Abstract
Colicins are cytotoxic proteins secreted by certain strains of Escherichia coli. Colicin M is unique among these toxins in that it acts in the periplasm and specifically inhibits murein biosynthesis by hydrolyzing the pyrophosphate linkage between bactoprenol and the murein precursor. We crystallized colicin M and determined the structure at 1.7Å resolution using x-ray crystallography. The protein has a novel structure composed of three domains with distinct functions. The N-domain is a short random coil and contains the exposed TonB box. The central domain includes a hydrophobic α-helix and binds presumably to the FhuA receptor. The C-domain is composed of a mixed α/β-fold and forms the phosphatase. The architectures of the individual modules show no similarity to known structures. Amino acid replacements in previously isolated inactive colicin M mutants are located in the phosphatase domain, which contains a number of surface-exposed residues conserved in predicted bacteriocins of other bacteria. The novel phosphatase domain displays no sequence similarity to known phosphatases. The N-terminal and central domains are not conserved among bacteriocins, which likely reflect the distinct import proteins required for the uptake of the various bacteriocins. The homology pattern supports our previous proposal that colicins evolved by combination of distinct functional domains.
Colicins are plasmid-encoded toxic proteins produced by Escherichia coli to kill E. coli strains that lack the respective colicin-encoding plasmid (1). Approximately half of the natural E. coli isolates produce one or more of these toxins. Colicins either specifically cleave DNA, rRNA, tRNA or form pores in the cytoplasmic membrane, thereby dissipating the electrochemical potential.
Colicins are not translocated uni-directionally like most other translocated bacterial proteins, i.e. from the cytoplasm to the cytoplasmic membrane, the periplasm, the outer membrane, and the environment of the cell, but are rather translocated in both directions, i.e. released by the producer cells and imported by colicin-sensitive cells. The mechanism of colicin import is highly specific and involves cognate outer membrane receptor proteins and an energy-coupled transport across the outer membrane. Energy driving the import across the outer membrane is provided by the proton motive force across the cytoplasmic membrane. Energy coupling between the outer and cytoplasmic membranes is mediated by either the Ton system or the Tol system. The Ton system consists of the three membrane proteins TonB, ExbB, and ExbD.
Colicin M belongs to the group of colicins whose uptake requires the Ton system (2, 3). These colicins contain a consensus sequence of seven residues at the N terminus, designated as the TonB box, which is also present in all outer membrane transport proteins that require the Ton system and the proton motive force for the import of substrates (4, 5). Both colicin M and its receptor FhuA contain the TonB box, and both are required for the import of colicin M (6, 7). The interaction of TonB with the TonB box of FhuA and colicin M was demonstrated by suppression of point mutations in TonB by point mutations in the TonB box of inactive FhuA and colicin M proteins. The suppressing TonB mutations are located in a three-stranded β-sheet that binds in parallel to the terminal β-strand formed by the TonB box of FhuA (8). Energy provided by the Ton complex is assumed to change the structure of FhuA so that colicin M is released from the FhuA-binding site and re-enters the periplasm through the open pore formed in FhuA. A two-step interaction with TonB has also been shown for the uptake of colicin B (9) and colicin Ia (10), which suggests a general mechanism in the uptake of TonB-dependent colicins.
Colicin M must enter cells from the outside to be toxic (11). Producer cells are protected by the colicin M immunity protein (12, 13). If producer cells are mutated and lack the immunity protein, they are not killed if they also have mutations in genes required for colicin M uptake, fhuA, tonB, exbB, or exbD.
Colicin M activity and immunity genes, and also virulence genes for various iron transport systems, complement resistance, and hemagglutination are encoded on large, self-transmissible pColBM plasmids (14). E. coli cells harboring colicin plasmids defend their ecological niche against cells lacking the colicin plasmid. In contrast to most colicin plasmids, pColBM plasmids lack a lysis gene. At most 10% of colicin M is released through an unknown mechanism (14, 15).
Among the colicins, colicin M has a unique mode of action. It inhibits murein and O-antigen biosynthesis (16–19) by interfering with the regeneration of bactoprenol, which translocates the precursors of murein (lipid II) and O-antigen from the cytoplasm across the cytoplasmic membrane into the periplasm. Upon incorporation of the precursors into murein and lipopolysaccharide, the terminal phosphate of the resulting bactoprenol pyrophosphate is released, and bactoprenol monophosphate re-enters the reaction cycle. Colicin M cleaves between bactoprenol and 1-pyrophospho-MurNAc-(pentapeptide)-GlcNAc (20), and the resulting bactoprenol does not serve as acceptor of the murein and O-antigen precursors.
Colicin M, composed of 271 amino acid residues, is the smallest colicin (21, 22). It consists of three functional domains found in all colicins as follows: the N-domain required for translocation (T) across the outer membrane, the central domain for binding to the receptor (R), and the C-domain in which the enzymatic activity (A) is located. These functional domains have been assigned based on the following findings. Mutations in the TonB box (residues 2–8) abolish colicin M uptake (6). Colicin M lacking a 5-kDa N-terminal fragment binds to cells via the receptor FhuA but does not enter cells (23), and this fragment kills cells when it is translocated into the periplasm via osmotic shock, which bypasses the uptake system (24). Removal of the C-terminal Lys-Arg residues by carboxypeptidase B inactivates colicin M (23). Finally, randomly generated inactive point mutants are located in the C-domain (6).
