Abstract
Microfibril-associated glycoprotein-1 (MAGP-1) is a small molecular weight component of the fibrillin-rich microfibril. Gene-targeted inactivation of MAGP-1 reveals a complex phenotype that includes increased body weight and size due to excess body fat, an altered wound healing response in bone and skin, and a bleeding diathesis. Elastic tissues rich in MAGP-1-containing microfibrils develop normally and show normal function. The penetrance of MAGP-1-null phenotypes is highly variable and mouse strain-dependent, suggesting the influence of modifier genes. MAGP-1 was found to bind active transforming growth factor-β (TGF-β) and BMP-7 with high affinity, suggesting that it may be an important modulator of microfibril-mediated growth factor signaling. Many of the phenotypic traits observed in MAGP-1-deficient mice are consistent with loss of TGF-β function and are generally opposite those associated with mutations in fibrillin-1 that result in enhanced TGF-β signaling. Increased body size and fat deposition in MAGP-1-mutant animals are particularly intriguing given the localization of obesity traits in humans to the region on chromosome 1 containing the MAGP-1 gene.
Microfibrils are important contributors to the structural integrity of tissues and participate in the assembly of elastic fibers during development. They also serve to bind and sequester growth factors in the extracellular matrix (ECM)3 (1, 2) and can directly signal cells through sequence motifs that interact with integrins and other cell-surface receptors (3–6). The major structural components of these microfibrils are the fibrillins, large glycoproteins rich in calcium binding epidermal growth factor-like domains (7), and the MAGPs, small, cysteine-rich proteins of unknown function. Other proteins can be localized to microfibrils, but it is not clear whether they are integral or associated proteins (8).
MAGP-1, a ∼20-kDa glycoprotein, is synthesized by most matrix-producing cells (9). The N-terminal half of the molecule contains sites for tyrosine sulfation and transglutaminase cross-linking as well as all of the tri- and tetrasaccharide O-linked sugars (10). The first 20 amino acids of the protein are enriched in acidic residues that, together with the sulfotyrosines, create a region of high negative charge capable of binding cationic proteins. The C-terminal half contains all of the molecule's thirteen cysteine residues and encodes a 54-amino acid sequence that defines a matrix-binding domain that targets MAGP-1 to the ECM. MAGP-1 binds to tropoelastin and type VI collagen (11, 12) and interacts with other molecules with defined structural roles in the ECM such as fibrillin-1 and -2 (13, 14), decorin (15), and biglycan (16). It does not, however, bind to the interstitial collagens I, III, or V (12). MAGP-1 also interacts with and facilitates the shedding of Notch1 (17), but there is no evidence for interaction with integrins.
MAGP-2 is the other member of the MAGP family and, like MAGP-1, is covalently associated with fibrillin-containing microfibrils (18). MAGP2 exhibits a more restricted tissue and developmental pattern of distribution, suggesting that it has a more specialized role than MAGP-1 in microfibril biology (19). Comparison with MAGP-1 shows a structural similarity confined to the two penultimate exons that encode the first six of the seven cysteine residues of the matrix binding domain. Sequences outside of this region, however, are dissimilar, suggesting that the two proteins have different functions (18).
The gene for MAGP-1 (MFAP2) is located on chromosome 1p36 in human and 4qD3 in mouse. Ortholog sequences are present in amphibians and teleost fishes. It is a relatively small gene in a gene-dense region of the genome with a compact minimal promoter contained within a ∼340-bp sequence immediately upstream of the transcription start site (20). In mammals, the gene is split into 9 exons, of which exon 1 is entirely non-coding. MAGP-1 isoforms arise through species-specific alternative splicing and show restricted patterns of expression relative to the canonical full-length form (21, 22). In mouse development, MAGP-1 mRNA is found in mesenchymal and connective tissue cells, where it is easily detected as early as day 8.5 of development (23).
First isolated from elastin-rich tissues and localized to microfibrils in elastic fibers, MAGP-1 was proposed to play a role in elastin fiber assembly. Functional binding studies identified distinct sites on MAGP-1 for binding to tropoelastin and fibrillin-1 and -2, consistent with it being a bridging molecule that binds tropoelastin onto the microfibril during elastin assembly (24). Recent studies with fibrillin mutant mice, however, suggest that microfibrils play little role in the initial stages of elastin assembly and, instead, contribute to tissue development and function through interactions with growth factors (25, 26).
To investigate the function of MAGP-1 we disrupted the Mfap2 gene in the mouse. We show that MAGP-1 deficiency leads to a multiorgan phenotype that is of variable penetrance and influenced by mouse genetic background. Rather than the phenotypes expected for loss of an elastic fiber protein, the MAGP-1 phenotypes manifest as bone abnormalities, increased fat deposition, bleeding diathesis, and impaired response to injury. These traits suggest that MAGP-1 is dispensable for elastic fiber assembly but important for other processes of tissue homeostasis or differentiation. Although no human disease has been linked to the MAGP-1 gene, the increased fat and body size associated with MAGP-1 deficiency in mice are consistent with the localization of obesity traits to the region on human chromosome 1 containing the MAGP-1 gene (27–29).
The traits that define the MAGP-1-null phenotype are, largely, opposite those described for fibrillin-1-mutant mice and for humans with Marfan syndrome. These differences are particularly interesting in that the fibrillins and MAGP-1 are binding partners and components of the same microfibril. In sum, they suggest contrasting biological functions for the two proteins.
EXPERIMENTAL PROCEDURES
Antibodies—An antibody to mouse MAGP-1 (mMAGP-1) was generated against a mouse MAGP-1-glutathione S-transferase fusion protein expressed in bacteria and first purified over glutathione-Sepharose 4B (Amersham Biosciences) followed by C4 reversed-phase high-pressure liquid chromatography. This expression construct contains exons 2–6 of MAGP-1 fused to the coding sequence for glutathione S-transferase. An antibody to mouse fibrillin-2 (mFib2Gly) was generated against a truncated fibrillin-2–6His construct expressed in bacteria and purified using nickel-nitrilotriacetic acid (Qiagen) followed by reversed-phase high-pressure liquid chromatography. This construct contains exon 10 of mouse fibrillin-2, which encodes the glycine-rich domain unique to this protein. The MRT6–17 antibody was previously described (30).
