Abstract
Microcin J25 (MccJ25) is a 21-residue plasmid-encoded ribosomally synthesized lariat-protoknot antibacterial peptide that targets bacterial RNA polymerase. MccJ25 consists of an 8-residue cycle followed by a 13-residue tail that loops back and threads through the cycle. We have performed systematic mutational scanning of MccJ25, constructing and analyzing more than 380 singly substituted derivatives of MccJ25. The results define residues important for production of MccJ25 (comprising synthesis of MccJ25 precursor, processing of MccJ25 precursor, export of mature MccJ25, and stability of mature MccJ25), inhibition of RNA polymerase, and inhibition of bacterial growth. The results show that only a small number of residues (three in the cycle and one in the threaded segment of the tail) are important for MccJ25 production. The results further show that only a small number of additional residues (two in the cycle and four in the threaded segment of the tail) are important for inhibition of transcription. The results open the way for design and construction of more potent MccJ25-based inhibitors of bacterial growth.
Microcins are a class of small (<10 kDa) ribosomally synthesized peptide antibiotics produced by Enterobacteriaceae (1). Some microcins undergo dramatic post-translational modifications by dedicated maturation machinery (2). Post-translationally modified microcins have unusual structures and target important cellular enzymes. The object of this study, microcin J25 (MccJ25),3 is a 21-amino acid peptide that inhibits the activity of Gram-negative bacterial RNA polymerase (RNAP) and that inhibits growth of Gram-negative bacteria (primarily or exclusively Gram-negative enterics) (3, 4). MccJ25 has an unusual lariat-protoknot (“threaded lasso”) structure, consisting of an eight-amino acid cycle with backbone-side chain lactam bond between the α-amino group of Gly1 and the γ-carboxyl group of Glu8, followed by a 13-amino acid tail that loops back and threads through the cycle (5-7). The threaded tail is trapped within the cycle by the aromatic side chains of MccJ25 residues Phe19 and Tyr20, which straddle the cycle (5-8). The unusual lariat-protoknot structure of MccJ25 results in exceptionally high thermal stability and exceptionally high resistance to denaturation by chaotropes and organic solvents (4, 8).
MccJ25 is produced by bacteria containing a plasmid-borne mcjABCD biosynthetic gene cluster (9). Mature MccJ25 is produced from a 58-amino acid ribosomally synthesized precursor, McjA, the product of the mcjA gene (9). Amino acid residues that become part of MccJ25 are located in the C-terminal portion of McjA. The maturation of McjA is catalyzed by McjB and McjC, the products of the mcjB and mcjC genes (10). McjC is homologous to amidotransferases of the asparagine-synthetase/glutamine-hydrolase class, which catalyze transfer of ammonia or an amine from an amide donor to a carboxyl acceptor; McjC thus probably participates in formation of the lactam bond between Gly1 and Glu8 (5-7). McjB is a putative protease and may participate in cleavage of the McjA precursor (11). McjD, the product of the mcjD gene, mediates the export of mature MccJ25 from the producing cells and also mediates resistance to MccJ25 of producing cells (9, 10).
Mutations that confer resistance to MccJ25 in nonproducing cells map to the rpoB and rpoC genes (12, 13), which encode the RNAP β and β′ subunits, respectively. RNAP purified from such rpoB or rpoC mutant cells is resistant to MccJ25 in vitro, establishing that RNAP is the functional cellular target of the drug (12). Amino acid substitutions in RNAP that lead to MccJ25 resistance involve residues located in the RNAP secondary channel, which mediates entry of transcription substrates, NTPs, to the RNAP active center. It has been proposed that MccJ25 inhibits transcription, at least in part, by binding within and obstructing the RNAP secondary channel, interfering with entry of NTPs (12-14). This proposal has received support from biochemical, kinetic, and single-molecule analyses as well as from structural modeling (13, 14).
The fact that MccJ25 is produced from a ribosomally synthesized precursor enables one to exploit the power of genetic engineering in order to produce a library of MccJ25 mutants that can be screened for desired biological properties. Here, we report the results of such an effort. We present an essentially complete collection of point mutations in the MccJ25 coding portion of the mcjA gene, and we determine the effects of amino acid substitutions resulting from these point mutations on production of MccJ25 (comprising synthesis of MccJ25 precursor, processing of MccJ25 precursor, export of mature MccJ25, and stability of mature MccJ25), on ability to inhibit bacterial RNAP in vitro, and on ability to inhibit the bacterial growth in culture. Our results establish that, despite the unusual lariat-protoknot structure of MccJ25, only three wild-type amino acid side chains of MccJ25 are strictly essential for production of MccJ25. In addition, our results define amino acid side chains of MccJ25 important for inhibition of bacterial RNAP and for inhibition of bacterial growth. The fact that only a small number of wild-type side chains of MccJ25 are strictly essential for production of MccJ25 suggests that it should be possible to construct substituted MccJ25 derivatives with higher potencies and/or broadened specificities in further rounds of mutagenesis.