Here we describe the crystal structure of colicin M solved at 1.7 Å. The functional domains of the novel structure are less separated from each other than observed in other colicins, e.g. colicins E3 (25) and Ia (26), but they can be recognized structurally. Homology analysis of colicin M-like proteins leads to four predicted bacteriocins with C-terminal domains similar to the phosphatase domain of colicin M, but with different receptor binding and translocation domains. The structure presented here forms a rational basis for the use of colicin M as a model protein to study protein import across the outer membrane and to characterize the unique phosphatase activity.
EXPERIMENTAL PROCEDURES
Preparation of Colicin M—E. coli BL21 fhuA was transformed with plasmid pMLD237, which encodes colicin M (cma gene) with an N-terminal His6 tag (generous gift of D. Mengin-Lecreulx, Université Paris-Sud, Orsay, France). Bacteria were grown in 800 ml of LB medium (10 g of tryptone, 5 g of yeast extract, 5 g of NaCl per liter) to an A578 nm of 0.5. Transcription of cma was induced by adding 1 mm isopropyl β-d-thiogalactopyranoside. Cultures were incubated for 3 h; cells were harvested by centrifugation, and the cell pellet was resuspended in 5 ml of buffer A (20 mm K2HPO4, 0.5 mm MgCl2, 150 mm NaCl, 2 mm mercaptoethanol, pH 7.9, supplemented with 1 mg of DNase). Cells were disrupted by repeated passage through a French press, and the soluble fraction containing colicin M was recovered after centrifugation for 1 h at 40,000 × g. Colicin M was purified by affinity chromatography on a nickel-nitrilotriacetic acid-agarose column. The protein solution was adsorbed to the column, and the column was then washed with buffer A supplemented with 20 mm imidazole. Colicin M was eluted with buffer A containing 200 mm imidazole. The entire isolation procedure was carried out at 4 °C. Fractions containing active colicin M were collected and examined by SDS-PAGE. Activity was tested by spotting 7 μl of 10-fold diluted solutions onto a nutrient agar plate seeded with the colicin M-sensitive E. coli strain Ab2847. Clear lysis zones were scored and expressed as the colicin titer. For example, a titer of 5 means that the colicin solution could be diluted 105-fold and still gave a clear lysis zone.
For crystallization, the buffer of the colicin M solution was changed to 10 mm Tris-HCl, 10 mm NaCl, 0.1% dodecyl β-d-maltoside, pH 7.4, by passage through a PD-10 desalting column (GE Healthcare). Colicin at 6 mg ml-1 was used for sitting-drop crystallization. Crystals of high resolution were obtained under two conditions as follows: 1) 0.4 μl of colicin solution supplemented with 0.4 μl of 20% polyethylene glycol 3350, 0.2 m potassium nitrate, 0.1 mm CaCl2, and 0.1 mm pyrophosphate (1.7 Å resolution); and 2) 0.3 μl of colicin solution supplemented with 0.4 μl of 0.1 m N-(2-acetamido) iminodiacetate, 12% polyethylene glycol 6000, 0.2 m MgCl2 (2.5 Å resolution). The reservoir was filled with 75 μl of the supplement solutions. Bushes of needle-shaped crystals were obtained. These crystals were isolated mechanically, placed into the above crystallization buffers supplemented with 12% polyethylene glycol 400, and flash-frozen in liquid nitrogen. Data were collected at the Swiss Light Source (Villigen, Switzerland) at beamline PXII at 100 K. Images were collected on a 225-mm MarCCD using 30% of the full beam intensity (400 mA). Data were processed and scaled using the XDS program package.
Derivative crystals were prepared from the same drop using Pt-salt I–IV of the Hampton platinium screen (Pt kit, Hampton Research). Crystals were soaked for 4 h in a solution containing the heavy atom at 2 mm concentration dissolved in the reservoir solution. Crystals were frozen according to the protocol established for the native crystals, and data were collected 10 eV above the theoretical platinium-edge at a wavelength 1.071 Å under conditions similar to the native crystal. Data were processed and reduced using the XDS program package (27). The anomalous contribution of each single derivative was initially estimated from the XDS scaling procedure.