Generation of MAGP-1-deficient Mice—A 5460-bp KpnI fragment of the murine Mfap2 gene (a gift from Dr. Jeffrey Bonadio) was subcloned into pBluescript. This subclone was digested with NdeI and BbsI to remove a 1264-bp internal fragment containing exons 3–6 of Mfap2. The linearized vector containing 5′- and 3′-flanking sequences was made blunt and ligated with a blunt-ended neomycin cassette from pGK-Neo-bpA. This resulted in a targeting vector with 1.2 kb of 5′ homology, 2.5 kb of 3′ homology, and the neomycin cassette oriented toward the 5′-end of Mfap2. The targeting vector (30 mg) was electroporated into 129/SvJ mouse embryonic stem (ES) cells that were then plated on irradiated mouse embryonic fibroblasts. Selection for ES cells incorporating the construct was by treatment with G418 for 5 days. G418-resistant ES colonies were screened by Southern blot of XbaI-digested genomic DNA.
Two ES clones positive for recombination were injected into C57Bl/6 blastocysts with one clone yielding chimeric mice with germ line transmission of the targeted allele. These mice were initially bred with Black Swiss mice (BSw, Taconic) to take advantage of the better parenting and large litter sizes found in this background. Once the line was stabilized, MAGP-targeted (MAGP-1-/-) mice were backcrossed for seven generations into the C57Bl/6 (Jackson Laboratories) strain background then outbred for seven generations back into BSw. All mice were housed in a pathogen-free animal facility, fed standard chow ad libitum, and treated following animal protocols approved by the Washington University Animal Studies Committee.
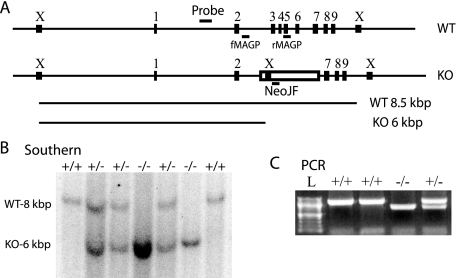
Genotype Analysis—Genomic DNA was obtained from mouse tails by a protocol consisting of protease K digestion, phenol/chloroform extraction, and ethanol/salt precipitation. For Southern blot analysis, DNA was digested with XbaI restriction enzyme (Roche Applied Science), separated by agarose gel electrophoresis, and transferred to Hybond N+ (Amersham Biosciences) membrane. MAGP-1 genomic probe was generated by PCR using primers gMAGP5 (5′-ATCCTGGACATAGCCTGTGC-3′) and gMAGP3 (5′-AACCACATGCGTTCAGCTC-3′) and mouse genomic DNA as template. The probe, a 330-bp product representing sequence 690 bp 5′ upstream of exon 2 of Mfap2 (Fig. 1A), was 32P-labeled and hybridized with membrane overnight at 65 °C. The membrane was washed and exposed to autoradiography film. The predicted band sizes are 8.5 and 6 kbp for wild-type and MAGP-1-/- fragments, respectively. For PCR analysis, DNA was mixed with genotyping primers NeoJF (5′-CTCGTGCTTTACGGTATCGC-3′), fMAGP (5′-ACAGTCTTGGCCTTTGAGGG-3′), rMAGP (5′-ACTGGAACTGCTCTTCAGGG-3′), 5 pmol each, to a 25-μl volume and added to Ready-To-Go PCR beads (Amersham Biosciences). Cycling parameters were 95 °C for 5 min followed by 30 cycles of 95 °C for 30 s, 62 °C for 30 s, 72 °C for 60 s, and a final extension of 72 °C for 10 min. The predicted band sizes were 1160 and 714 bp for wild-type and MAGP-1-/- product, respectively (Fig. 1A).
FIGURE 1.
Generation of MAGP-1 deficient mice. A, schematic of the Mfap2 locus in wild-type (WT) and MAGP-1-/- (KO) animals demonstrating mRNA disruption and genotyping strategies. Numbered rectangles correspond to Mfap2 exons 1–9, the open rectangle corresponds to the inserted neomycin resistance cassette. Short horizontal lines indicate the location of the Southern blot probe and PCR primers, boxes marked with “X” indicate XbaI restriction sites. Long horizontal lines represent fragments hybridizing to the Southern blot probe, 8.5 and 6 kbp for wild-type (WT) and MAGP-1-/- (KO), respectively. All distances, except the length of PCR primers, are to scale. B, XbaI-digested genomic DNA samples from a MAGP-1+/- cross were separated by agarose gel electrophoresis and detected by Southern blot. Resulting genotypes are indicated above each lane. C, genomic DNA was submitted to PCR with genotyping primers and separated by agarose gel electrophoresis. The predicted band sizes are 1160 and 714 bp for wild-type and MAGP-1-/-, respectively. L, 100-bp ladder.
RNA Preparation and Northern Analysis—Total RNA was isolated from mouse lung using the isothiocyanate-acid phenol method. The integrity of the RNA was checked by electrophoretic separation of 15 μg of total RNA fractionated in 1.2% agarose, 2.2 m formaldehyde gels and visualized by ethidium bromide staining; migration distances of 18 S and 28 S rRNA were noted. For Northern analysis, RNA was transferred to Hybond N+, hybridized with radioactively labeled full-length mouse MAGP-1A cDNA, and exposed to autoradiography film.
Semi-quantitative Reverse Transcription-PCR of Alternatively Spliced MAGP-1 mRNAs—To ascertain the relative levels of full-length (MAGP-1A) and alternatively spliced (MAGP-1D) mRNAs, a semi-quantitative reverse transcription-PCR was performed in which total lung RNA obtained from newborn animals was used as template. The MAGP-1D splice form found in mice is composed of exons 1 and 2 spliced to exons 7–9 (21). First strand cDNA was synthesized from 1 μg of total RNA using 0.5 μg of oligo(dT) as primer and 2.5 units of SuperScript II reverse transcriptase (Invitrogen) at 23 °C for 10 min followed by 50 min at 50 °C. 2 μl of cDNA was used as a template for a 25-μl PCR reaction, with 10 pmol each of the primers OMM-5 (5′-CGCACTAGAGCACAGCGCAGAC-3′) and OMM-UTR (5′-CACAGCTCCTGGCACAGGAGGC-3′), 0.2 mm each of the four dNTPs, and 0.5 unit of TaqDNA polymerase (Roche Applied Science). To ensure that the PCR reactions were in the exponential phase of amplification and would accurately reflect the relative levels of the alternatively spliced mRNAs, after an initial denaturation step of 2 min at 94 °C, only 20 cycles of denaturation at 94 °C for 1 min, annealing at 57 °C for 1 min, and extension at 72 °C for 1 min were performed. PCR products were then separated in 1.2% agarose gels, transferred to Hybond N+, and detected by Southern blot using mouse MAGP-1A cDNA as a probe. Identities of the hybridizing bands were ascertained by repeating the PCR using 40 cycles of amplification and DNA sequencing of the ethidium bromide-visualized products.
