Abstract
We previously reported that shear stress induces phosphorylation and disassembly of keratin intermediate filaments (IFs). Shear stress also induces a time- and strain-dependent degradation of keratin IFs, and the current study examines the mechanisms involved in degradation of keratin proteins in human A549 cells exposed to 0–24 h of shear stress (7.5–30 dynes/cm2). Ubiquitin was found to be covalently associated with keratin proteins immunoprecipitated from shear-stressed cells, and pretreatment with the proteasomal inhibitor MG132 prevented the degradation of the keratin IF network. Importantly, phosphorylation of K8 Ser-73 is required for the shear stress-mediated ubiquitination, disassembly, and degradation of the keratin IF network. Immunofluorescence microscopy revealed that shear stress caused the thin array of keratin fibrils observed in control cells to be reorganized into a perinuclear aggregate, known as an aggresome, and that ubiquitin was also associated with this structure. Finally, the E2 enzymes, UbcH5b, -c, and Ubc3, but not E2-25K are required for the shear stress-mediated ubiquitin-proteasomal degradation of keratin proteins. These data suggest that shear stress promotes the disassembly and degradation of the keratin IF network via phosphorylation and the ubiquitin-proteasome pathway.
Mechanical stimuli are important modulators of cellular function; this is especially true in the mechanically ventilated lungs of patients with acute lung injury. In these patients, shear stress is created by the cyclic opening and closing of the edematous, surfactant-depleted, collapsed alveoli. It has been previously demonstrated that physical forces, such as shear stress, induce rapid and global changes in the patterns of intermediate filaments (IFs)2 (1–4).
Keratin IFs are the major cytoskeletal component of epithelial cells and are assembled as obligate heteropolymers of type I (K9-K20) and type II (K1-K8) IF proteins (5–7). Lung alveolar epithelial cells (AEC) express primarily K8 and K18 with variable levels of K7 and K19 (1, 8, 9). The prototype structure of all IF proteins, including keratins, consists of a structurally conserved central coiled-coil α-helix termed the “rod” that is flanked by non-α-helical N-terminal “head” and C-terminal “tail” domains (6). Most of the structural heterogeneity of the different keratins resides in their head and tail domains, which contain the sites for several post-translational modifications including phosphorylation and ubiquitination.
The ubiquitin-proteasome pathway (UPP) (10) is involved in regulating the cell cycle, signal transduction, differentiation, and stress response. Most of these functions are mediated by the conditional turnover of regulatory and structural proteins (10–12). The process begins with activation of ubiquitin by the ubiquitin-activating enzyme (E1) (10), followed by transfer of ubiquitin to E2, an ubiquitin-conjugating enzyme. E2 shuttles the ubiquitin molecule to the substrate-specific ubiquitin ligase (E3), which then delivers the ubiquitin to the substrate to be degraded. Polyubiquitin chain formation continues by the conjugation of subsequent ubiquitin moieties to the attached ubiquitin, and the substrate-ubiquitin conjugate is then degraded by the 26 S proteasome in an ATP-dependent manner. Isopeptidases cleave the ubiquitin chain, and the single ubiquitin molecules are recycled (10).
We examined the assembly state of keratin IFs in human A549 cells and found that shear stress results in the disassembly and degradation of keratin proteins via the ubiquitin-proteasome pathway. Inhibition of the proteasome prevents the degradation of keratin proteins in shear-stressed cells. Interestingly, recognition of the keratin protein for ubiquitination, and subsequent degradation, appears to be dependent upon the phosphorylation of K8 Ser-73 via protein kinase C (1). Thus, phosphorylated keratin IF proteins are targeted for degradation via the ubiquitin-proteasome pathway in A549 cells in response to a mechanical stimulus.
EXPERIMENTAL PROCEDURES
Culture of AEC—Human alveolar epithelial (A549) cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 2 mm l-glutamine, 50 μg/ml gentamicin, 100 units/ml penicillin, 100 μg/ml streptomycin. Cells were incubated in a humidified atmosphere of 5% CO2/95% air at 37 °C. An in vitro model for shear stress was used to examine the effect of physical forces on keratin IFs in A549 cells, as previously described (1, 13). A549 cells were cultured on 25 × 75 × 1 mm glass slides and subjected to continuous laminar flow to generate shear stress (7.5, 15, and 30 dynes/cm2) using the Flexcell Streamer Device.
Immunofluorescence—A549 cells grown on glass slides were rinsed 3× in PBS and fixed in either methanol (-20 °C) for 4 min or 3.5% formaldehyde at room temperature for 7 min. Following formaldehyde fixation, cells were permeabilized with 0.05% Tween 20 for 5 min, washed 3× with PBS, and processed for indirect immunofluorescence as previously described (13, 14). Following staining, the glass slides were washed in PBS and mounted in gelvatol containing 100 mg/ml Dabco (1,4-diazabicyclo [2.2.2] octane; Aldrich Chemical (14)). Images of fixed, stained preparations were taken with a Zeiss LSM 510 microscope (Carl Zeiss) (14).