Data Collection and Structure Solution, Refinement, and Analysis—Five different data sets were collected and included in the automated structure solution version of SHARP/autoSHARP (28). Heavy atom sites were obtained by the SHELX program package included in autoSHARP and further refined by the SHARP algorithm. Initial experimental phases were calculated using SHARP and transferred to the solvent flattening procedures Solomon and DM as included in the program package. Phases were further improved using Pirate from the CCP4 program package (29), and an initial model was built at 2.9 Å resolution using Buccaneer. This model was further refined by manual and automatic rebuilding using the PHENIX program package at a resolution of 2.4 Å. A final model was obtained after several rounds of manual rebuilding and refinement using Coot (30) and Refmac from the CCP4i package and includes 536 residues and 144 water molecules (Table 1). Crystals in space group C2221 were obtained, and data were collected under essentially the same conditions as described for crystal form I. The structure of crystal form II was solved by molecular replacement using MOLREP from the CCP4 program package. The structure geometry has been checked using the Procheck program, and values are described in Table 1. Pictures were generated in DINO and Pymol using surface representations from MSMS (Michel Sanner, The Scripts Research Institute).
TABLE 1.
Summary of data collection and phasing and refinement statistics
| Crystal form I | Crystal form II | |
|---|---|---|
| Data collectiona | ||
| Wavelength (Å) | 0.962 | 0.934 |
| Space group | P42212 | C2221 |
| Resolution (Å) | 40 to 2.5 (2.65 to 2.5) | 40 to 1.7 (1.8 to 1.7) |
| Cell constants (Å) | a = 119.9, c = 96.2 | a = 50.5, b = 108.7, c = 224.9 |
| Unique reflections | 23,282 (3858) | 67,044 (10,264) |
| Redundancy | 5.9 (6.3) | 5.6 (5.1) |
| Completeness (%) | 93.2 (98) | 97.7 (93.8) |
| Rmerge (%) | 17.5 (39.8) | 12.3 (45.2) |
| I/σ(I) | 8 (2.7) | 9.9 (2.4) |
| Wilson B-factor | 32 | 22.5 |
| Phasing statistics | ||
| FOMb after SHARP | (40 to 2.8) | 0.31 |
| FOM after DM | (40 to 2.8) | 0.89 |
| Refinement statisticsa | ||
| Space group | P42212 | C2221 |
| Resolution (Å) | 40 to 2.5 (2.57 to 2.5) | 40 to 1.7 (1.74 to 1.7) |
| Rcryst (%) | 0.23 (0.28) | 0.23 (0.28) |
| Rfree (%) | 0.29 (0.34) | 0.26 (0.36) |
| Non-hydrogen atoms | 4280 | 4710 |
| Waters | 144 | 493 |
| Ligand I (nitrate) | 5 | |
| Ligand II (magnesium) | 2 | |
| Mean B-value (Å2) | 29 | 11 |
| r.m.s.d.c of bond length (Å2) | 0.01 | 0.016 |
| r.m.s.d. of angle (degree) | 1.25 | 1.8 |
| Model quality | ||
| Residues in most favored region (%) | 521 (97.2) | 510 (95.1) |
| Residues in most allowed region (%) | 13 (2.4) | 14 (2.6) |
| Residues in outlier region (%) | 2 (0.4) | 6 (2.2) |
Numbers in parentheses refer to the highest resolution shell
FOM indicates figure of merit
r.m.s.d. indicates root mean square deviation
RESULTS AND DISCUSSION
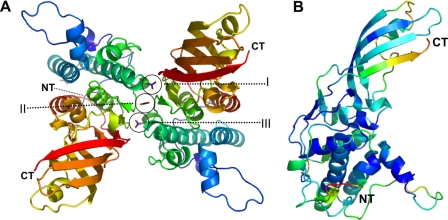
Structure Solution and Comparison of Independent Models Derived from Two Crystal Forms—Colicin M was overproduced and purified to electrophoretic homogeneity and further used for crystallization. Crystals were obtained in two different forms. Crystal form I (CFI)2 occurred in space group P42212 with two molecules in the asymmetric unit (Fig. 1A) and a solvent content of 57%, and crystal form II (CFII) occurred in space group C2221 with two molecules in the asymmetric unit and a solvent content of 51%.
FIGURE 1.
Characteristics of the colicin M structure. A, colicin M dimer (crystal form I) stabilized by three nitrate molecules (I–III) derived from the crystallization buffer. The nitrate molecules are attached to the backbone of the protein, thereby increasing the intermolecular surface. B, B-factor distribution (blue, low; red, high) of colicin M reveals a well ordered core molecule in which only the N terminus (marked NT) and C terminus (marked CT) show a higher mobility. Figures were prepared using Pymol.
The crystal structure was solved by the MIR technique using form I crystals and four different platinium derivatives for experimental phasing at 2.8 Å resolution. The structure in crystal form II was solved by molecular replacement using the completed structure of crystal form I as the search model. Both crystal forms allowed the unambiguous tracing of the entire protein (residues 2–271).
In CFII, five nitrate molecules from the crystallization buffer are visible in the electron density map, three of which (I, II, and III) are aligned in a row between dimers forming an interface for inter-chain contacts (see Fig. 1A). Nitrate molecules I and III are related by noncrystallographic symmetry, and both are clamped by Ser-145 and Arg-88 of the two adjacent protein molecules. Nitrate molecule II is located in-between these two molecules and ligated by the backbone nitrogens of Lys-120 and Gln-121 of both protein molecules and two water molecules in addition, which are related by 2-fold noncrystallographic symmetry to achieve a 6-fold symmetric coordination sphere (details not shown).