Bone and Fat Phenotypes in MAGP-1-/- Mice—Three- or six-month-old male BSw mice were weighed prior to euthanization with isoflurane. After dissecting into the peritoneum, both epididymal fat pads were isolated and weighed after removal of the testis and epididymis. Percent body weight of epididymal fat was calculated as % = 100 * [fat pad weight (g)/body weight (g)].
Limb bone lesions in adult mice were assessed by palpation. The mice were then sacrificed and dissected to remove the majority of soft tissue. For full skeletal preparation, the skeleton was cleared of remaining soft tissue in potassium hydroxide (2%), stained with alizarin red (0.015%), and preserved in 1:1 95% ethyl alcohol:glycerol.
Dermal Wounding Assay—Three-month-old male BSw mice were sedated with 1.0% isoflurane, shaved dorsally, and immobilized under a dissecting microscope. A 6-mm biopsy punch was used to excise a full-thickness piece of dorsal skin. The wound area was photographed using a dissecting microscope equipped with a QICAM Fast 1394 camera and Q-Capture software (QImaging). The mice were then allowed to recover in individual cages. Images were taken immediately after the initial surgery and during the healing process on sedated mice to monitor wound size. Using ImageJ software (National Institutes of Health), borders of each wound were outlined manually, and the interior area was measured and expressed as the percentage of the area of an ideal 6-mm-diameter circle at the same magnification.
Tissue Preparation/Histology—All dissected tissues were fixed in 10% formalin in phosphate-buffered saline (PBS). Lungs were first inflated to 25 cm of water with fixative and tied off at the trachea before processing with the other tissues. Dorsal skin was pressed flat between two pieces of laboratory wipe and two histology sponges in a histology cassette for the fixation step. Bone was decalcified in 14% EDTA, 10% NH4OH, pH 8.0, for 2 weeks after fixation, with solution changed regularly until the bone was pliable. Fixed tissues were dehydrated for 20 min each in 30%, 50%, and 70% ethyl alcohol and then paraffin-embedded by the Washington University School of Medicine Histology and Microscopy Core Facility. Sections, 5 μm thick, were obtained and processed for hematoxylin & eosin staining. Electron microscopy of arterial sections was as described by Davis (31).
Analysis of Vascular Physiology—Fourteen-week-old BSw male mice were used for blood pressure, heart rate, and vascular pressure/diameter measurements as previously described (32). Briefly, animals were sedated with 1.0% isoflurane and then restrained on a heated holder to maintain body temperature. A Millar pressure transducer was inserted into the right carotid artery and moved to the aorta where heart rate and systolic and diastolic blood pressure were monitored. The aorta was then dissected from the animal and separated into ascending, thoracic, and abdominal segments that were removed along with the left carotid artery to physiological buffer. Vessels were cannulated and subjected to transmural pressures from 0 to 175 mmHg at intervals of 25 mmHg. Outer diameters were recorded at each pressure value as measured by video imaging of the vessel.
Desmosine Analysis—Aortae were dissected from neonatal mice and hydrolyzed overnight in 6 n constant boiling HCl. Hydrolysates were dried, resuspended in ultrapure water, filtered to remove insoluble material, and analyzed for desmosine content using a radioimmunoassay (33).
Generation of Neonatal Skin Fibroblasts and Indirect Immunofluorescence—Neonatal mice were euthanized with carbon dioxide. Pups were transferred to a tissue culture hood and rinsed once with water and twice with 70% ethanol. The skin was removed to a sterile p100 tissue culture dish, minced well with a razor, allowed to attach, and cultured in growth media (Dulbecco's modified essential growth medium supplemented with 10 units/ml penicillin, 10 units/ml streptomycin, 1 μg/ml gentamycin, 2 mm sodium pyruvate, non-essential amino acids (Mediatech), and 10% fetal bovine serum (BioMeda)). Explanted mouse skin fibroblasts were harvested by trypsin/EDTA (Mediatech).
Mouse skin fibroblasts were plated on a Permanox LabTek chamber slide (Nalge Nunc). Cells were rinsed three times with PBS and fixed with cold (-20 °C) methanol twice for 2 min apiece. After rinsing three times with PBS, cells were incubated 10 min in blocking buffer (PBS, pH 7.4, 0.1% Tween 20, 1% coldwater fish gelatin) followed by 1 h in primary antibody solution (1:500 primary antibody in blocking buffer). Cells were washed three times with blocking buffer then incubated 1 h with a 1:1000 dilution of Alexa 594-conjugated secondary antibody. Finally, the cells were washed three times in blocking buffer, three times in PBS, mounted on a slide using Gel/Mount (BioMeda), and visualized using indirect immunofluorescence.
Cloning and Expression of Mouse MAGP-1 Protein—An SphI restriction site was added to the 5′-end, and an SacI restriction site was added to the 3′-end of mouse Mfap2 (IMAGE clone 7496676) using PCR amplification with the Mfap2 primers listed in Table 1. The PCR product was cut with SphI and SacI and purified then ligated into similarly cut and purified pQE31 (Qiagen). The ligation product was used to transform the M15 strain of Escherichia coli, and a colony producing MAGP-1 protein was selected. The sequence of the resulting clone was verified. The resulting protein included a 6-His tag (MRGSHHHHHHTDPHA) at the N terminus of the full-length mature protein (QGQYDLD...). For large scale expression and purification, 6HIS-muMAGP-1A was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside and purified using denaturing conditions on nickel-nitrilotriacetic acid agarose according to the manufacturer's instructions (Qiagen, Valencia, CA). The nickel-purified protein was desalted using a 1.5 × 35 cm Sephadex G-25 column (GE-Healthcare) then lyophilized. The N terminus of the purified protein was verified by sequencing using an ABI 473A protein sequencer.
TABLE 1.