Protein Isolation, Immunoblotting, and Immunoprecipitation Analysis—Total cell lysates were obtained by solubilizing A549 cells in Laemmli sample buffer. Keratin-enriched cytoskeletal preparations were made according to previously published procedures (1). In brief, Triton X-100 (TX-100) soluble or TX-100 insoluble fractions were prepared by treating cells for 10 min at 4 °C with buffer containing 1% TX-100, 5 mm EDTA, and a protease inhibitor mix (1 mm phenylmethylsulfonyl fluoride, 10 μm leupeptin, 10 μm pepstatin, and 25 μg/ml aprotinin) in PBS, pH 7.4, followed by centrifugation (16,000 × g, 10 min). The supernatant was collected as the soluble fraction. The pellet was homogenized in 1 ml of 10 mm Tris-HCl, pH 7.6, 140 mm NaCl, 1.5 m KCl, 5 mm EDTA, 0.5% Triton X-100, and the protease inhibitor mix. After 30 min (4 °C), this homogenate was centrifuged (16,000 × g; 10 min), and the pellet (insoluble fraction) was rehomogenized with 5 mm EDTA in PBS, pH 7.4, and centrifuged again. The resulting Triton X-100 insoluble fraction was dissolved in Laemmli sample buffer containing 1% β-mercaptoethanol, sonicated, and boiled for 5 min (15). Proteins were separated by 10% SDS-PAGE, transferred to nitrocellulose membranes, and blotted with either anti-K8/18 monoclonal antibody (Research Diagnostics, Inc.; 1:500 in TBS) or anti-ubiquitin (Sigma, 1:100 in TBS). Membranes were washed three times with TBS containing 0.1% of Tween for 30 min, then incubated with secondary antibodies coupled to horseradish peroxidase (in dilutions recommended by supplier), and visualized using enhanced chemiluminescence (Amersham Biosciences).
For immunoprecipitation studies, cells were solubilized in immunoprecipitation buffer (50 mm Tris-HCl, pH 8.0, 150 mm NaCl, 1% Triton X-100, 5 mm EDTA, 1 mm phenylmethylsulfonyl fluoride) and 1% SDS was added lysates or fractions. After dilution with IP buffer to a final SDS concentration of 0.1%, the samples were processed for immunoprecipitation as described by Ridge et al. (1), using an anti-K8 antibody coupled to protein A/G-Sepharose. The beads were washed once with radioimmune precipitation assay buffer and twice with PBS (3 mm EDTA). Proteins were solubilized in 3× Laemmli sample buffer and then prepared for immunoblotting as described above.
Pulse Chase and Immunoprecipitation Protocol—A459 cells cultured on glass slides at 60–70% confluence were incubated for 2 h in l-methionine/l-cysteine-free medium (Invitrogen) at 37 °C with 5% CO2. Subsequently, cells were labeled for 30 min with a mixture of [35S]methionine/cysteine (200 μCi/ml; EaseTag™ mix, PerkinElmer Life Sciences) and then exposed to shear stress (30 dynes/cm2) in the presence of pre-warmed Dulbecco's modified Eagle's medium supplemented with 5 mm unlabeled l-methionine/l-cysteine. At the indicated times, the cells were washed twice with cold phosphate-buffered saline, collected, and then an equal number of cells were resuspended in identical volumes of TX-100 containing buffer and centrifuged at low speeds to obtain soluble and insoluble fractions. Keratin 8/18 was immunoprecipitated from both fractions as described above in the presence of serine protease inhibitors (Roche Applied Science). And then pre-absorbed for 1 h with protein-A/G plus and incubated with anti-K8 and K18 antibodies overnight at 20 °C followed by a 3-h incubation with protein-A/G-agarose beads (Santa Cruz Biotechnology). Next, the agarose beads were washed five times with modified lysis buffer containing 0.1% digitonin and then mixed with a modified 2× loading sample buffer (2.5% SDS, 0.1 m dithiothreitol, 0.06 m Tris base, 20% glycerol, 0.008 m EDTA, pH 6.8). Proteins were separated on 7.5% or 10% SDS-polyacrylamide gels. Gels were stained, fixed in 50% methanol, 10% acetic acid, then treated with Fluorenhance (Amersham Biosciences) for 30 min, dried, and exposed to either x-ray film (Hyperfilm; (Eastman Kodak Co.) at 80 °C for 16–24 h or put on an intensifying screen for quantification with a PhosphorImager (Amersham Biosciences). Samples were also taken for scintillation counting.
Transient Transfections of Dominant Negative (DN) Constructs—A549 cells were transiently transfected with ∼2.5 μg of a cDNA coding the dominant-negative UbcH5a, -b, -c, Ubc3, and E2–25K (DN-UbH5a, DN-UbH5b, DN-UbH5c, DN-Ubc3, or DN-E2–25K, respectively). All constructs were tagged with His. The Amaxa Nucleofector apparatus (Amaxa, Cologne, Germany) was used for transfection. 2.5 μg of plasmid DNA was mixed with 0.1 ml of cell suspension, transferred to a 2.0 mm electroporation cuvette and nucleofected. DNA quantity, cell concentration, and buffer volume were kept constant for all experiments. After electroporation, cells were immediately transferred to 2.0 ml of complete medium, and cultured at 37 °C until analysis. Viability of cells immediately after transfection was determined by trypan blue exclusion.
Miscellaneous—Lactate dehydrogenase release was measured using a commercially available assay (Cytotoxicity Detection kit, Roche Pharmaceuticals, Indianapolis, IN). Protein content was determined according to Bradford using a commercial dye reagent (Bio-Rad).
Statistical Analysis—Comparisons were performed using the unpaired Student's t test. One-way analysis of variance with Tukey's test was used to analyze the data. p < 0.05 values were considered significant.