We calculated the difference between the two structures of CFI and CFII to be 1.1 Å root mean square deviation (for 260 superimposed residues), and between the two independent monomers of CFI (for 270 superimposed Cα) and of CFII (for 270 superimposed Cα) to be 0.4 and 0.35 Å, respectively. These results indicate that the monomeric structures in crystal forms I and II are almost identical but differ slightly between the two crystal forms. The differences between the two crystal forms originate mainly in the N-terminal part (residues 28–38) and are caused by different crystal contacts. However, these differences are also reflected by the B-factor distribution of the model, which in addition indicates a higher flexibility at the C terminus (Fig. 1B). Because the resolution and the overall B-factor distribution of CFII is better (1.7 and 2.5 Å, average Cα-B-factors 11 and 14), chain A of CFII (RCSB ID code rcsb047783, PDB entry 3DA4) was used for the following discussion of the structure.
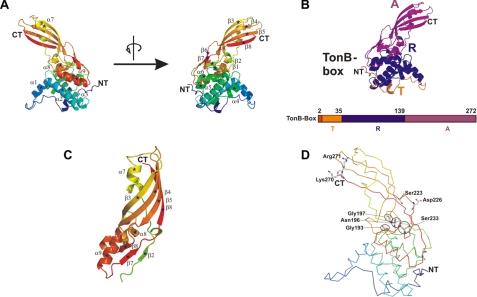
For the following discussion of the colicin M structure and function, we will use the following nomenclature: 1) the N-domain includes residues 1–35 and is involved in translocation of colicin M across the outer membrane; 2) the central domain (residues 36–140) contains the colicin M-binding site to the FhuA outer membrane receptor protein; and 3) the C-domain (residues 141–271) includes the phosphatase.
Overall Structure of Colicin M—The overall architecture of colicin M shows a unique fold that has not been observed in other proteins (Fig. 1A). As in the crystal structures of other colicins (1), the functional domains of colicin M form distinct structural entities connected by a few residues (Fig. 2, B–D) but are less well separated from each other than in, for example, colicins Ia (26) and E3 (25). In colicin B, only the pore-forming domain forms a distinct structure separated from the N-terminal lobe, which contains the receptor binding and translocation domains (31). This weak association between single domains of colicin M might facilitate unfolding during import across the outer membrane.
FIGURE 2.
Crystal structure of colicin M. A, structure of the protein from the N to C terminus, colored from blue to red, shown in two different views related by 180° rotation. Secondary structure elements are marked (α1–α9 and β1–β8). B, succession of the three functional domains, color-coded in orange (N-domain, translocation domain, T), blue (central receptor binding domain, R), and magenta (C-domain, activity domain, A). The TonB box is shown in red. C, structure of the C-domain of colicin M. The secondary structure elements are indicated as in the entire structure in A. D, mutated residues that result in inactive colicin M are indicated (6).
The N-domain—The N-terminal domain displays a higher degree of flexibility than the rest of the molecule, as revealed by B-factor analysis (Fig. 1B) and lack of secondary structure. The flexibility is further underlined by the occurrence of 6 proline residues within only 40 residues, which supposedly disrupt defined secondary structures. The TonB box (residues 2–7) is followed by a long and unstructured random coil that forms a loop-like structure (residues 8–35) and wraps around almost the entire intermediate domain. Such a disordered motif has also been proposed for several other colicins, but it was only rarely determined because of the disorder of this part in the crystal lattices (1). Notably, the presence of an unstructured N-terminal sequence in proteins designed to unfold during translocation has also been observed for proteins destined to the mitochondrial matrix and to the proteasomal cavity (35). A high flexibility is probably the prerequisite as the N-terminal part of colicin M must interact with the periplasmically localized TonB protein (see also Fig. 5) to initiate translocation.
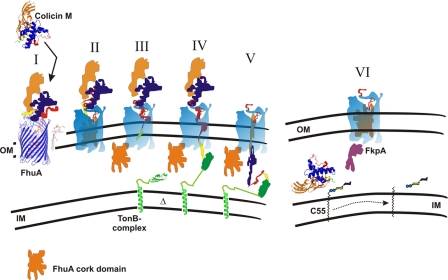
FIGURE 5.
Tentative model of colicin M uptake across the outer membrane (OM) of E. coli. I, colicin M (crystal structure) binds to the FhuA protein, whose crystal structure is shown. II, colicin M partially unfolds while bound to FhuA. III, N-terminal domain (red) with the TonB box (yellow) enters the pore in FhuA, which is formed by interaction of FhuA with energized TonB (green), leading to movement of the globular domain (cork) out of FhuA. IV, TonB box of colicin M interacts with TonB, and colicin M unfolds further and enters the periplasm through the FhuA pore. VI, in the periplasm, colicin M is refolded with the help of the FkpA chaperon and cleaves the pyrophosphate bond between C55 isoprenoid and the murein precursor. IM, inner membrane; Δ, membrane potential. For the sake of clarity, the ExbB and ExbD proteins associated with TonB and required for TonB activity are not shown.