Phenotypes observed in mixed strain and Black Swiss (BSw) MAGP-1-deficient mice Penetrance of phenotypes is described qualitatively. “Increased Fat Deposition” was scored as >2% epididymal fat; “Bone Lesions” represents the presence of at least one palpable limb lesion.
| Phenotype | Penetrance in mixed | Penetrance in BSw |
|---|---|---|
| Increased body length | High | Moderate |
| Increased body weight | High | High |
| Increased fat deposition | High | High |
| Male sterility | Moderate | Very low |
| Kyphosis/scoliosis | Moderate | Moderate |
| Bone Lesions | Low | Low |
Direct Binding Enzyme-linked Immunosorbent Assay—Costar 96-well plates (Costar 3590) were coated with 35 μl of 0.5 μg/ml recombinant human TGF-β1 (R&D Systems) or 5 μg/ml for BMP-7 (a gift from Dr. Keith Hruska, Washington University School of Medicine), recombinant bovine tropoelastin (34), and bovine serum albumin diluted in 10 mm carbonate buffer, pH 9.6. The plates were washed and blocked with PBS-milk-gelatin (PBS, 1% nonfat dry milk, 1% fish gelatin, and 0.01% Tween 20). Purified full-length, 6-His tagged MAGP-1 was diluted in PBS-milk-gelatin to various concentrations then added to the plates (50 μl per well) and incubated overnight at 4 °C. The plates were washed with PBS and incubated for 1 h at room temperature with rabbit polyclonal anti-bovine MAGP-1 antiserum (1:1000 dilution). Bound antibody was detected with horseradish peroxidase-conjugated secondary antibodies (1:2000 diluted donkey anti-rabbit IgG, horseradish peroxidase-linked whole antibody (GE Healthcare)). The amount of secondary antibody bound in each well was determined colorimetrically with the peroxidase substrate 2,2′-azino-di-(3-ethyl-benzthiaaoline-6-sulfonate (KPL Laboratories) at 410 nm.
Surface Plasmon Resonance Analysis—Analysis of the interaction between mouse MAGP-1, TGF-β1, and BMP-7 was performed on a Biacore X. A CM-5 sensor chip (Biacore BR-1000-140) was activated with 100 μl of 0.2 m 1-ethyl-3(3-dimethylaminopropyl)carbodiimide, 0.05 m N-hydroxysuccinimide, washed with 100 μl of water then coupled with 100 μl of 100 μg/ml MAGP-1 in 10 mm sodium acetate, pH 4.5. This coupling was ∼8700 relative units. Excess reactive sites were blocked with 70 μl of 1 m ethanolamine, and the chip was precycled with 200 μl of HBST (10 mm HEPES, 150 mm NaCl, 0.01% Tween 20) 100 μl of 0.2 m glycine, pH 2.3, and 200 μl of HBST. The analytes used for this study were carrier-free human recombinant TGF-β1 (R&D Systems), carrier-free active BMP-7, and lysozyme (Sigma). Lysozyme was used as the control because of its similarity to TGF-β1 and BMP-7 in size, pI, hydrophobicity, and overall amino acid composition. The analytes were dissolved in HBST at the designated concentrations, and 100 μl was injected for each analysis. Association and dissociation curves were monitored for 2000 s before the chip was regenerated with 50 μl of 0.2 m glycine, pH 2.3. After regeneration, the baseline was allowed to stabilize for 180 s before the next injection. The curves were analyzed using BIAevaluation version 4.1.
RESULTS
MAGP-1-deficient Mice Are Viable and Fertile—Homologous recombination was used to disrupt the MAGP-1 gene (Mfap2) by replacing exons 3–6 with a neomycin resistance cassette (Fig. 1A). Deletion of these exons removes 83 of 185 residues, including the signal sequence, which results in a functional knock-out of MAGP-1. Germ line transmission of the mutant Mfap2 allele was confirmed by Southern blot and PCR genotyping analysis. Southern blots show a shorter 6-kbp XbaI fragment hybridizing to the probe in the targeted allele as opposed to the 8.5-kbp fragment in the wild-type allele, consistent with the additional XbaI site present in the neomycin cassette (Fig. 1B). PCR used a common forward primer in intron 2 (fMAGP) and reverse primers located in exon 5 (rMAGP) and the neomycin cassette (NeoJF) exclusive to each allele. Different sized PCR products indicated the presence of the wild-type or targeted allele of Mfap2 (Fig. 1C).
Mice heterozygous for the mutation had no apparent phenotype when compared with their wild-type littermates. Breeding of heterozygote mice resulted in a Mendelian distribution of progeny, indicating that Mfap2 was not an essential gene for embryonic development in the mouse. Homozygous mutant mice survive weaning, live a normal lifespan, are fertile, and give normal sized litters.
Northern blotting of tissue from wild-type and homozygous mutant mice confirmed that full-length MAGP-1 mRNA was produced only in wild-type mice (Fig. 2A). Two bands were identified in wild-type animals but were absent from the knock-out. The major band, which migrates faster than 18 S rRNA, is estimated to be 1.3 kbp and is consistent with the previously published size of the full-length transcript (MAGP-1A) (23). The minor, slower migrating band is visible only on an overexposed autoradiogram and is consistent with unprocessed transcript.
FIGURE 2.
No full-length MAGP-1A transcript in MAGP-1-/- mice. A, total RNA from mouse lung was separated by agarose/formaldehyde electrophoresis and detected by Northern blot with a MAGP-1A probe. “18S” and “28S” represent migration distance of 18 S and 28 S rRNA, respectively. As expected, no mRNA for full-length MAGP-1A was detected. B, total lung RNA was reverse transcribed and PCR-amplified using mouse MAGP-1-specific primers. Exponential phase products were separated by agarose gel electrophoresis and detected by Southern blot. Indicated are transcripts for MAGP-1A and MAGP-1D mRNA, as confirmed by sequencing of parallel samples amplified past the exponential phase.
RNA expression was analyzed further using reverse transcription-PCR/Southern blotting. RNA from wild-type and homozygous mutant mouse lung was used as a template for reverse transcription-PCR using primers located in exons 1 and 9 of Mfap2. PCR products representing the exponential phase of amplification were detected by Southern blot and identified by sequencing (Fig. 2B). This technique revealed full-length transcript (MAGP-1A) at 1 kbp in wild-type animals as well as a smaller RNA at 700 bp that corresponds to the alternate splice form MAGP-1D (exons 1–2 spliced to exons 7–9) (35), albeit at levels <0.1% of MAGP-1A. The smaller, but not the full-length, transcript was also found in homozygous mutant animals, suggesting that skipping occurred around the neomycin-resistance cassette to produce the MAGP-1D isoform. Because MAGP-1D is expressed at extremely low levels, lacks a signal sequence, and will not be secreted, the animal homozygous for this targeted allele is essentially a functional knock-out for MAGP-1.