RESULTS
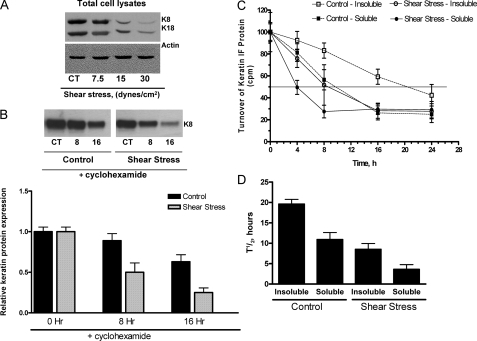
Effect of Shear Stress on Keratin Protein Turnover—A549 cells exposed to shear stress (7.5, 15, and 30 dynes/cm2) for 16 h exhibit a strain-dependent decrease in K8 and K18 protein levels (Fig. 1A). This decrease in protein abundance may be a reflection of increased degradation or changes in the rate of transcription/translation of the protein. To test for increased degradation, we examined the effect of shear stress on the stability of keratin proteins in the absence of new protein synthesis by adding cycloheximide (CHX). A549 cells were exposed to shear stress (30 dynes/cm2) for 0, 8, and 16 h in the presence CHX (μm); at the end of each time point total cell lysates were prepared. Under control conditions, keratin protein levels were reduced by ∼10 and 40% after 8 and 16 h, respectively. In contrast, A549 cells exposed to shear stress had a ∼50 and 75% decrease in keratin protein levels after 8 and 16 h, respectively (Fig. 1B) indicating that keratin proteins were degraded following exposure to prolonged shear stress. To determine whether the rates of degradation were different for soluble or insoluble (particles or filaments) keratin proteins, pulse chase experiments were performed. A549 cells were labeled for 30 min with [35S]methionine and chased with medium containing a 10-fold excess of unlabeled methionine for 0, 4, 8, 16, 24 h. A549 cells were simultaneously exposed to shear stress (30 dynes/cm2) during the chase. Proteins were separated into TX-100 soluble and insoluble fractions followed by immunoprecipitation of keratin using an anti-keratin 8 antibody. As shown in Fig. 1C, under control conditions the TX-100 insoluble, filamentous keratin protein (open squares, dashed line) has a half-life of ∼20 h, which is in agreement with previous reports. The TX-100 soluble, non-filamentous keratin protein has a half-life of ∼10 h (Fig. 1C, closed square, dashed line). Shear stress significantly alters the half-life of both insoluble and soluble keratin proteins, 8 and 4 h, respectively, as compared with control (Fig. 1C, open and closed circles, solid lines).
FIGURE 1.
Effects of mechanical strain on keratin IF proteins. A, total cell lysates were prepared from A549 cells exposed to shear stress (7.5, 15, and 30 dynes/cm2) for 24 h. Equal amounts of protein were separated by 7.5% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-K8 and -K18 antibodies. A representative immunoblot shows a strain-dependent decrease in both K8 and K18 protein abundance as compared with static control conditions (n = 4). B, A549 cells were pretreated with cycloheximide (50 μm) and then exposed to 30 dynes/cm2 for 8 and 16 h and compared with static control cells. Total cell lysates were prepared, and equal amounts of protein were separated by 10% SDS-PAGE, transferred to nitrocellulose and immunoblotted with anti-K8 antibodies. A representative immunoblot shows that a time-dependent decrease in keratin protein abundance (n = 3). C and D, rate of newly synthesized keratin in control and shear-stressed A549 cells. Control and shear-stressed A549 cells were metabolically labeled using [35S]methionine and chased for indicated time periods in medium containing cold methionine. Keratin proteins were immunoprecipitated from Triton X-100 soluble and insoluble fractions, analyzed by SDS-PAGE, and the radioactive label incorporated per μg of keratin was determined by scintillation counting and densitometric scanning. C, decrease in keratin-specific label shown in half-logarithmic scale; D, calculated half-life keratin. Bars represent mean ± S.D. (n = 3).
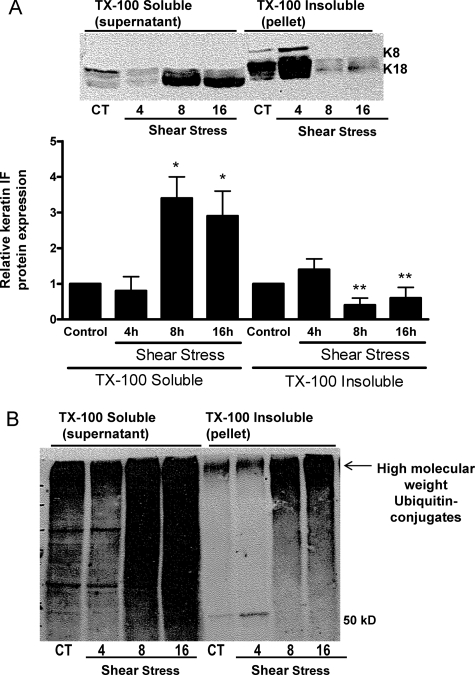
A549 cells exposed to shear stress (30 dynes/cm2) for 0, 4, 8, and 16 h exhibited a time-dependent decrease in the amount of TX-100 insoluble (i.e. pelletable, filamentous) K8 and K18; which coincided with significant increases in the TX-100 soluble (i.e. non-filamentous, disassembled) keratin 8 and 18 protein levels detected in the supernatant (Fig. 2A; see “Experimental Procedures”). These results are in agreement with our previously published report (1). As shown in Fig. 2B, when the membrane was re-blotted with an anti-ubiquitin antibody, a high molecular weight smear, representing ubiquitinated keratin, was observed in TX-100 insoluble fraction (filamentous protein) extracted from A549 cells exposed to shear stress for either 8 or 16 h (Fig. 2B). However, the strongest ubiquitin signal was associated with the TX-100 soluble fraction (non-filamentous protein) extracted from shear-stressed A549 cells. The ubiquitinated conjugates migrated primarily as a high molecular weight smear, shown by the arrow, although lower molecular weight bands were also detected in TX-100 soluble fraction (Fig. 2B).
FIGURE 2.