EXPERIMENTAL PROCEDURES
Mutagenesis—Plasmid pTUC202, which carries the MccJ25 biosynthetic gene cluster (9), was used as the template for site-directed mutagenesis. Mutagenesis was performed with the QuikChange site-directed mutagenesis kit (Stratagene) in the presence of 6% DMSO. For each targeted codon, a pair of ∼40-nt oligonucleotide primers introducing all possible substitutions in the first and the second position and specific substitution to G or C into the third position of the codon were used. Mutagenized plasmid DNA was introduced by transformation into MAX Efficiency DH5α-competent cells (Invitrogen). Cells were plated on LB agar plates containing 200 μg/ml chloramphenicol and cultured overnight at 37 °C. Plasmids were prepared from individual transformants, sequences were determined, and clones with desired substitutions were selected.
MccJ25 Derivatives—Transformants of Escherichia coli strain DH5α (Invitrogen) harboring the pTUC202 derivative of interest were cultured for 18 h at 37 °C in 500 ml of M9 minimal medium (15) containing 1 mm thiamine, 2% glucose, and 200 μg/ml chloramphenicol. The culture supernatant was obtained by centrifugation for 30 min at 4 °C at 4000 × g, and 500 ml of the culture supernatant was loaded onto a 35-ml Sep-Pak C8 cartridge (Waters) prewashed with 5 volumes of 100% acetonitrile and 20 volumes of 0.1% trifluoroacetic acid. After loading, the cartridge was washed with 20 volumes of 0.1% trifluoroacetic acid and was eluted successively with 5, 10, 20, and 30% acetonitrile in 0.1% trifluoroacetic acid. Fractions containing MccJ25 derivatives (typically the fraction with 30% acetonitrile) were concentrated by lyophilization, dissolved in 1.5% trifluoroacetic acid, and loaded onto a 3-ml C18 cartridge (Waters), prewashed with 5 volumes of 100% acetonitrile and 20 volumes of 0.1% trifluoroacetic acid. The cartridge was washed successively with 20 volumes of 10 and 20% acetonitrile in 0.1% trifluoroacetic acid and was eluted successively with 23, 25, 27, and 29% acetonitrile with 0.1% trifluoroacetic acid. Fractions containing MccJ25 derivatives (typically fractions with 27 or 29% acetonitrile) were concentrated by lyophilization and dissolved in water with 0.1% trifluoroacetic acid. Typical yields were ∼1 mg/liter of cultured medium.
Culture Supernatants—Transformants of E. coli strain DH5α harboring pTUC202 derivatives were cultured overnight at 37 °C in 5 ml of M9 minimal medium (15) containing 1 mm thiamine, 2% glucose, and 200 μg/ml chloramphenicol. Culture supernatants were prepared by centrifugation for 10 min at 4 °C at 4000 × g and were stored at -20 °C.
Analysis of Production—To 90 μl of culture supernatant (prepared as above) was added 10 μl of 1% trifluoroacetic acid. The sample was loaded onto a C18 ZipTip (Millipore) preequilibrated per instructions of the manufacturer, and was eluted in 5 μl of 50% acetonitrile in 0.1% trifluoroacetic acid. Samples were analyzed by matrix-assisted laser desorption ionization (MALDI) mass spectrometry on a PE Biosystems DE-PRO mass spectrometer in a positive mode using α-cyanohydroxycinnamic acid as matrix. Mass spectra of culture supernatants of cells harboring wild-type pTUC202 exhibited a MALDI-mass spectrometry peak corresponding to mature MccJ25 (m/z = 2108 [M + H+]) that exceeded the background by ∼100 times. Mass spectra of culture supernatants of cells harboring pTUC202 mutants that exhibited unequivocal peaks with expected m/z values (2002-2237 [M + H+], depending on sequence of the mutant) that exceeded the background by at least 3 times were scored as positives.