MAGP-1-/- Phenotypes Are Strain-dependent—Initially, the MAGP-1-/- mice were in a complex mixed background with strain contributions from 129/SvJ, C57Bl/6, and BSw. Several phenotypes were noticed in mixed strain animals, including increased body size, bone abnormalities, male sterility associated with testicular hydrocele and inverted seminal vesicles, and increased peritoneal and retroperitoneal fat. All of these phenotypes were expressed with variable penetrance (Table 1).
In an attempt to stabilize the phenotypes, mixed strain mice were inbred for seven generations into the C57Bl/6 background. Knock-outs in the C57 background, however, no longer displayed several of the traits identified in the mixed MAGP-1-/- line, including enhanced body size, bone, and fat phenotypes. These phenotypic differences between the mixed and C57 backgrounds suggest a strain-dependent response to the MAGP-1-/- allele. This was confirmed by breeding the mutation for seven generations into the BSw background, which restored most of the original phenotypes, including bone abnormalities and large body size.
MAGP-1 Deficiency Results in Bone Lesions—The mixed MAGP-1-/- line showed a spectrum of bone abnormalities, including infrequent mild kyphosis and scoliosis, abnormal rib cage formation, and lesions of the long bone (Fig. 3A). Rib deformation and scoliosis returned at a similar moderate penetrance in the BSw animals and were often present in the same mouse. No syndactyly, polydactyly, or arachnodactyly were seen.
FIGURE 3.
Bone phenotypes in MAGP-1-/- mice. A, the skeleton of an affected MAGP-1-/- mixed strain mouse shows mild kyphosis of the spine as indicated by the arrowhead. Bone lesions on the front and hind limbs are indicated with arrows. B, a hematoxylin & eosin-stained cross-section of unaffected, mid-shaft decalcified tibial bone from a 3-month-old MAGP-1-/- mouse showing intact cortical bone (CO) surrounding bone marrow (MA). C, low power section through a lesional area. D, higher magnification of a lesional region showing new bone formation with chondrocytes (CR) and trabecular bond (TR) associated with a cortical bone (CO) fracture common to the lesional areas. E, section from lesional area showing varying stages of bone formation with chondrocytes and trabecular bone outside of cortical bone. Resorption of cortical bone (arrowhead) and formation of new trabecular bone is occurring in the marrow cavity that is largely devoid of marrow cells. Scale bar = 100 μm. F–I, micro-computed tomography of legs excised from 10-month-old wild-type (F and H) and MAGP-1-/- (G and I) mice viewed medially. Focal areas of low bone density in MAGP-1-/- animals are indicated with arrows. Scale bar = 1 mm.
As young as 3 months of age, mixed background and BSw mice were identified with substantial endochondral bone lesions. Histological analysis showed that the lesions were most often associated with bone fractures, suggesting abnormal callus formation. Unlike the tibia and fibula of the contralateral leg, which contained normal cortical bone and marrow (Fig. 3B), the tibial and fibular lesional areas exhibited many stages of bone formation, including the presence of cartilage and trabecular bone, both inside and outside of the cortical bone (Fig. 3, C–E). Also present in the lesional areas were chondrocytes and osteoblasts as well as tartrate-resistant acid phosphatase-positive osteoclasts. None of these bone-remodeling cells were found in the similar location in normal tibial bone.
To address whether there was a mechanical basis for the predisposition of MAGP-1-/- bones to fracture, the legs of wild-type and MAGP-1-/- BSw mice were examined by micro-computed tomography. Measurements of femur length and mid-shaft thickness of cortical bone showed no differences between the genotypes when normalized to body weight. However, with high frequency the MAGP-1-/- tibias showed focal areas of reduced bone density visualized as “holes” on the micro-computed tomography image (Fig. 3, F–I). These areas could constitute weak points in the bone that lead to fracture. Small lesions such as these, or lesions located elsewhere, will go unnoticed when the limbs are examined by palpation, meaning that our low penetrance estimate for bone lesions is likely an underestimate (Table 1). None of these bone phenotypes were observed in wild-type BSw mice or in the C57 animals.
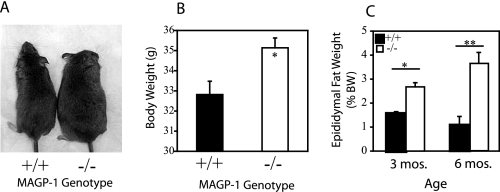
MAGP-1-/- Animals Are Larger and Have Increased Fat Deposition—Mixed strain MAGP-1-/- mice had increased body size and weight relative to wild-type animals in the same background (Fig. 4A). Specific differences were also documented in bone length and body fat in this background. C57 MAGP-1-/- mice showed no differences in body size, weight, or fat content relative to wild types. In contrast, the body weight of BSw MAGP-1-/- animals was significantly elevated compared with wild-type BSw siblings (Fig. 4B).
FIGURE 4.
MAGP-1 deficiency leads to increased body size and fat deposition. A, side-by-side comparison of representative wild-type (left) and MAGP-1-/- (right) mixed strain mice demonstrates that MAGP-1-/- mice are larger than age-matched wild-types. B, graph showing that BSw MAGP-1-/- male mice have significantly higher body weight at 3 months of age than age-matched BSw wild-type animals. *, p < 0.01 for wild-type versus MAGP-1-/-; error bars indicate mean ± S.E. (n = 34). C, BSw MAGP-1-/- mice have increased fat deposition relative to wild-types. Epididymal fat pads were harvested from 3- and 6-month-old BSw male mice. Fat pad ratio was calculated as fat pad weight (g) divided by body weight (BW, g) and expressed as a percentage. * and ** indicate p < 0.02 and p < 1 × 10-6, respectively, for wild-type versus MAGP-1-/-; error bars indicate mean ± S.E. (n = 38).
One contribution to the increased size is an increase in overall adiposity found in the mixed strain and BSw MAGP-1-/- mice. As a representative measure of fat deposition, epididymal fat pads were harvested from 3- and 6-month-old male BSw mice and weighed. At both adult ages, MAGP-1-/- mice had significantly higher body fat percentages compared with wild types, even after normalization to an increased body weight (Fig. 4C). This phenotype of increased fat deposition was highly penetrant (Table 1).
Elastin-rich Tissues Have Normal Structure and Function in MAGP-1-/- Animals—Because MAGP-1 is an integral component of microfibrils associated with elastic fibers, we expected major changes in the architecture and physiology of elastic tissues (skin, lung, and aorta) in the MAGP-1-/- mouse. Surprisingly, a thorough investigation of these tissues demonstrated that this was not the case in any of the genetic backgrounds. The skin of MAGP-1-/- mice was grossly normal with no apparent skin laxity, a trait present in many other elastic fiber-deficient mouse models and in humans with elastic fiber gene mutations. Histology of MAGP-1-/- skin sections showed the proper cellular composition of dermis and epidermis, although subdermal fat was noticeably increased. Inspection of the lungs from knock-out animals also failed to reveal any defects; MAGP-1-/- lungs have proper alveolar and airway structure, with no indication of an emphysematous phenotype (Fig. 5).