Effects of mechanical strain on keratin IF solubility. AEC were exposed to shear stress (30 dynes/cm2) for 4, 8, and 16 h and compared with static control cells. Keratin-enriched cytoskeletal preparations were separated into Triton X-100 soluble and insoluble fractions. Equal amounts of protein were separated by 10% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-K8 and -K18, anti-ubiquitin antibodies. A, representative immunoblot shows an increased solubility of both K8 and K18 upon exposure to shear stress (n = 4). B, representative immunoblot shows formation of high molecular weight ubiquitin conjugates in lysates prepared from shear-stressed A549 cells.
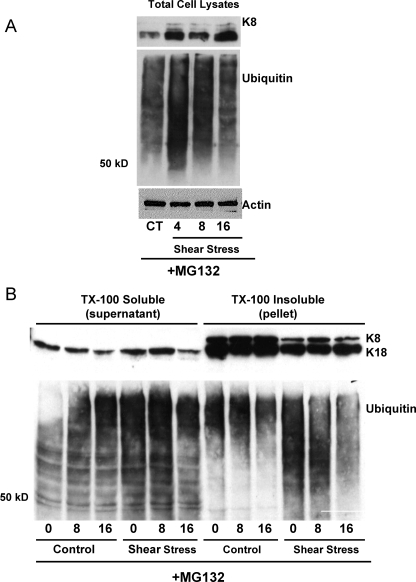
To determine if keratin IF proteins were degraded via the UPP, A549 cells were treated with proteasome inhibitor MG132 (50 μm) and subsequently exposed to shear stress. Total cell lysates (Fig. 3A) and keratin proteins fractionated in TX-100 soluble and insoluble fractions (Fig. 3B), separated on SDS-PAGE, and immunoblotted with antibodies against K8/K18 and ubiquitin were analyzed. The shear stress-induced degradation of keratin 8 and 18 was prevented in A549 cells incubated with MG132.
FIGURE 3.
Effects of MG132 on keratin IF solubility in response to shear stress. A, total cell lysates were prepared from A549 cells exposed to shear stress (30 dynes/cm2) for 0, 4, 8, and 16 h in the presence of MG132, 50 μm. Equal amounts of protein were separated by 7.5% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-K8 and -ubiquitin antibodies. A representative immunoblot shows that MG132 prevented the shear stress-induced decrease in keratin protein abundance (n = 3). B, keratin-enriched cytoskeletal preparations were separated into Triton X-100 soluble and insoluble fractions. Equal amounts of protein were separated by 10% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-K8 and -K18, and anti-ubiquitin antibodies. A representative immunoblot shows that MG132 prevents the shear stress-induced disassembly and degradation of K8 and K18 (n = 4) and enabled the formation of high molecular weight ubiquitin conjugates in lysates prepared from both control and shear-stressed A549 cells.
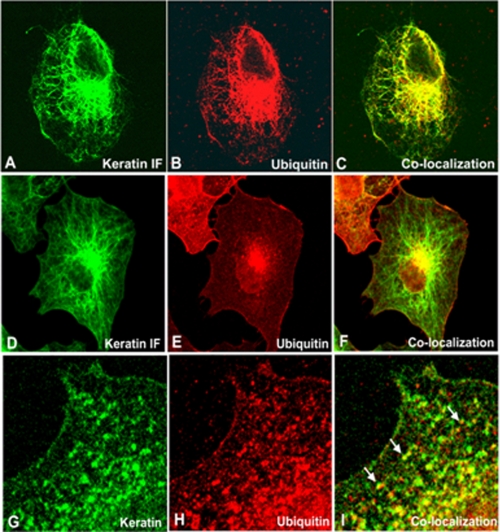
Effect of Mechanical Stimuli on the Keratin IF Network in A549 Cells—We previously observed that shear stress induces changes in the keratin IF network (1). To determine whether ubiquitin is associated with keratin proteins, A549 cells exposed to shear stress were fixed and processed for indirect immunofluorescence using anti-K8/K18 and anti-ubiquitin antibodies. In cells exposed to shear stress for 16 h, ubiquitin was associated with a filamentous staining pattern which co-localized with keratin IFs (Fig. 4, panels A–C). Between 16 and 24 h, we observed the formation of Mallory bodies/aggresomes and both keratin and ubiquitin proteins were co-localized in this perinuclear structure (Fig. 4, panels D–F). By 36 h, the keratin IF network was extensively disassembled and the non-filamentous keratin particles or “dots” co-localized with ubiquitin (Fig. 4, panels G–I). Keratin and ubiquitin were not associated in control cells (data not shown).
FIGURE 4.
Effects of shear stress on keratin IF network. Cells were exposed to shear stress (30 dynes/cm2), fixed, and processed for indirect immunofluorescence using anti-K8/K18 and anti-ubiquitin. In cells exposed to continuous shear stress for 16 h, ubiquitin staining revealed a filamentous pattern that co-localized with keratin IFs (panels A–C). At ∼24 h, we observed the formation of aggresomes/Mallory bodies. Both keratin and ubiquitin proteins were co-localized in this perinuclear structure (panels D–F). By ∼36 h, the keratin IF network was extensively disassembled and the non-filamentous keratin particles or “dots” co-localized with ubiquitin (panels G–I).