Analysis of Inhibition of RNAP—The fluorescence-detected abortive initiation assay of Ref. 13 was adapted for high throughput analysis of inhibition of RNAP by culture supernatants containing MccJ25 derivatives. Assays were performed in Corning black flat-bottom 96-well plates (Corning Glass). To each well, was added, successively, 43 μl of culture supernatant (prepared as above), 1 μl of 5 μm E. coli RNAP holoenzyme, 1 μl of 1 μm promoter DNA fragment (DNA fragment lacUV5-12 of Ref. 16), 1 μl of 1 m Tris-HCl (pH 8.0), and 0.5 μl of 1 m MgCl2. Following 15 min at 37 °C, 0.5 μl of 1 mg/ml heparin (Sigma) and 0.5 μl of 2.5 mm (γ-AmNS)UTP (Molecular Probes, Inc.) were added. Following a further 2-min incubation at 37 °C, RNA synthesis was initiated by the addition of 2.5 μl of 10 mm ApA (Sigma), and fluorescence emission intensity was monitored for 15 min at 37 °C using a GENios Pro multimode scanner (TECAN, Inc.). The reaction rate was determined from the quantity of (γ-AmNS)UTP consumed within time t, as in Ref. 17),
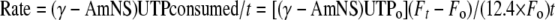 |
(Eq.1) |
where (γ-AmNS)UTP0 is the quantity of (γ-AmNS)UTP at time 0, F0 is the fluorescence emission intensity at time 0, and Ft is the fluorescence emission intensity at time t. Reaction rates with culture supernatants containing MccJ25 derivatives were compared with reaction rates of positive controls (culture supernatants from cells harboring pTUC202) and of negative controls (culture supernatants from nontransformed DH5α cells). Since assays are performed using unfractionated culture supernatants, results depend on quantities of MccJ25 derivatives in culture supernatants as well as on intrinsic inhibitory activities of MccJ25 derivatives. Therefore, assays must be considered as qualitative, plus-or-minus assays. In this work, all culture supernatants exhibiting unequivocally above-background inhibitory activities (i.e. inhibitory activities >20% of the inhibitory activity of wild-type MccJ25) are scored as positives. Results reported are based on at least two independent determinations (with full agreement among determinations in >95% of cases).
Analysis of Inhibition of Bacterial Growth—E. coli strain DH5α or Shigella flexneri clinical isolate BK 12440 (provided by Dr. B. Kreisworth, Public Health Research Institute Inc., Newark, NJ) were cultured overnight at 37 °C in 5 ml of M9 medium (15). Aliquots (50 μl; ∼1 × 109 cells) were mixed with 3 ml of LB top agar (0.8% LB agar) at 45 °C and were applied as overlays to plates containing 25 ml of standard LB agar. After hardening of overlays, 5-μl drops of culture supernatants containing MccJ25 derivatives (prepared as above) or purified MccJ25 were deposited on surfaces of overlays, drops were allowed to dry, plates were incubated for 5-8 h at 37 °C, and plates were examined for the presence of growth inhibition zones. Results with culture supernatants containing MccJ25 derivative were compared with results of positive controls (culture supernatants from cells harboring pTUC202) and of negative controls (culture supernatants from nontransformed DH5α cells). For each derivative, microbiological tests were performed at least three times, with no conflicting results between the tests. Since most assays are performed using unfractionated culture supernatants, results depend on quantities of MccJ25 derivatives in culture supernatants as well as on intrinsic inhibitory activities of MccJ25 derivatives. Therefore, these assays must be considered as qualitative, plus-or-minus assays. In this work, all culture supernatants exhibiting unequivocally inhibitory activities (determined as the presence of anabiosis haloes) are scored as positives.
RESULTS
Systematic Mutational Scan of MccJ25—We constructed all possible single-amino acid substitutions of residues 1-7 and 9-21 of MccJ25. These residues comprise seven of eight residues of the MccJ25 cycle and all 13 residues of the MccJ25 tail. In addition, we constructed one substitution of residue 8 of MccJ25, the side chain of which forms the lactam bond that defines the MccJ25 cycle (5-7) and which previously has been shown not to tolerate nonconservative substitutions, constructing a conservative substitution of Glu (which contains a side chain δ-carboxyl) by Asp (which contains a side chain β-carboxyl). In total, we constructed and analyzed 381 single-amino acid substitutions of MccJ25.
Substitutions were constructed by mutagenesis of the mcjA gene, which encodes the MccJ25 precursor (9), on a plasmid-borne mcjABCD biosynthetic gene cluster (9), followed by expression of the mutant mcjABCD biosynthetic gene cluster (see “Experimental Procedures”).
Determinants for Production, Maturation, and Stability of MccJ25—To assess effects of substitutions on production of MccJ25 (comprising synthesis of MccJ25 precursor, processing of MccJ25 precursor, export of mature MccJ25, and stability of mature MccJ25), we prepared culture supernatants of cells harboring substituted mcjABCD biosynthetic gene clusters, and we assayed culture supernatants for the presence of substituted mature MccJ25 by use of MALDI-MS. We tested 381 single-amino acid substitutions of MccJ25, comprising all possible single-amino acid substitutions at residues 1-7 and 9-21 of MccJ25 and one single-amino acid substitution at residue 8 of MccJ25.