FIGURE 5.
MAGP-1-/- mice have proper elastic tissue structure and normal cardiovascular physiology. A, histology of lung and ascending aorta from adult BSw mice shows no obvious difference between wild-type and MAGP-1-/- genotypes. Scale bar = 100 μm. Normal wall structure in MAGP-/- mice is also visualized using electron microscopy (Aorta-EM). The lumen of the vessel is at the top. Scale bar = 10 μm.
Major blood vessels are also unaffected in MAGP-1-/- animals. The aortic trunk appeared grossly normal with no sign of stenosis, dilation, or tortuosity, and there was no histological difference in the appearance, number of lamellar layers, or thickness of the aortic wall (Fig. 5). The lamellar ultrastructure of the ascending aorta was also normal; it displayed thick, continuous elastin deposition between properly oriented vascular smooth muscle cells.
As a functional measurement of the performance of the vascular system in the MAGP-1-/- animals, we determined both blood pressure and vascular compliance. MAGP-1-/- mice showed no difference from wild types in heart rate (655 ± 9, 621 ± 19 bpm for heart rate) or blood pressure (Fig. 6A). Pressure/diameter relationships as an indication of vascular compliance also showed no difference between genotypes for the four main arterial segments (Fig. 6B). Finally, desmosine levels in wild-type and MAGP-1-/- vessels were compared as a measurement of elastin content. No difference was found between the genotypes (1815 ± 189 and 1688 ± 125 pmol of desmosine per mg of protein for wild-type and MAGP-1-/-, respectively, n = 12), suggesting that elastin cross-linking proceeds normally in the absence of MAGP-1.
FIGURE 6.
Cardiovascular physiology in MAGP-1-/- mice. A, systolic and diastolic blood pressures are similar for wild-type and MAGP-1-/- genotypes. B, the pressure/diameter relationship of MAGP-1-/- arterial vessels is comparable to that of wild-type, showing that vascular compliance is not affected by the absence of MAGP-1. Data are plotted as outer diameter of vessel versus applied transmural pressure. No data point showed a significant difference between genotypes; error bars indicate mean ± S.E. (n = 11).
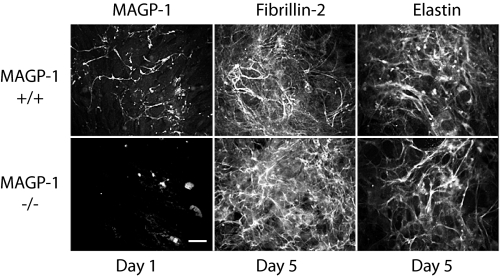
Skin Fibroblasts from MAGP-1-/- Mice Assemble an Elastin Matrix—To confirm proper elastic fiber assembly in the absence of MAGP-1, we used an in vitro model of elastic fiber formation. Mouse skin fibroblasts were explanted from the skin of neonatal BSw MAGP-1-/- and wild-type mice and were grown as primary cultures. Wild-type mouse skin fibroblasts deposited detectable MAGP-1 matrix 1 day after reaching confluence, but at this time point, no fibrillin-2 or tropoelastin was evident (Fig. 7). Between 5 and 7 days post-confluency extensive fibrillin-2 and tropoelastin networks were apparent in the extracellular matrix. Cells from MAGP-1-/- animals did not produce a MAGP-1 matrix, confirming inactivation of the MAGP-1 gene, but did produce fibrillin-2 and tropoelastin (Fig. 7). The similar appearance of fibrillin-2 and elastin in the extracellular matrix of both cell types shows that the microfibrillar and elastin fiber networks form normally in the absence of MAGP-1. We cannot exclude the possibility, however, that MAGP-2 compensates for MAGP-1 in facilitating elastic fiber assembly in these cells.
FIGURE 7.
MAGP-1-deficient skin fibroblasts produce an elastin-containing matrix lacking MAGP-1. Mouse skin fibroblasts from newborn wild-type and MAGP-1-/- mice were examined by indirect immunofluorescence using antibodies to MAGP-1, fibrillin-2, and elastin. MAGP-1 was detected in the ECM of wild-type cells as early as 1 day post confluency. By day 7, extensive fibrillin-1 and tropoelastin networks were apparent in the ECM. Fibrillin-1 and tropoelastin production followed a similar time course in MAGP-1-/- cells. As expected, cells from MAGP-1-/- animals do not produce a MAGP-1 matrix, confirming inactivation of the MAGP-1 gene. The similar appearance of fibrillin-2 and elastin in the extracellular matrix of both cell types shows that the microfibrillar and elastin fiber networks form normally in the absence of MAGP-1. Scale bar = 100 μm.
Delayed Wound Healing and Bleeding Diathesis in MAGP-1-/- Animals—To assess a possible role for MAGP-1 in tissue repair, we compared the healing of excisional dermal wounds in BSw MAGP-1-/- and wild-type mice. MAGP-1-/- animals have a delay in the wound healing process compared with wild-types, with a lag period of ∼7 days (Fig. 8A). Differences were most evident in the early phase (first 3 days) of wound closure. A more detailed characterization of this phenotype is currently in progress.
FIGURE 8.
MAGP-1 deficiency leads to a delay in the wound healing and prolonged bleeding. A, excisional wounds 6 mm in diameter were made on the dorsum of 3-month-old BSw male mice. Healing was followed by calculating wound size as percentage of the original 6-mm wound. **, p < 0.01 for wild-type versus MAGP-1-/-; error bars indicate mean ± S.E. (n = 27). B, MAGP-1 deficiency results in delayed thrombotic occlusion of the carotid artery. Blood flow in the common carotid artery was monitored continuously with an ultrasonic flow probe. Local endothelial injury was induced by application of a 540 nm laser beam to the carotid artery followed by injection of rose Bengal dye into the lateral tail vein. Shown are the times from the onset of injury until the cessation of flow. Tail vein bleeding time was determined by making a transverse incision over a lateral vein and immersing the tail in normal saline (37 °C) in a hand-held test tube. The time from the incision to the cessation of bleeding was recorded as the bleeding time. **, p < 0.01 for +/+ versus -/-. Error bars indicate mean ± S.E. (n = 24).