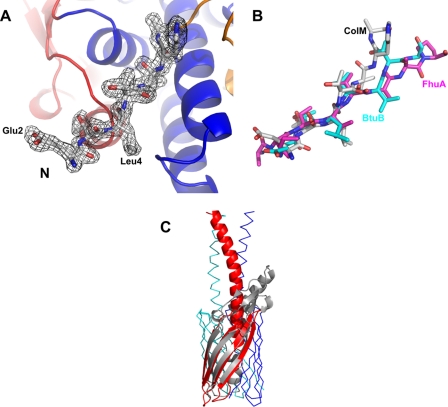
The TonB box (Fig. 2B, residues 2–8) is exposed on the surface and thus may be accessible to interact with TonB. This N-terminal arrangement is stabilized only by a small number of weak interactions, mainly H-bonds to the backbone of the following central domain, and may be detached to interact with TonB. Genetic data support the involvement of the TonB box in interaction with TonB. Mutations in the TonB box (L4N, L4P, V6R, V6G, and V6E) inactivate colicin M. Sensitivity to colicin M is restored in cells in which the mutation V6R is combined with the chromosomal TonB mutation Q160L. Suppression of the V6R mutation by the Q160L mutation suggests interaction of colicin M with TonB through regions in which these mutations are localized (6). In FhuA, the TonB box is not visible (32, 33), but co-crystals of FhuA with a C-proximal fragment of TonB (residues 158–235) reveal a FhuA N-proximal β-strand (TonB box) bound to a three-stranded β-sheet of TonB (8). A similar structure of a TonB fragment bound to the BtuB vitamin B12 transporter has been determined (34). In colicin M, the TonB box displays an elongated and well structured motif (Fig. 3A) that can be overlaid onto the structures of FhuA and BtuB bound to the TonB fragment (Fig. 3B). In colicin B, the TonB box is folded back and attached to the N-terminal lobe (31). The TonB box of colicin Ia interacts along one surface of the N-proximal helical sheet (26).
FIGURE 3.
Similarities of colicin M to other protein structures. A, simulated annealed 2Fo - Fc map of the N-terminal TonB-box domain. B, superposition of the N termini of colicin M (white), FhuA (magenta; in complex with a TonB fragment; PDB entry 2GRX), and BtuB (cyan; in complex with a TonB fragment; PDB entry 2GSK). C, superposition of the C-terminal domain of colicin M (gray) with the membrane-spanning part of the outer membrane transporter Hia of H. influenzae (red, blue, and green) (39).
Central Domain—The central domain formed by residues 36–140 is globular and entirely α-helical (Figs. 1 and 2). The domain consists of six helices (α1–α6) of different lengths; five of these helices are wrapped around the longest central helix, α3, formed between residues Pro-70 and Leu-92. Residues Leu-36 to Gln-46 of α1 (Fig. 2A and Fig. 4A, LLVQVVYSFFQ) are strongly hydrophobic and extend outward from the globular domain. It is particularly appealing to hypothesize that this helix attaches colicin M to the cytoplasmic membrane in close proximity to the substrate, undecaprenyl-PP-MurNAc(pentapeptide)-GlcNAc, or binds to a portion of the C55 polyisoprenoid. A functional importance of this helix is supported by its conservation among homologous bacteriocins of colicin M (Fig. 4A). In the predicted receptor binding and translocation domains, only this region is somewhat conserved. α3 and α6 form the hydrophobic core of the central domain, which displays the lowest B-factors within the entire protein (Fig. 1B). Although there are no strong ionic interactions discernible between the central domain and the C-domain (Fig. 2C), a small β-sheet (β1, β2, β6, and β7) connects the two domains by hydrogen bridges and may contribute to the overall stability of the protein. A mutant in this region, R115C, is defective in colicin M uptake (6).
FIGURE 4.
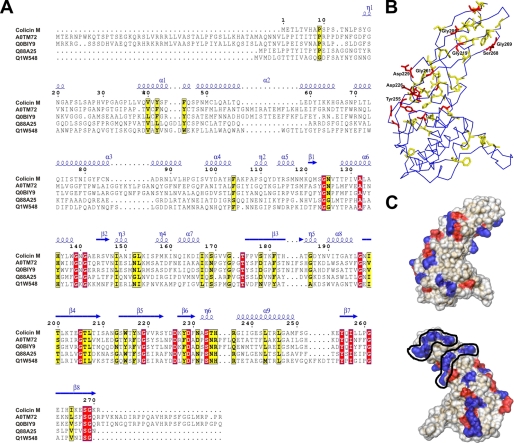
A, comparison of the colicin M amino acid sequence with the sequences of predicted bacteriocins of Burkholderia amifaria MC40-6 (accession number AOTM72), Burkholderia cepacia (QOBIY7), Pseudomonas syringae pv. tomato (Q88A25), and Pseudomonas aeruginosa (Q1W548). Amino acids that occur in all bacteriocins are shaded red, and those that occur in most but not all strains are shaded yellow. B, localization of the conserved amino acids in the colicin M structure. C, distribution of surface-exposed positively charged (blue) and negatively charged (red) amino acids.α1–α9,α-helices;β1–β8,β-strands; andη1–η6, interhelical regions.