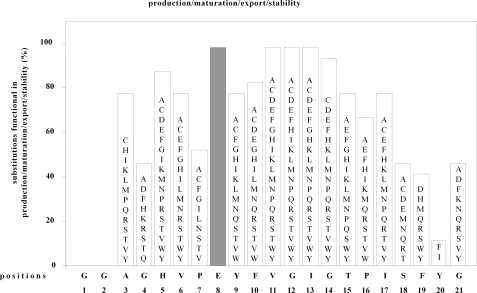
Unequivocal mass ions corresponding to expected masses of mutant mature MccJ25 derivatives were detected for 242 of the 381 tested mutants (64%; Fig. 1). It is striking that such a high fraction, nearly two-thirds, of mutant MccJ25 derivatives is competent for production, maturation, export, and stability.
FIGURE 1.
Effects of single-amino acid substitutions in MccJ25 on production, maturation, export, and stability of MccJ25. Data are presented for 380 single-amino acid substitution derivatives of MccJ25, comprising all possible substitutions at positions 1-7 and 9-21. The sequence of MccJ25 is shown at the bottom. The heights of bars indicate the percentage of tested amino acid substitutions that do not prevent production, maturation, export, and stability of MccJ25. Letters within bars list amino acid substitutions that do not prevent production, maturation, and stability of MccJ25. (Position 8 (gray bar) was not subjected to saturation mutagenesis in this work; see “Results.”)
The results indicate that wild-type side chains are strictly essential for production/maturation/export/stability only for three residues of MccJ25 (i.e. the two residues that form the lactam bond that defines the MccJ25 cycle (Gly1 and Glu8) and one adjacent residue (Gly2)) (Fig. 1 and data not shown). For these residues, no tested substitutions are compatible with production/maturation/export/stability. The results indicate that the wild-type side chain is important, albeit not strictly essential, at one residue (i.e. the distal aromatic residue of the pair of aromatic residues proposed to trap the threaded segment of the MccJ25 tail within the MccJ25 cycle: Tyr20) (Fig. 1). For this residue, only two tested substitutions, Tyr20 > Phe20 and Tyr20 > Ile20, are compatible with production/maturation/export/stability. Interestingly, the proximal aromatic residue of the pair of aromatic residues proposed to trap the threaded segment of the MccJ25 tail within the MccJ25 cycle, Phe19, is considerably much more tolerant of substitution (with eight substitutions being tolerated; Fig. 1).
Wild-type side chains at residues 11-14, located within the loop segment of the MccJ25 tail, are totally (positions 11-13) or nearly totally (position 14) dispensable for MccJ25 production/maturation/export/stability, consistent with previous results indicating that these residues can be deleted, substituted, or chemically modified without loss of function (13, 18).4
In summary, the results for production/maturation/export/stability indicate that there is a strict requirement for wild-type side chains at and adjacent to the site of proteolytic cleavage of the MccJ25 precursor (residues 1 and 2), at and adjacent to the site of cyclization of the MccJ25 precursor (residues 1, 2, and 8), and at the distal aromatic residue that locks the threaded segment of the MccJ25 threaded tail within the MccJ25 cycle. Side chains of the N-terminal half of the MccJ25 tail, the segment that forms the loop of the MccJ25 tail, are unimportant. Perhaps surprisingly, side chains of three of eight residues within the MccJ25 cycle also are relatively unimportant.
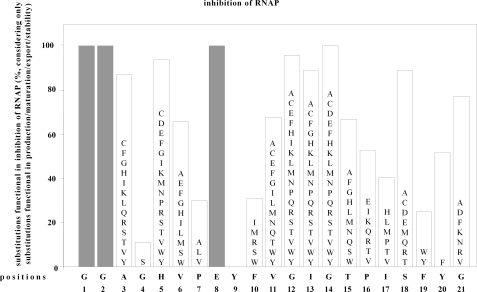
Determinants of MccJ25 for Inhibition of RNAP—To assess the effects of substitutions on inhibition of RNAP by MccJ25, we prepared culture supernatants of cells harboring substituted mcjABCD biosynthetic gene clusters, and we assayed culture supernatants for the ability to inhibit RNAP in vitro. We tested 242 single-amino acid substitutions of MccJ25, comprising all single-amino acid substitutions found to be compatible with production/maturation/export/stability of MccJ25 (see above). The results indicate that 155 of the 242 substituted MccJ25 derivatives competent for production/maturation/export/stability also are competent for inhibition of RNAP (64%; Fig. 2).
FIGURE 2.
Effects of single-amino acid substitutions in MccJ25 on inhibition of RNAP. Data are presented for 242 single-amino acid substitution derivatives of MccJ25, comprising all substitutions shown to be competent for production/maturation/export/stability (see Fig. 1). The sequence of MccJ25 is shown at the bottom. The heights of bars indicate the percentage of tested amino acid substitutions that do not prevent inhibition of RNAP by MccJ25. Letters within bars list amino acid substitutions that do not prevent inhibition of RNAP by MccJ25. (Positions 1, 2, and 8 (gray bars) were not analyzed, since no single-amino acid substitution derivatives at these positions were shown to be competent for production/maturation/export/stability (see Fig. 1).)