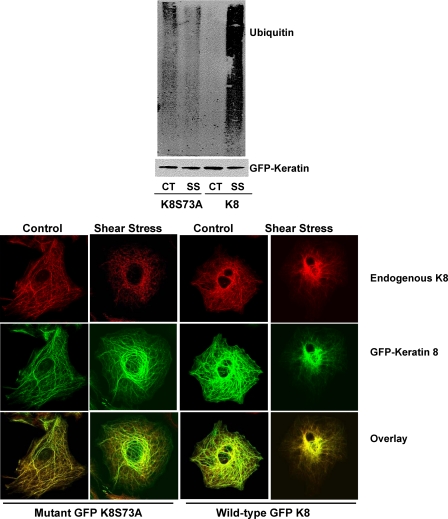
Phosphorylation of K8 Ser-73 Acts as a Signal Recognition for Ubiquitin Conjugation—Previously, we reported that the phosphorylation of K8 pSer-73, but not K8 pSer-23 or K8 pSer-43, was associated with the shear stress-mediated disassembly of the keratin IF network (1). To determine whether the phosphorylation of K8 Ser-73 was required for substrate recognition by ubiquitin A549 cells were stably transfected GFP-K8 or GFP-K8 S73A constructs. These cells were exposed to shear stress (30 dynes/cm2, 16 h), the GFP-tagged K8 was immunoprecipitated, proteins were separated by SDS-PAGE, transferred, and immunoblotted with anti-K8 and anti-ubiquitin antibodies. As shown in Fig. 5 (upper panel), ubiquitin was not covalently associated with GFP-tagged K8S73A in either control or shear-stressed cells. As anticipated, the wild-type GFP-tagged K8 was covalently associated with ubiquitin in cells exposed to shear stress.
FIGURE 5.
A549 cells were transiently transfected with GFP-tagged K8 (wild-type) or GFP-tagged K8 S73A mutant. Upper panel, wild-type and mutant cells were exposed to shear stress (30 dynes/cm2; 16 h) GFP was immunoprecipitated by dilution of whole cell extracts with radioimmune precipitation assay buffer using a rabbit polyclonal anti-GFP antibody and recovered with protein A/G-Sepharose. Proteins were separated on 10% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with an anti-keratin 8 antibody and an anti-ubiquitin antibody. A representative immunoblot shows that the K8S73A mutant did not covalently attach to ubiquitin following exposure to shear stress, but that the wild-type keratin was covalently associated with ubiquitin following exposure to shear stress (n = 4). Lower panel, cells were exposed to shear stress (30 dynes/cm2), fixed, and processed for indirect immunofluorescence using anti-K8/K18 and direct immunofluorescence for GFP. In GFP-K8S73A and GFP-K8 cells exposed to shear stress for ∼16 h, the endogenous keratin IF network (red channel) was disassembled; In the GFP-K8S73A cells, the exogenously expressed GFP-keratin IF network was not disassembled/degraded following exposure to shear stress, but in the GFP-K8 cells, the exogenously expressed keratin IF network was disassembled.
In Fig. 5 (lower panel), we examined whether expression of the mutant GFP-tagged K8S73A affected the keratin IF network in either control or shear stress cells as compared with GFP-tagged K8. Stably transfected A549 cells exposed to shear stress were fixed and processed for indirect immunofluorescence using anti-K8 and direct immunofluorescence for GFP. In GFP-K8S73A cells exposed to shear stress for ∼16 h, the endogenous keratin IF network (red channel) was disassembled; in contrast the GFP-K8S73A IF network was not affected following exposure to shear stress. In the GFP-K8 cells exposed to shear stress both the endogenous keratin (red channel) and GFP-K8 (green channel) IF networks were disassembled, as compared with static control cells.
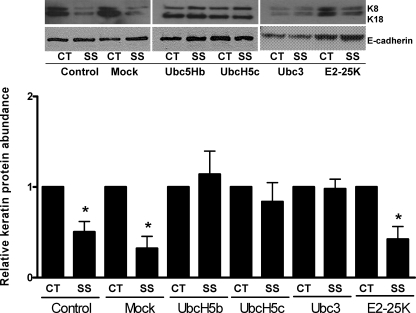
Ubiquitin Carrier Proteins Involved in Shear Stress-mediated Degradation of Keratin IF Proteins—To study the role of UbcH5a, -b, -c, Ubc3, and E2–25K in the degradation of keratin IF proteins, we tested whether DN constructs of the enzymes had an effect on the shear stressed-induced degradation of keratin IF proteins in A549 cells. As shown in Fig. 6, transient expression of DN Ubc3, UbcH5b, UbcH5c, and to a lesser extent UbcH5a (data not shown), significantly inhibit the shear stress-mediated degradation of keratin IF proteins. In A549 cells transiently transfected with the DN E2–25K or mock-transfected, the keratin IF proteins are degraded following exposure to shear stress. Transfection efficiency was assessed by the expression of His protein (data not shown). Cells transfected with the dominant negative ligase constructs had no changes in cell death, as measured by lactate dehydrogenase levels (data not shown), as compared with non-transfected control cells.
FIGURE 6.
A549 cells were transiently transfected with empty vector (mock) or dominant negative constructs expressing UbcH5b, -c, Ubc3, or E2–25K. Twenty-four hours following transfection, cells were exposed to shear stress (30dynes/cm2, 16 h). Total cell lysates were prepared, and equal amounts of protein were separated by 7.5% SDS-PAGE, transferred to nitrocellulose, and immunoblotted with anti-K8 and -E-cadherin antibodies. A representative immunoblot shows that UbcH5b, -c, and Ubc3, but not E2–25K, prevents the shear stress-induced degradation of K8 and K18. Bars represent mean ± S.D. (n = 4).
DISCUSSION
The physiological significance of keratin IF networks becomes evident in disease conditions, such as acute respiratory distress syndrome (ARDS), wherein the loss of the mechanical integrity of the lung epithelium results in flooding of lung airspaces with plasma, impaired gas exchange, and consequent patient mortality. Mechanical ventilation is often required to manage patients with acute respiratory failure of different origins. However, mechanical ventilation itself can also cause or exacerbate lung injury, resulting in ventilator induced lung injury (VILI) (16, 17). The mechanisms associated with VILI have not been fully elucidated but it is believed that the opening of a fluid-filled, collapsed airway generates a shear force across the epithelium (18). These harmful mechanoforces within the lung are borne by the lung epithelial cells (1, 13). The shear stress gradient generated under these conditions has been estimated to be between 5 and 325 dynes/cm2 (18). Hence in this study we used a shear stress of 7.5–30 dyn/cm2 to examine mechanisms involved in the degradation of the keratin IF network to approximate the in vivo pathophysiologic conditions. It is important to note that the shear stresses used in our study are much greater than in vivo normal physiological levels (no shear stress for normal alveolar epithelial cells and 5–7 dyn/cm2 on endothelial cells).