Another interesting phenotype that was fully penetrant in both the C57 and BSw backgrounds was a bleeding diathesis characterized by delayed thrombotic occlusion following carotid injury as well as prolonged bleeding from a tail vein incision (Fig. 8B). Normal occlusion times were restored when recombinant MAGP-1 was infused into deficient animals prior to vessel wounding. Blood coagulation was normal in these animals as assessed by activated partial thromboplastin time and prothrombin time. Platelet number was lower in MAGP-1-deficient mice, but the platelets showed normal aggregation properties in response to various agonists. MAGP-1 was not found in normal platelets or in the plasma of wild-type mice. A full characterization of this phenotype can be found in a separate publication (36).
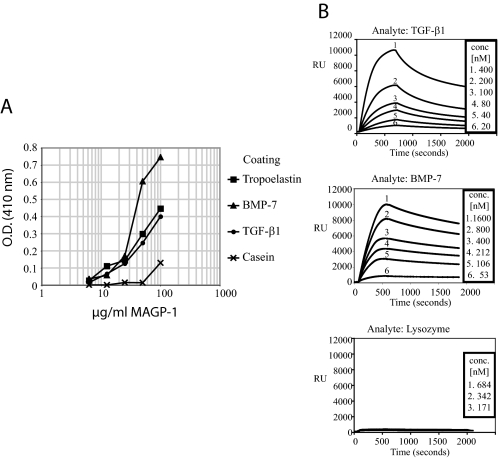
MAGP-1 Binds Active TGF-β1 and BMP-7—Many of the phenotypes observed in the MAGP-1-deficient mice suggest misregulation of TGF-β signaling pathways. Other laboratories have shown that pro-BMPs and the large latent complex of TGF-β bind to purified fibrillin-1 (1, 37–39) and localize to microfibrils in the ECM (40, 41). To determine if MAGP-1 also interacts with TGF-β family members, we performed direct solid phase binding assays with active TGF-β1 and BMP-7. Fig. 9A shows that MAGP-1 bound to both proteins in a dose-dependent fashion at concentrations equal to binding to tropoelastin, the positive control. Issues with solubility of MAGP-1 prevent extension of the curves to higher concentrations past the linear region of the binding curve.
FIGURE 9.
MAGP-1 interacts with active TGF-β and BMP-7. A, solid phase binding assay demonstrating an interaction between MAGP-1 and active TGF-β and BMP-7. The growth factors were coated onto wells of a microtiter plate and incubated with dilutions of recombinant MAGP-1. Bound MAGP-1 was detected using a MAGP-1-specific antibody after thorough washing to remove unbound protein. Wells coated with casein and tropoelastin served as negative and positive controls, respectively. B, surface plasmon resonance experiments with immobilized MAGP-1 and BMP-7 and TGF-β1 in solution. Sensorgrams show binding of each growth factor at various concentrations to MAGP-1. TGF-β1 interacted with MAGP-1 with an affinity of 19.9 ± 6 nm (S.E., n = 16) and a dissociation rate of 4.0 × 10-3 (1/s) (C, top). BMP-7 bound to MAGP-1 with a KD of 10.5 ± 4.3 nm (S.E., n = 13) (C, middle). The dissociation rate constant was 2.6 × 10-4 (1/s). Lysozyme showed no measurable interaction with the MAGP-1-coated sensor chip (C, bottom).
Surface plasmon resonance technology was used to determine the affinity of the interaction between MAGP-1 and TGF-β1 or BMP-7 (Fig. 9B). TGF-β1 binding to MAGP-1 was dose-dependent with an affinity of 19.9 ± 6 nm (S.E., n = 16) and a dissociation rate of 4.0 × 10-3 (1/s) (Fig. 9B, top). BMP-7 bound to MAGP-1 with a KD of 10.5 ± 4.3 nm (S.E., n = 13) (Fig. 9B, middle). The dissociation rate constant was 2.6 × 10-4 (1/s). It is interesting that both TGF-β1 and BMP-7 bind MAGP with similar affinities even though the dissociation rate is nearly 10-fold different. Lysozyme showed no measurable interaction with the MAGP-1-coated sensor chip (Fig. 9B, bottom).
DISCUSSION
MAGP-1 was first identified as a component of elastic fiber microfibrils (42) and together with the fibrillins was thought to be necessary for elastin assembly (43). Hence, inactivation of the MAGP-1 gene was expected to primarily affect elastin-rich tissues such as skin, vasculature, and lung. Analysis of elastic tissues in MAGP-1-/- animals, however, showed an elastin content similar to wild-type animals. There was no apparent skin laxity and no noticeable defects in lung structure. Vascular development was normal and blood pressure and vascular compliance were similar to wild-type animals. Ultrastructural analysis of the aortic wall in MAGP-1-/- mice found no difference in fiber orientation, elastin density, or lamellar number when compared with wild-type samples. Taken together, these data suggest that MAGP-1 does not play an essential role in elastic fiber assembly. This conclusion was supported by in vitro studies documenting the formation of an extensive elastic fiber network in cultured MAGP-1-/- cells.
The absence of a detectable vascular phenotype in the MAGP-1-/- mouse is in contrast to changes that occur when MAGP-1 levels are modified in the zebrafish. Knock-down of MAGP-1 in zebrafish using MAGP-1 morpholinos results in vascular defects that include dilated vessels in the brain and the eyes, irregular lumens of axial vessels, and a dilated caudal vein with altered venous plexus formation (44). Although these studies suggest that MAGP-1 is required for maintaining the structural integrity of the vessel wall and for venous plexus morphogenesis in the fish, it is not clear why similar phenotypes were not found in the mouse. It is also interesting that fibrillin-1 and MAGP-1 morphant embryos exhibit overlapping vascular defects in zebrafish, which is clearly not the case in the mouse.
The multiorgan spectrum of phenotypes in the MAGP-1-/- mouse shows interesting contrasts with phenotypes associated with fibrillin-1 mutations. Depending on the genetic background, bone abnormalities in MAGP-1-/- mice include a tendency to bone fractures and abnormal callus formation. There is also an increase in total skeletal size, but this is in proportion to an increase in body size. This is in contrast to the disproportionate overgrowth of long bones associated with fibrillin-1 mutations in humans with Marfan syndrome (45) and in mice underexpressing fibrillin-1 (46). Another interesting trait of MAGP-1-/- animals is increased adiposity. Marfan syndrome individuals, in contrast, tend to have muscle and fat hypoplasia and are unable to increase muscle mass despite physical exercise (45, 47). Mutations in fibrillin-1 also lead to heart valve disease and aortic aneurysms (46, 48), whereas MAGP-1-/- mice have normal blood vessels and normal cardiovascular function. Other notable differences include a bleeding diathesis and slower wound healing in MAGP-1-/- mice not seen in humans or reported in mice with fibrillin-1 mutations. These differences are interesting given that fibrillin-1 and MAGP-1 are components of the same microfibril. Together, they suggest contrasting biological functions for the two proteins.