The results indicate that the wild-type side chain of one residue of MccJ25 is strictly essential for inhibition of RNAP (i.e. the wild-type side chain of the residue immediately following the MccJ25 cycle (Tyr9)) (Fig. 2). For this residue, no tested substitutions are compatible with inhibition of RNAP. The results indicate that the wild-type side chains of five additional residues are important, albeit not strictly essential (i.e. the wild-type side chains of two residues within the MccJ25 cycle (Gly4 and Pro7), one residue nearly immediately following the MccJ25 cycle (Phe10), and the proximal aromatic residue of the pair of aromatic residues proposed to trap the threaded segment of the MccJ25 tail within the MccJ25 cycle (Phe19)) (Fig. 2). For each of these residues, <40% of tested substitutions, typically substitutions conservative with respect to size and/or conservative with respect to polarity, are compatible with inhibition of RNAP. We infer that the wild-type side chains of these residues (Gly4, Pro7, Tyr9, Phe10, and Phe19) are likely to make direct interactions with RNAP in the RNAP-MccJ25 complex. We note that the wild-type side chains of these residues all are hydrophobic and in three cases are aromatic, and we suggest that RNAP-MccJ25 interactions involve predominantly hydrophobic interactions.
Wild-type side chains at residues 11-15, located within the loop segment of the MccJ25 tail, are totally, or nearly totally, dispensable for inhibition of RNAP, consistent with previous results indicating that these residues can be deleted, substituted, or chemically modified without loss of function (13, 18).4
Determinants of MccJ25 for Inhibition of Bacterial Growth—To assess effects of substitutions on inhibition of bacterial growth by MccJ25, we prepared culture supernatants of cells harboring substituted mcjABCD biosynthetic gene clusters, and we assayed culture supernatants for the ability to inhibit bacterial growth in culture. We tested 155 single-amino acid substitutions of MccJ25, comprising all single-amino acid substitutions found to be compatible with production/maturation/export/stability of MccJ25 and compatible with inhibition of RNAP by MccJ25 (see above).
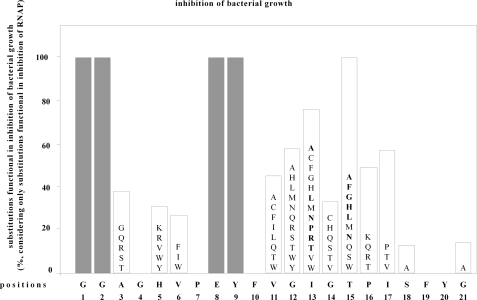
The results indicate that 70 of the 155 MccJ25 derivatives competent for production/maturation/export/stability and competent for inhibition of RNAP also are competent for inhibition of bacterial growth (45%) (Fig. 3). We infer that the 70 MccJ25 derivatives are able to permeate bacterial cells in order to inhibit RNAP inside bacterial cells.
FIGURE 3.
Effects of single-amino acid substitutions in MccJ25 mutants on permeation into bacterial cells and inhibition of bacterial growth. Data are presented for 155 single-amino acid substitution derivatives of MccJ25, comprising all substitutions shown to be competent for production/maturation/export/stability and competent for inhibition of RNAP (see Figs. 1 and 2). The sequence of MccJ25 is shown at the bottom. The heights of bars indicate the percentage of tested amino acid substitutions that do not prevent permeation into bacterial cells and inhibition of bacterial growth. Letters within bars list amino acid substitutions that do not prevent permeation into bacterial cells and inhibition of bacterial growth. Letters in boldface type denote amino acid substitutions that result in apparent inhibitory activities higher than that of wild-type MccJ25. (Positions 1, 2, 8, and 9 (gray bars) were not analyzed, since no single-amino acid substitution derivatives at these positions were shown to be competent for production/maturation/export/stability and competent for inhibition of RNAP (see Figs. 1 and 2).)
The results indicate that wild-type side chains of six residues of MccJ25 are strictly essential for permeation into bacterial cells in order to inhibit RNAP inside bacterial cells: i.e. the wild-type side chains of two residues within the MccJ25 cycle (Gly4 and Pro7), the two residues immediately following the MccJ25 cycle (Tyr9 and Phe10), and the pair of aromatic residues proposed to trap the threaded segment of the MccJ25 tail within the MccJ25 cycle (Phe19 and Tyr20) (Fig. 3). For each of these residues, no tested substitution is compatible with permeation into bacterial cells. Wild-type side chains of six additional residues of MccJ25 are important for permeation into bacterial cells in order to inhibit RNAP inside bacterial cells: three residues of the MccJ25 cycle (Ala3, His5, and Val6), two residues of the MccJ25 tail (Gly14 and Ser18), and the MccJ25 C-terminal residue (Gly21) (Fig. 3). For each of these residues, <40% of tested substitutions are compatible with permeation into bacterial cells.