We have previously reported that shear stress alters the assembly state of the keratin IF network in alveolar epithelial cells via a PKC-dependent phosphorylation of K8 pSer 73 (1). Changes in the assembly state of IF networks may have a significant impact on signaling networks that regulate responses to changes in the cellular environment. Specifically, the assembly state of IFs is important in the determination and maintenance of cell shape, transmission of mechanical stress, and targeting of molecules between the nucleus and cytoplasm (19, 20). Traditionally, the IF network has been viewed as a static architecture, based on the low solubility and detergent extractability of IF proteins. Several reports have now demonstrated that IFs are highly dynamic cellular structures as reflected by their assembly/disassembly states throughout the cell cycle (4, 21, 22). While details regarding the mechanisms regulating the assembly dynamics of keratin IFs are not known, there are a number of reports that demonstrate the significance of phosphorylation and ubiquitination in regulating the assembly dynamics of keratin IFs (4, 21–24).
Our studies demonstrate that shear stress effects the assembly state of the keratin IF network (Figs. 2 and 4). As shown in Fig. 2, A549 cells exposed to shear stress for as little as 8 h had an increase in keratin protein levels in the soluble fraction, indicating that shear stress promotes the disassembly of the IF network. The rate of keratin IF network disassembly and degradation is dependent upon the degree of shear stress and duration (1, 13); increased levels of shear stress accelerate the rate of disassembly and degradation of keratin proteins (Fig. 1A). Further, it appears that soluble keratin protein is degraded at an accelerated rate as compared with insoluble, filamentous keratin protein (Fig. 1C). Interestingly, the TX-100 soluble fraction had a stronger keratin ubiquitin-conjugation signal, as compared with the TX-100 insoluble fraction (Fig. 2B). These results suggest that soluble keratin may be more efficiently degraded via the UPP, as compared with the insoluble, filamentous keratin protein. To eliminate the possibility that keratin proteins were not themselves ubiquitinated but were bound to an ubiquitinated protein, we lysed cells in buffer containing 1% SDS and boiled before immunoprecipitation (Fig. 5). After these strong denaturing conditions, ubiquitin-labeled protein was still found in keratin immunoprecipitates, suggesting that ubiquitin is covalently attached to keratin proteins. We also observed by immunofluorescent microscopy that the keratin IF network was co-localized with ubiquitin several hours following exposure to shear stress. Ubiquitin exhibited a filamentous staining pattern that co-localized with keratin IFs; this pattern of staining was observed in A549 cells 8–16 h after exposure to shear stress (Fig. 4, panels A–C). We also observed the formation of aggresomes or Mallory bodies in A549 cells exposed to shear stress; these are perinuclear structures that contain both keratin and ubiquitin (Fig. 4, panels D–F). In fact, a common pathological feature of many IF-related diseases is the accumulation of intracytoplasmic inclusion bodies or aggresomes (25) consisting of modified IF proteins (26, 27). Examples include the neurodegenerative diseases such as, amyotrophic lateral sclerosis and Parkinson disease (28), as well as alcoholic hepatitis and other liver disorders that result in the formation of Mallory bodies (MBs) (29–31). These perinuclear structures consist predominantly of K8 and variable amounts of K18 assembled in a nonfilamentous manner (32). In addition to keratin, non-keratin components such as ubiquitin have also been identified as constituents of MBs (28, 33–35). In patients with interstitial pneumonitis, MB-like structures were observed (36–39). The pathogenetic mechanisms responsible for the formation of these structures are as yet unclear. Post-translational modifications of keratin IF proteins, such as phosphorylation (40–43), ubiquitination (44, 45), and cross-linking (46), may play major roles in MB formation. These structures may form when the proteasome capacity is exceeded (47) or inhibited, resulting in the subsequent accumulation of ubiquitinated proteins within aggresomes. In our studies, Mallory bodies/aggresomes were expressed in ∼30% of the cells exposed to shear stress.
In this study, we show that UbcH5 family members and Ubc3 participate in the degradation of keratin IF proteins. Dominant negative Ubc3, UbcH5b, and -c, and to a lesser extent UbcH5a, was shown to prevent the shear stress-mediated degradation of keratin IF proteins in human A549 cells (Fig. 6). Members of the UbcH5 family are highly homologous. UbcH5b and -c are 97% identical at the amino acid level, and with a difference of only four amino acids, it is likely that UbcH5b and -c have similar biologic activities. Although UbcH5a is more diverse than UbcH5b and -c with only being 89 and 88% identical at the amino acid level, respectively (48, 49), these differences may account the differences in sensitivity in the degradation of keratin IF proteins. The various E2s associated with keratin IF protein degradation may reflect the different cellular functions associated with ubiquitin E2s. Although the precise biological role of many of the ubiquitin E2s are not well defined in mammalian cells, there is accumulating evidence that E2s have unique functions. For example, Ubc3 is required for the G1-S transition, Ubc2 is central to DNA repair, while Ubc 4/5 is required for viability in Saccharomyces cerevisiae (50). These functions may depend upon the specificity for the individual E3s. The E3 associated with keratin IF protein degradation has not been identified.