C-domain—The C-domain forms an elongated mixed α/β structure. The most obvious structural feature is the open β-barrel formed by strands β3/β4/β5/β8 (Fig. 2C). This barrel has α/β-extensions on one side of the domain. Our search for structurally similar domains yielded the β-domain of the autotransporter Hia from Haemophilus influenzae. In Fig. 3C, the C-terminal domain of colicin M (gray) is superimposed with the outer membrane-spanning part of Hia (red) (39). Although the extensions of colicin M do not fit into the trimeric model of the Hia transporter, a structural analogy in the barrel domains is obvious. However, charged and hydrophilic residues on colicin M are exposed to the outer surface of the barrel, which is atypical for outer membrane β-barrels. Although the secretion of the protein via an autotransporter pathway might be possible, less than 10% of colicin M is released by cells; therefore, it seems unlikely that colicin M is secreted through such a designed export mechanism. Moreover, we assume that the low amount of protein is released by lysis of a small portion of the colicin M-producing cells. A possible mechanism of the β-sheet in colicin M import seems also rather unlikely. The C-domain would first have to integrate as a β-barrel into the outer membrane and then enter the periplasm to act as a phosphatase. These considerations argue against a function of the C-domain in import or export of colicin M across the outer membrane, as the β-barrel of Hia may function in the export of the Hia adhesin. Our view onto the import pathway is given in Fig. 5.
Several patches of positively charged amino acids appear on the surface (Fig. 4C). Two positively charged patches on the C-domain (Lys-168, Lys-270, Arg-271, and Lys-180, Lys-202, Arg-222) are striking considering possible interactions with the double negatively charged substrate. A functional role of Lys-270 and Arg-271 is supported by the finding that their release by carboxypeptidase B inactivates colicin M (23). However, these residues are not well conserved among the predicted bacteriocins (Fig. 4A); therefore, the cleavage mechanisms might be different. Up to now, colicin M is the only colicin with a biochemically demonstrated phosphatase activity.
Primary Structure Analysis and Sequence Comparison—The current genomic data bases contain no deduced amino acid sequences similar to colicin M for which a phosphatase activity is predicted. However, four open reading frames from pathogenic bacteria are homologous to colicin M (Fig. 4, A and B). Interestingly, only the region of colicin M assigned to the phosphatase domain shows similarity to the bacteriocins. Sequence identity includes residues 124–270 and thus begins somewhat earlier than the domain assignment based on the crystal structure where the C-domain starts with residue 141. Residues Asp-226 and Asp-229 are conserved among all these bacteriocins, and a D226N mutation inactivates colicin M (6). Mutations in two additional conserved and functionally important sites, G197S/G197D/G197V and S233A/S233T, that result in inactive colicin M were previously identified by random mutagenesis and site-directed mutagenesis, respectively (6). These findings suggest that the predicted bacteriocins assume structures that resemble the colicin M phosphatase domain, have phosphatase activity, and kill cells similarly to colicin M.
Glycine residues are typically among the most-conserved residues and are located between secondary structure elements to ensure structural conservation (Gly-139, Gly-141, and Gly-197 in colicin M). It has been noted previously that colicins contain an unusually large number of glycine residues. Colicin M contains 25 glycine residues (9.2% of all residues), in a range similar to other colicins (1). These small and flexible residues are equally distributed over the whole protein. Eight of the 25 glycine residues are conserved among the five bacteriocins (Fig. 4A). Conservation of the glycine residues suggests that they are important to maintain a geometric flexibility during passage through the outer membrane. These residues might smoothly guide colicins during translocation across the outer membrane, possibly through a channel protein that potentially strongly restricts the conformational degree of freedom. The hydrophobic core could be maintained by the conserved residues Tyr-255 and Ile-257, which point toward the interior of the hydrophobic domain.
The receptor binding (central domains) and translocation domains (N-domains) show little similarity to the N-terminal and central domains of the bacteriocins (Fig. 4A). Presumably, these distinct domains facilitate the import of the phosphatase domain by import proteins of strains that strongly differ from those of E. coli. The distinct receptor binding and translocation domains of bacteriocins reflect the distinct uptake proteins in the various bacteria. The different degrees of similarity among the receptor binding and translocation domains and the phosphatase domain support our previous proposal that colicins evolved by horizontal gene transfer of plasmids and exchange of functional domains (3, 36). The phosphatase domain evolved from a single ancestor gene that was fused to distinct receptor binding and translocation domain genes.