The apparent inhibitory activities, as estimated from the diameters of growth inhibition zones, differed for different substituted MccJ25 derivatives; 37 substitutions resulted in apparent inhibitory activities less than or comparable with that of wild-type MccJ25 (10 substitutions in the MccJ25 cycle and 27 substitutions in the MccJ25 tail; substitutions in normal type in Fig. 3), whereas 12 substitutions resulted in apparent inhibitory activities higher than that of wild-type MccJ25 (12 substitutions at residues 13 and 15 in the MccJ25 tail; substitutions in boldface type in Fig. 3). In principle, these differences in apparent inhibitory activities may reflect differences in quantities of MccJ25 derivatives in culture supernatants or may reflect differences in intrinsic inhibitory activities (for example, differences in potencies of inhibition of RNAP and/or differences in efficiencies of entry into bacterial cells). As a first step to distinguish between these possibilities, we purified [Gly15]MccJ25, a substituted MccJ25 derivative that exhibited higher apparent inhibitory activity than wild-type MccJ25, and we assessed the ability of purified [Gly15]MccJ25 to inhibit RNAP and to inhibit bacterial growth (Table 1). The results indicate that purified [Gly15]MccJ25 exhibits a potency of inhibition of RNAP comparable with that of purified wild-type MccJ25 (top of Table 1) and exhibits a potency of inhibition of bacterial growth higher than that of purified wild-type MccJ25 (both in assays with E. coli and in assays with a clinical isolate of S. flexneri; bottom of Table 1). We infer that [Gly15]MccJ25 has a higher intrinsic inhibitory activity with respect to bacterial growth than MccJ25, most likely attributable to a higher efficiency of entry into bacterial cells.
TABLE 1.
Properties of superfunctional MccJ25 derivative [Gly15]MccJ25
| MccJ25 derivative | Inhibition of RNAP (IC50)a |
|---|---|
| μm | |
| MccJ25 | 2.5 |
| [Gly15]MccJ25 | 2.5 |
|
MccJ25 derivative
|
Inhibition of bacterial growth (inhibition zone
diameter)b
|
||
|---|---|---|---|
| E. coli DH5α | S. flexneri BK 12440 | ||
| mm | |||
| MccJ25 | 3 ± 1 | 8 ± 1.5 | |
| [Gly15]MccJ25 | 9 ± 1 | 13 ± 1.7 | |
IC50 is the concentration of MccJ25 derivative resulting in 50% inhibition of RNAP.
Mean results of three independent measurements with S.D. values are presented.
DISCUSSION
In this work, we report the results of structure-activity analysis of MccJ25 using a comprehensive panel of MccJ25 point mutants obtained from saturation mutagenesis.
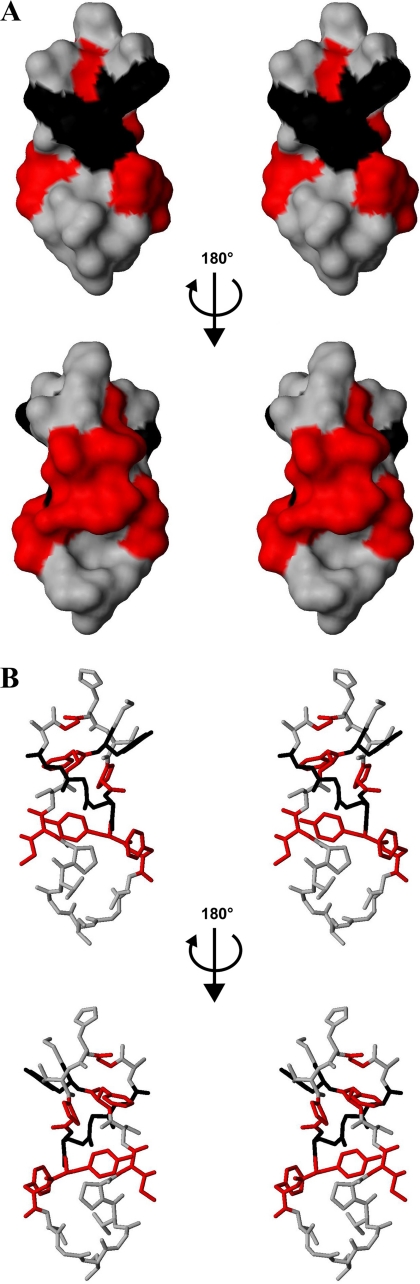
Of the 381 single-amino acid substitutions analyzed, 242 were shown to be compatible with production of MccJ25 (comprising synthesis of MccJ25 precursor, processing of MccJ25 precursor, export of mature MccJ25, and stability of mature MccJ25) (Fig. 1). Inspection of the lariat-protoknot (threaded lasso) covalent structure of MccJ25 suggests that the residues that form the lactam linkage of the MccJ25 cycle and at least one of the two aromatic residues that lock the threaded MccJ25 tail within the MccJ25 cycle may be critical for production of MccJ25, whereas residues in the loop segment of the MccJ25 tail may be less critical. Our findings support these expectations. The residues that form the lactam linkage (Gly1 and Glu8) and one immediately adjacent residue (Gly2) are the sole residues for which no non-wild-type side chains are tolerated in production of MccJ25, and the distal aromatic residue of the pair of aromatic residues that lock the threaded MccJ25 tail within the MccJ25 cycle (Tyr20) is the sole additional residue for which nearly no non-wild-type side chains are tolerated. In the three-dimensional structure of MccJ25 (5-7), these residues form a discrete, continuous surface determinant on one face of MccJ25 (Fig. 4, black residues). We propose that the MccJ25 maturation machinery (McjB and McjC; see Refs. 9 and 11) recognizes and interacts with this surface determinant during processing of MccJ25 precursor.