It has been shown that both the assembly and organization of IFs are regulated by post-translational modifications, especially phosphorylation. Phosphorylation is known to occur within the head and tail domains, which are responsible for most of the structural heterogeneity and presumed tissue specific functions of IF proteins. In the case of keratin 8, a number of in vivo phosphorylation sites have been mapped and these play essential roles in regulating filament assembly and disassembly in vivo (51). We have previously demonstrated that PKCδ phosphorylates K8pSer-73 and that this phosphorylation event is responsible for the disassembly of keratin IF proteins (1). In this report we demonstrate that phosphorylation of K8 Ser-73 is required for the ubiquitination of keratin protein following shear stress in A549 cells. Mutation of this site, K8 S73A, prevented ubiquitin from being covalently attached to keratin protein. Taken together, these results suggest that shear stress activates a signal transduction cascade that leads to the activation of protein kinase Cδ, which phosphorylates keratin 8 at Ser-73(1). These events may signal the activation of ubiquitin by the ubiquitin activating enzyme, E1, followed by the transfer of ubiquitin to the ubiquitin-conjugating enzyme, E2. Specifically, the E2s involved in keratin degradation are from the UbcH5/Ubc3 families. The E2 shuttles the ubiquitin molecule to a keratin-specific ubiquitin ligase, E3, which has yet to be identified. The E3 delivers the ubiquitin to the phosphorylated keratin, which has been targeted for degradation. It is possible that additional sites on K18 may require post-translational modifications for this protein to be degraded. The polyubiquitinated keratin protein is then degraded by the 26 S proteasome.
Acknowledgments
We thank Drs. Ed Kuczmarski and Alik Sitikov for thoughtful advice and careful editing of this work.
This work was supported, in whole or in part, by National Institutes of Health Grants P01-HL71643 (to R. G. D. and K. M. R.), R01-HL079190 (to K. M. R.), and GM 36806 (to R. G. D.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: IF, intermediate filament; UPP, ubiquitin-proteasome pathway; AEC, alveolar epithelial cells; PBS, phosphate-buffered saline; GFP, green fluorescent protein; TX-100, Triton X-100; DN, dominant negative.
References
- 1.Ridge, K., Linz, L., Flitney, F., Kuczmarski, E., Chou, Y., Omary, M., Sznajder, J., and Goldman, R. (2005) J. Biol. Chem. 280 30400-30405 [DOI] [PubMed] [Google Scholar]
- 2.Helmke, B. P., Goldman, R. D., and Davies, P. F. (2000) Circ. Res. 86 745-752 [DOI] [PubMed] [Google Scholar]
- 3.Helmke, B. P., Thakker, D. B., Goldman, R. D., and Davies, P. F. (2001) Biophys. J. 80 184-194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yoon, K. H., Yoon, M., Moir, R. D., Khuon, S., Flitney, F. W., and Goldman, R. D. (2001) J. Cell Biol. 153 503-516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schweizer, J., Bowden, P., Coulombe, P., Langbein, L., Lane, E., Magin, T., Maltais, L., Omary, M., Parry, D., Rogers, M., and Wright, M. (2006) J. Cell Biol. 174 169-174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrmann, H., and Aebi, U. (2000) Curr. Opin. Cell Biol. 12 79-90 [DOI] [PubMed] [Google Scholar]
- 7.Coulombe, P. A., and Omary, M. B. (2002) Curr. Opin. Cell Biol. 14 110-122 [DOI] [PubMed] [Google Scholar]
- 8.Paine, R., Ben-Ze'ev, A., Farmer, S. R., and Brody, J. S. (1988) Dev. Biol. 129 505-515 [DOI] [PubMed] [Google Scholar]
- 9.Woodcock-Mitchell, J., Rannels, S. R., Mitchell, J., Rannels, D. E., and Low, R. B. (1989) Am. Rev. Respir. Dis. 139 343-351 [DOI] [PubMed] [Google Scholar]
- 10.Hershko, A., and Ciechanover, A. (1998) Annu. Rev. Biochem. 67 425-479 [DOI] [PubMed] [Google Scholar]
- 11.Ciechanover, A., Orian, A., and Schwartz, A. L. (2000) J. Cell. Biochem. 34 (suppl.) 40-51 [DOI] [PubMed] [Google Scholar]
- 12.Shang, F., Gong, X., and Taylor, A. (1997) J. Biol. Chem. 272 23086-23093 [DOI] [PubMed] [Google Scholar]
- 13.Sivaramakrishnan, S., DeGiulio, J. V., Lorand, L., Goldman, R. D., and Ridge, K. M. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 889-894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yoon, M., Moir, R. D., Prahlad, V., and Goldman, R. D. (1998) J. Cell Biol. 143 147-157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 16.Lecuona, E., Saldias, F., Comellas, A., Ridge, K., Guerrero, C., and Sznajder, J. I. (1999) Am. J. Respir. Crit. Care Med. 159 603-609 [DOI] [PubMed] [Google Scholar]
- 17.Sznajder, J. I., Ridge, K. M., Saumon, G., and Dreyfuss, D. (1998) in Pulmonary Edema (Matthay, M., and Ingbar, D., eds) Vol. 13, pp. 413-430, Marcel Dekker [Google Scholar]