Tentative Model of Colicin M Import—The import mechanism of colicins has been studied extensively but was never completely unraveled (1). Here we present how we envisage colicin M import based on the crystal structure (Fig. 5). It is unlikely that colicin M can pass through the outer membrane in the compact structure obtained by x-ray crystallography. Rather, as suggested for the import of other colicins, it partially unfolds during translocation across the outer membrane. Upon binding to FhuA, colicin M alters its structure and becomes trypsin-sensitive, in contrast to unbound colicin M, which is trypsin-resistant (37). Because the TonB box of FhuA is also required for colicin M uptake, it is likely that TonB first interacts with FhuA and later with colicin M. Interaction with FhuA may open the pore in the FhuA β-barrel, into which the N terminus of colicin M can enter until TonB is reached and the interaction between TonB and FhuA is replaced by the binding of colicin M to TonB. The FhuA pore is opened by the proton motive force of the cytoplasmic membrane through interaction of FhuA with the energy-transducing TonB-ExbB-ExbD protein complex (4). Evidence has been provided that TonB-dependent colicins enter cells through the receptor protein to which they bind and not through a co-receptor, as has been shown for some Tol-dependent colicins (1). A recent study on colicin B is particularly revealing (38). Cysteine residues engineered into the globular domain (cork) of FepA become exposed upon binding of colicin B to FepA. Exposure requires TonB and the TonB box of FepA. Because some of the cysteine derivatives react poorly with biotin maleimide, the authors concluded that the cork does not entirely leave the FepA β-barrel and colicin B must unfold to enter the periplasm. Alternatively, the entire cork moves out of the β-barrel, and some of the cysteine residues display low reactivity for other reasons, such as binding to a protein. We assume that colicin M also enters the periplasm through the FhuA pore because the TonB box of FhuA is required for colicin M uptake (and not for binding), and extensive searches for mutants resistant to colicin M revealed mutations only in fhuA, tonB, exbB, and exbD. In addition, “colicin M-tolerant” mutants (24, 37) are mutated in fkpA, which encodes a periplasmic chaperone (40). FkpA is essential for colicin M activity. Once the entire colicin M or its activity domain has entered the periplasm, FkpA presumably refolds the partially unfolded colicin M. Colicin M can then interact with the cytoplasmic membrane where its substrate is located. The C55-isoprenoid of bactoprenol is inserted in the lipid bilayer. The pyrophosphate bond between bactoprenol and the murein precursor is at the surface of the bilayer and can be approached by colicin M and cleaved. It is likely that one of the hydrophobic helices of colicin M, in particular helix α1, attaches the protein to the cytoplasmic membrane. This idea is supported by the fixation of the colicin M immunity protein through its N-terminal end to the cytoplasmic membrane (12). In this way, the two proteins come into close contact, which facilitates protection of colicin M-producing strains by the immunity protein. The three-dimensional movement of both proteins in the periplasm is reduced to a two-dimensional movement along the surface of the cytoplasmic membrane. Because colicin M must pass only the outer membrane, it is particularly suited to study import of a protein into the periplasm.
Acknowledgments
We thank Dr. Reinhard Albrecht for very helpful contributions in protein crystallization. We acknowledge Dr. Ehmke Pohl from Beamline PX10 at the Swiss Light Source, Villigen, Switzerland, for excellent technical assistance and the Beamline support, and Karen A. Brune for critically reading the manuscript. This work was carried out in the Department of Protein Evolution, and the support of Dr. Andrei Lupas, head of the department, is greatly acknowledged.
The atomic coordinates and structure factors (code 3DA4) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: CFI, crystal form I; CFII, crystal form II; PDB, Protein Data Bank.
References
- 1.Cascales, C., Buchanan, S. K., Duché, D., Kleanthous, C., Lloubés, R., Postle, K., Riley, M., Slatin, S., and Cavard, D. (2007) Microb. Mol. Biol. Rev. 71 158-229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Braun, V. (1995) FEMS Microbiol. Rev. 16 295-307 [DOI] [PubMed] [Google Scholar]
- 3.Braun, V., Patzer, S. I., and Hantke, K. (2002) Biochimie (Paris) 84 365-380 [DOI] [PubMed] [Google Scholar]