FIGURE 4.
Locations of residues important for production/maturation/export/stability of MccJ25 and for inhibition of RNAP by MccJ25. Shown are locations of residues shown here to be important for production/maturation/export/stability of MccJ25 (residues 1, 2, 8, and 20; black) and for inhibition of RNAP by MccJ25 (residues 4, 7, 9, 10, 17, and 19; red) on the three-dimensional structure of MccJ25 (see Ref. 5; see also Refs. 3 and 4). A, stereoviews in solvent-accessible surface representation. B, stereoviews in stick representation.
Of the 242 substituted MccJ25 derivatives competent for production of MccJ25, 155 substituted MccJ25 derivatives also are competent for inhibition of RNAP in vitro (Fig. 2). The residue immediately following the MccJ25 cycle (Tyr9) is the sole residue for which no non-wild-type side chain is tolerated in inhibition of RNAP; two residues of the MccJ25 cycle (Gly4 and Pro7), one residue nearly immediately following the MccJ25 cycle (Phe10), and the proximal aromatic residue of the pair of aromatic residues that lock the threaded MccJ25 tail within the MccJ25 cycle (Phe19) are the sole additional residues for which <40% of non-wild-type side chains are tolerated. In the three-dimensional structure of MccJ25 (5-7), these residues form a discrete, continuous surface determinant on one face of MccJ25 (Fig. 4, red residues), the face opposite the face with the determinant for production of MccJ25 (Fig. 4, red residues and black residues). We propose that the surface determinant comprising Gly4, Pro7, Tyr9, Phe10, and Phe19 of MccJ25 makes direct interactions with RNAP in the RNAP-MccJ25 complex (a proposal consistent with the provisional model for the structure of the RNAP-MccJ25 complex in Ref. 13). We note that the wild-type side chains of Gly4, Pro7, Tyr9, Phe10, and Phe19 of MccJ25 all are hydrophobic and in three cases are aromatic, and we suggest that RNAP-MccJ25 interactions involve predominantly hydrophobic interactions.
Of the 155 substituted MccJ25 derivatives competent for production of MccJ25 and competent for inhibition of RNAP by MccJ25 in vitro, 70 substituted MccJ25 derivatives also are competent for inhibition of bacterial growth by MccJ25 in culture and thus apparently are competent to permeate bacterial cells in order to inhibit RNAP inside bacterial cells (Fig. 3).
Two residues of the MccJ25 cycle (Gly4 and Pro7), one residue nearly immediately following the MccJ25 cycle (Phe10), and the pair of aromatic residues that lock the threaded MccJ25 tail within the MccJ25 cycle (Phe19 and Tyr20) are the sole residues for which no non-wild-type side chains are tolerated in permeation into bacterial cells. Three residues of the MccJ25 cycle (Ala3, His5, and Val6), and three residues of the MccJ25 tail (Gly14, Ser18, and Gly21) are the sole additional residues at which <40% or fewer of tested substitutions are compatible with permeation into bacterial cells (Fig. 4). The residues at which substitutions affect the ability to permeate bacterial cells do not form a single surface determinant (possibly reflecting the fact that, during permeation of MccJ25 into bacterial cells, MccJ25 makes successive interactions with import complexes in the cell outer membrane and with import complexes in the cell inner membrane; see Refs. 19 and 20).
In summary, our results show that only a small number of wild-type side chains of MccJ25 are strictly essential for MccJ25 production and function. Therefore, despite its small size and complex structure, MccJ25 can be regarded as an attractive platform for engineering antibacterials with higher potencies and/or broadened specificities by further rounds of mutagenesis.