- 18.Bilek, A. M., Dee, K. C., and Gaver, D. P., III (2003) J. Appl. Physiol. 94 770-783 [DOI] [PubMed] [Google Scholar]
- 19.Ku, N. O., Zhou, X., Toivola, D. M., and Omary, M. B. (1999) Am. J. Physiol. 277 G1108-G1137 [DOI] [PubMed] [Google Scholar]
- 20.Chang, L., and Goldman, R. D. (2004) Nat. Rev. Mol. Cell Biol. 5 601-613 [DOI] [PubMed] [Google Scholar]
- 21.Toivola, D. M., Goldman, R. D., Garrod, D. R., and Eriksson, J. E. (1997) J. Cell Sci. 110 23-33 [DOI] [PubMed] [Google Scholar]
- 22.Ku, N. O., and Omary, M. B. (1997) J. Biol. Chem. 272 7556-7564 [DOI] [PubMed] [Google Scholar]
- 23.Ku, N., Liao, J., Chou, C., and Omary, M. (1996) Cancer Metastasis Rev. 15 429-444 [DOI] [PubMed] [Google Scholar]
- 24.Lee, J. C., Schickling, O., Stegh, A. H., Oshima, R. G., Dinsdale, D., Cohen, G. M., and Peter, M. E. (2002) J. Cell Biol. 158 1051-1066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kopito, R. R. (2000) Trends Cell Biol. 10 524-530 [DOI] [PubMed] [Google Scholar]
- 26.Zatloukal, K., French, S. W., Stumptner, C., Strnad, P., Harada, M., Toivola, D. M., Cadrin, M., and Omary, M. B. (2007) Special Issue-Intermediate Filaments 313 2033-2049 [DOI] [PubMed] [Google Scholar]
- 27.Omary, M. B., Coulombe, P. A., and McLean, W. H. I. (2004) N. Engl. J. Med. 351 2087-2100 [DOI] [PubMed] [Google Scholar]
- 28.Lowe, J., Blanchard, A., Morrell, K., Lennox, G., Reynolds, L., Billett, M., Landon, M., and Mayer, R. J. (1988) J. Pathol. 155 9-15 [DOI] [PubMed] [Google Scholar]
- 29.Julien, J. (1997) Trends Cell Biol. 7 243-249 [DOI] [PubMed] [Google Scholar]
- 30.Jellinger, K. (1990) Adv. Neurol. 53 1-16 [PubMed] [Google Scholar]
- 31.Ku, N. O., Michie, S. A., Soetikno, R. M., Resurreccion, E. Z., Broome, R. L., Oshima, R. G., and Omary, M. B. (1996) J. Clin. Investig. 98 1034-1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hazan, R., Denk, H., Franke, W. W., Lackinger, E., and Schiller, D. L. (1986) Lab. Investig. 54 543-553 [PubMed] [Google Scholar]
- 33.Zatloukal, K., Denk, H., Spurej, G., Lackinger, E., Preisegger, K. H., and Franke, W. W. (1990) Lab. Investig. 62 427-434 [PubMed] [Google Scholar]
- 34.Zatloukal, K., Spurej, G., Rainer, I., Lackinger, E., and Denk, H. (1990) Hepatology 11 652-661 [DOI] [PubMed] [Google Scholar]
- 35.Preisegger, K. H., Zatloukal, K., Spurej, G., Riegelnegg, D., and Denk, H. (1992) Lab. Investig. 66 193-199 [PubMed] [Google Scholar]
- 36.Nonomura, A., Saito, K., Kono, N., and Ohta, G. (1986) Acta Pathologica Japonica 36 1669-1677 [DOI] [PubMed] [Google Scholar]
- 37.Nonomura, A., Kono, N., and Ohta, G. (1986) Acta Pathologica Japonica 36 869-878 [DOI] [PubMed] [Google Scholar]
- 38.Shimizu, S., Kobayashi, H., Watanabe, H., and Ohnishi, Y. (1986) Acta Pathologica Japonica 36 105-112 [DOI] [PubMed] [Google Scholar]
- 39.Michel, R. P., Limacher, J. J., and Kimoff, R. J. (1982) Human Pathol. 13 81-85 [DOI] [PubMed] [Google Scholar]
- 40.Cadrin, M., and Martinoli, M. G. (1995) Biochem. Cell Biol. 73 627-634 [DOI] [PubMed] [Google Scholar]
- 41.Cadrin, M., Anderson, N. M., Aasheim, L. H., Kawahara, H., Franks, D. J., and French, S. W. (1995) Lab. Investig. 72 453-460 [PubMed] [Google Scholar]
- 42.Cadrin, M., French, S. W., and Wong, P. T. (1991) Exp. Mol. Pathol. 55 170-179 [DOI] [PubMed] [Google Scholar]
- 43.Salmhofer, H., Rainer, I., Zatloukal, K., and Denk, H. (1994) Hepatology 20 731-740 [PubMed] [Google Scholar]
- 44.Ku, N. O., and Omary, M. B. (2000) J. Cell Biol. 149 547-552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ohta, M., Marceau, N., Perry, G., Manetto, V., Gambetti, P., Autilio-Gambetti, L., Metuzals, J., Kawahara, H., Cadrin, M., and French, S. W. (1988) Lab. Investig. 59 848-856 [PubMed] [Google Scholar]
- 46.Zatloukal, K., Fesus, L., Denk, H., Tarcsa, E., Spurej, G., and Bock, G. (1992) Lab. Investig. 66 774-777 [PubMed] [Google Scholar]
- 47.Johnston, J. A., Ward, C. L., and Kopito, R. R. (1998) J. Cell Biol. 143 1883-1898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Scheffner, M., Huibregtse, J. M., and Howley, P. M. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 8797-8801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jensen, J. P., Bates, P. W., Yang, M., Vierstra, R. D., and Weissman, A. M. (1995) J. Biol. Chem. 270 30408-30414 [DOI] [PubMed] [Google Scholar]
- 50.Pickart, C. M. (2001) Annu. Rev. Biochem. 70 503-533 [DOI] [PubMed] [Google Scholar]
- 51.Omary, M. B., NO, K., Tao, G. Z., Toivola, D. M., and Liao, J. (2006) Trends Biochemical Sci. 31 383-394 [DOI] [PubMed] [Google Scholar]