- 4.Postle, K., and Larsen, R. A. (2007) Biometals 20 453-465 [DOI] [PubMed] [Google Scholar]
- 5.Chu, B. C., Peacock, R. S., and Vogel, H. J. (2007) Biometals 20 467-483 [DOI] [PubMed] [Google Scholar]
- 6.Pilsl, H., Glaser, C., Gross, P., Killmann, H., Ölschläger, T., and Braun, V. (1993) Mol. Gen. Genet. 240 103-112 [DOI] [PubMed] [Google Scholar]
- 7.Schöffler, H., and Braun, V. (1989) Mol. Gen. Genet. 217 378-383 [DOI] [PubMed] [Google Scholar]
- 8.Pawelek, P. D., Croteau, N., Ng-Thow-Hing, C., Khursigara, C. M., Moiseeva, N., Allaire, M., and Coulton, J. W. (2006) Science 312 1399-1402 [DOI] [PubMed] [Google Scholar]
- 9.Mende, J., and Braun, V. (1990) Mol. Microbiol. 4 1523-1533 [DOI] [PubMed] [Google Scholar]
- 10.Buchanan, S. K., Lukacik, P., Grizot, S., Ghirlando, R., Mu Ali, M., Barnard, T. J., Jakes, K. S., Kienker, P. K., and Esser, L. (2007) EMBO J. 26 2594-2604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Harkness, R. E., and Braun, V. (1990) Mol. Gen. Genet. 222 37-40 [DOI] [PubMed] [Google Scholar]
- 12.Gross, P., and Braun, V. (1996) Mol. Gen. Genet. 251 388-396 [DOI] [PubMed] [Google Scholar]
- 13.Ölschläger, T., Turba, A., and Braun, V. (1991) Mol. Microbiol. 5 1105-1111 [DOI] [PubMed] [Google Scholar]
- 14.Johnson, T. J., Johnson, S. J., and Nolan, L. K. (2006) J. Bacteriol. 188 5975-5983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Thumm, G.,Ölschläger, T., and Braun, V. (1988) Plasmid 20 75-82 [DOI] [PubMed] [Google Scholar]
- 16.Schaller, K., Höltje, J.-V., and Braun, V. (1982) J. Bacteriol. 152 994-1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Schaller, K., Krauel, A., and Braun, V. (1982) J. Bacteriol. 147 135-139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Harkness, R. E., and Braun, V. (1989) J. Biol. Chem. 264 6177-6182 [PubMed] [Google Scholar]
- 19.Harkness, R. E., and Braun, V. (1989) J. Biol. Chem. 264 14717-14722 [Google Scholar]
- 20.El Ghachi, M., Bouhss, A., Berreteau, J. H., Tuze, T., Auger, G., Blanot, D., and Mengin-Lecreulx, D. (2006) J. Biol. Chem. 281 22761-22772 [DOI] [PubMed] [Google Scholar]
- 21.Braun, V., Schaller, K., and Wabl, S. A. (1974) Antimicrob. Agents Chemother. 5 520-533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Köck, J.,Ölschläger, T., Kamp, R. M., and Braun, V. (1987) J. Bacteriol. 169 3358-3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dreher, R., Braun, V., and Wittmann-Liebold, B. (1985) Arch. Microbiol. 140 343-346 [DOI] [PubMed] [Google Scholar]
- 24.Braun, V., Frenz, J., Hantke, K., and Schaller, K. (1980) J. Bacteriol. 142 162-168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soelaiman, S., Jakes, K., Wu, N., Li, C., and Shoham, M. (2001) Mol. Cell 8 1053-1062 [DOI] [PubMed] [Google Scholar]
- 26.Wiener, M., Freyman, D., Ghosh, P., and Stroud, R. M. (1997) Nature 385 461-464 [DOI] [PubMed] [Google Scholar]
- 27.Kabsch, W. (1993) J. Appl. Crystallogr. 26 795-800 [Google Scholar]
- 28.Vonrhein, C., Blanc, E., Roversi, P., and Bricogne, G. (2007) Methods Mol. Biol. 364 215-230 [DOI] [PubMed] [Google Scholar]
- 29.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50 760-76315299374 [Google Scholar]
- 30.Emsley, P., and Cowtan, K. (2004) Acta Crystallogr. Sect. D. Biol. Crystallogr. 60 2126-2132 [DOI] [PubMed] [Google Scholar]
- 31.Hilsenbeck, J. L., Park, H., Chen, G., Youn, B., Postle, K., and Kang, C. (2004) Mol. Microbiol. 51 711-720 [DOI] [PubMed] [Google Scholar]
- 32.Ferguson, A. D., Hofmann, E., Coulton, J. W., Diederichs, K., and Welte, W. (1998) Science 282 2215-2219 [DOI] [PubMed] [Google Scholar]
- 33.Locher, K. P., Rees, B., Koebnik, R., Mitschler, A., Moulinier, L., Rosenbusch, J. P., and Moras, D. (1998) Cell 95 771-778 [DOI] [PubMed] [Google Scholar]
- 34.Shultis, D. D., Purdy, M. D., Banchs, C. N., and Wiener, M. C. (2006) Science 312 1396-1399 [DOI] [PubMed] [Google Scholar]
- 35.Wilcox, A. J., Choy, J., Bustamante, C., and Matouschek, A. (2005) Proc. Natl. Acad. Sci. U. S. A. 102 15435-15440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roos, U., Harkness, R. E., and Braun, V. (1989) Mol. Microbiol. 3 891-902 [DOI] [PubMed] [Google Scholar]
- 37.Schaller, K., Dreher, R., and Braun, V. (1981) J. Bacteriol. 146 54-63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Devenathan, S., and Postle, K. (2007) Mol. Microbiol. 65 441-453 [DOI] [PubMed] [Google Scholar]
- 39.Meng, G., Surana, N. K., St. Geme, J. W., III, and Waksman, G. (2006) EMBO J. 25 2297-2304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hullmann, J., Patzer, S. I., Römer, C., Hantke, K., and Braun, V. (2008) Mol. Microbiol. 69 926-937 [DOI] [PMC free article] [PubMed] [Google Scholar]