This work was supported, in whole or in part, by National Institutes of Health Grants GM41376 (to R. H. E.) and GM64530 (to K. S.) and by a NIAID North-east Biodefense center U54-AI057158-Lipkin Grant (to K. S.). This work was also supported by a Howard Hughes Medical Institute Investigatorship (to R. H. E.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Author's Choice—Final version full access.
Footnotes
The abbreviations used are: MccJ25, microcin J25; RNAP, RNA polymerase; (γ-AmNS)UTP, uridine 5′-triphosphoro-γ-l-(5-sulfonaic acid)naphthylamidate.
J. Mukhopadhyay and R. H. Ebright, unpublished results.
References
- 1.Asensio, C., and Perez-Diaz, J. C. (1976) Biochem. Biophys. Res. Commun. 69 7-14 [DOI] [PubMed] [Google Scholar]
- 2.Severinov, K., Semenova, E., Kazakov, A., Kazakov, T., and Gelfand, M. S. (2007) Mol. Microbiol. 65 1380-1394 [DOI] [PubMed] [Google Scholar]
- 3.Salomon, R. A., and Farias, R. N. (1992) J. Bacteriol. 174 7428-7435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blond, A., Peduzzi, J., Goulard, C., Chiuchiolo, M. J., Barthelemy, M., Prigent, Y., Salomón, R. A., Farías, R. N., Moreno, F., and Rebuffat, S. (1999) Eur. J. Biochem. 259 747-755 [DOI] [PubMed] [Google Scholar]
- 5.Wilson, K. A., Kalkum, M., Ottesen, J., Yuzenkova, J., Chait, B. T., Landick, R., Muir, T., Severinov, K., and Darst, S. A. (2003) J. Am. Chem. Soc. 125 12475-12483 [DOI] [PubMed] [Google Scholar]
- 6.Rosengren, K. J., Clark, R. J., Daly, N. L., Goransson, U., Jones, A., and Craik, D. J. (2003) J. Am. Chem. Soc. 125 12464-12474 [DOI] [PubMed] [Google Scholar]
- 7.Bayro, M. J., Mukhopadhyay, J., Swapna, G. V., Huang, J. Y., Ma, L. C., Sineva, E., Dawson, P. E., Gaetano, T. M., and Ebright, R. H. (2003) J. Am. Chem. Soc. 125 12382-12383 [DOI] [PubMed] [Google Scholar]
- 8.Rosengren, K. J., Blond, A., Afonso, C., Tabet, J. C., Rebuffat, S., and Craik, D. J. (2004) Biochemistry 43 4696-4702 [DOI] [PubMed] [Google Scholar]
- 9.Solbiati, J. O., Ciaccio, M., Farias, R. N., and Salomon, R. A. (1996) J. Bacteriol. 178 3661-3663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solbiati, J. O., Ciaccio, M., Farias, R. N., Gonzalez-Pastor, J. E., Moreno, F., and Salomon, R. A. (1999) J. Bacteriol. 181 2659-2662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duquesne, S., Destoumieux-Garzon, D., Zirah, S., Goulard, C., Peduzzi, J., and Rebuffat, S. (2007) Chem. Biol. 14 793-803 [DOI] [PubMed] [Google Scholar]
- 12.Yuzenkova, Y., Delgado, M., Nechaev, S., Savalia, D., Epshtein, V., Artsimovitch, I., Landick, R., Farias, R. N., Salomon, R., and Severinov, K. (2002) J. Biol. Chem. 277 50867-50875 [DOI] [PubMed] [Google Scholar]
- 13.Mukhopadhyay, J., Sineva, E., Knight, J., Levy, R. M., and Ebright, R. H. (2004) Mol. Cell 14 739-751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Adelman, K., Yuzenkova, J., La Porta, A., Zenkin, N., Lee, J., Lis, J. T., Borukhov, S., Wang, M. D., and Severinov, K. (2004) Mol. Cell 16 753-762 [DOI] [PubMed] [Google Scholar]
- 15.Miller, J. H. (1992) A Short Course in Bacterial Genetics, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 16.Mukhopadhyay, J., Kapanidis, A. N., Mekler, V., Kortkhonjia, E., Ebright, Y. W., and Ebright, R. H. (2001) Cell 106 453-463 [DOI] [PubMed] [Google Scholar]
- 17.Schlageck, J. G., Baughman, M., and Yarbrough L. R. (1979) J. Biol. Chem. 254 12074-12077 [PubMed] [Google Scholar]
- 18.Semenova, E., Yuzenkova, Y., Peduzzi, J., Rebuffat, S., and Severinov, K. (2005) J. Bacteriol. 187 3859-3863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Salomon, R. A., and Farias, R. N. (1993) J. Bacteriol. 175 7741-7742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Braun, V., Patzer, S. I., and Hantke, K. (2002) Biochimie (Paris) 84 365-380 [DOI] [PubMed] [Google Scholar]