mol. Cell. Biol. 25 6964-6979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Muller, S., Berger, M., Lehembre, F., Seeler, J. S., Haupt, Y., and Dejean, A. (2000) J. Biol. Chem. 275 13321-13329 [DOI] [PubMed] [Google Scholar]
- 56.Desterro, J. M., Rodriguez, M. S., and Hay, R. T. (1998) Mol. Cell 2 233-239 [DOI] [PubMed] [Google Scholar]
- 57.Hietakangas, V., Anckar, J., Blomster, H. A., Fujimoto, M., Palvimo, J. J., Nakai, A., and Sistonen, L. (2006) Proc. Natl. Acad. Sci. U. S. A. 103 45-50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin, J. Y., Ohshima, T., and Shimotohno, K. (2004) FEBS Lett. 573 15-18 [DOI] [PubMed] [Google Scholar]
- 59.Verger, A., Perdomo, J., and Crossley, M. (2003) EMBO Rep. 4 137-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pascual, G., Fong, A. L., Ogawa, S., Gamliel, A., Li, A. C., Perissi, V., Rose, D. W., Willson, T. M., Rosenfeld, M. G., and Glass, C. K. (2005) Nature 437 759-763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang, S. H., and Sharrocks, A. D. (2004) Mol. Cell 13 611-617 [DOI] [PubMed] [Google Scholar]
- 62.Girdwood, D., Bumpass, D., Baughan, O. A., Thain, A., Anderson, L. A., Snowden, A. W., Garcia-Wilson, E., Perkins, N. D., and Hay, R. T. (2003) Mol. Cell 11 1043-1054 [DOI] [PubMed] [Google Scholar]
- 63.David, G., Neptune, M. A., and DePinho, R. A. (2002) J. Biol. Chem. 277 23658-23663 [DOI] [PubMed] [Google Scholar]
- 64.Agalioti, T., Lomvardas, S., Parekh, B., Yie, J., Maniatis, T., and Thanos, D. (2000) Cell 103 667-678 [DOI] [PubMed] [Google Scholar]
- 65.Ross, S., Best, J. L., Zon, L. I., and Gill, G. (2002) Mol. Cell 10 831-842 [DOI] [PubMed] [Google Scholar]
- 66.Dobreva, G., Dambacher, J., and Grosschedl, R. (2003) Genes Dev. 17 3048-3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.