Phenotypes associated with mutations in microfibrillar proteins have been attributed to an inability of the mutant protein to bind and sequester growth factors, particularly TGF-β family members (26, 49–51). In evaluating MAGP-1 for biological function, we found that it can bind active, processed forms of BMP-7 and active TGF-β, which is in contrast to the latent forms of these growth factors that bind to the fibrillins (1, 38, 39). Furthermore, functional mapping studies show that MAGP-1 binds to fibrillin-1 in the same general region where pro-BMP and latent TGF-β bind (13, 52), suggesting that MAGP-1 may be an important modulator of fibrillin-mediated growth factor signaling by regulating how much, and perhaps which, growth factors bind to the microfibril. This raises the interesting possibility that the contrasting phenotypes observed for fibrillin-1 and MAGP-1 insufficiency arise as a result of their abilities to bind different forms of TGF-β growth factors. Alternatively, MAGP-1 could serve to sequester or concentrate active TGF-β or BMP close to reactive cells, could function to remove active growth factor from the cellular milieu, or could block binding of latent TGF-βs to fibrillin. Given the complex structure of the microfibril, it is important to consider that mutations within the fibrillin protein will have disruptive effects on microfibrils generally. Thus, a particular phenotype may not be directly related to changes in fibrillin but could be due to functional disruption of other proteins that are normally associated with fibrillin, such as MAGP-1.
TGF-β family members are known negative regulators of chondrocyte (53), muscle (54), and fat (55) cell differentiation. Hence, the excess fat, bone-associated cartilage, and muscle seen in the MAGP-1-/- mice are consistent with loss of TGF-β suppression of growth and/or differentiation of these cell types. TGF-β is also released by platelets during clotting and is an important regulator of wound healing (56, 57): two other processes that are affected in the MAGP-1-/- mouse. Studies by Cohn et al. (47) showed that excess TGF-β signaling is responsible for the lack of muscle regeneration in fibrillin-1-deficient mice, another example of how fibrillin-1 and MAGP-1 loss of function results in contrasting effects.
Although the phenotypic traits evident in the MAGP-1-/- mice are consistent with a function for MAGP-1 in growth factor regulation, we cannot exclude a possible direct effect of MAGP-1 independent of its role as a microfibrillar protein. The spectrum of MAGP-1 phenotypes, bone, fat, bleeding, and wound healing, resembles in many ways what has been found with deficiency of matricellular proteins. This family of proteins has also been termed modulatory adhesion molecules, because they serve primarily a cell modulatory rather than a direct structural role in the ECM. Matricellular deficiency in the mouse has led to a variety of phenotypes, including altered wound healing (58, 59), adiposity (58, 60), and foreign body response (61) as well as predisposition to bleeding diathesis (62). More studies are required to elucidate the molecular mechanisms behind MAGP-1 function.
No human disease has yet been directly associated with mutations in the MAGP-1 gene. However, recent studies have linked obesity phenotypes to a locus at 1p36 that includes MFAP2 and many other genes. In one study, a genome scan identified 1p36 as containing a quantitative trait locus for the phenotypes of obesity itself (27), whereas the second study used intermediate phenotypes to link 1p36 to obesity-associated with hypertension (29). Although we did not see hypertension in the MAGP-1-/- mice, we did see increased body size and enhanced adipogenesis, suggesting that MAGP-1 may contribute to obesity in humans. Obesity is a multifactorial trait that is influenced by a complex interplay of genes and environment (63). Furthermore, obesity is characterized by etiologic heterogeneity; meaning distinct sets of genes may contribute to its development in different individuals (29). This complexity in humans is similar to what we find in mice where many of the MAGP-1-/- phenotypic traits are mouse strain-dependent and incompletely penetrating. Therefore, definition of the loci and genes that account for variable expression of Mfap2 phenotypes in mice may eventually provide insights into genetic factors that may influence MFAP2 phenotypes in humans.
Acknowledgments
We thank Dave Schetler and Chris Ciliberto for technical assistance, Dr. Matthew Silva and Blaine Christensen for assistance with micro-computed tomography imaging, Dr. Barry Starcher for desmosine assays, Dr. Hideki Sugitani and Dr. Adrian Shifren for helpful discussion, and the Washington University School of Medicine Histology and Microscopy Core Facility for tissue processing. We also thank Dr. Douglas Tollefsen and Dr. Cristina Vicente for assistance with characterizing the bleeding phenotype, Dr. Jeffrey Arbeit and Robert Neumann for assistance with the dermal wound studies, and Dr. Keith Hruska for providing active BMP-7.
This work was supported, in whole or in part, by National Institutes of Health Grants HL53325, HL71960, HL74138 (to R. P. M.), HL54049 (to R. A. P.), and T32 HL007873 (to J. S. W.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: ECM, extracellular matrix; ES, embryonic stem cell; PBS, phosphate-buffered saline; TGF, transforming growth factor.
References
- 1.Sengle, G., Charbonneau, N. L., Ono, R. N., Sasaki, T., Alvarez, J., Keene, D. R., Bachinger, H. P., and Sakai, L. Y. (2008) J. Biol. Chem. 283 13874-13888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Charbonneau, N. L., Ono, R. N., Corson, G. M., Keene, D. R., and Sakai, L. Y. (2004) Birth Defects Res. C. Embryo Today 72 37-50 [DOI] [PubMed] [Google Scholar]
- 3.Ritty, T. M., Broekelmann, T., Tisdale, C., Milewicz, D. M., and Mecham, R. P. (1999) J. Biol. Chem. 274 8933-8940 [DOI] [PubMed] [Google Scholar]
- 4.Sakamoto, H., Broekelmann, T., Cheresh, D. A., Ramirez, F., Rosenbloom, J., and Mecham, R. P. (1996) J. Biol. Chem. 271 4916-4922 [PubMed] [Google Scholar]
- 5.Withers, A. P., Burl, S., Faed, M. J. W., D'Arrigo, C., and Boxer, M. (1994) J. Med. Genet. 31 174 [Google Scholar]
- 6.Pfaff, M., Reinhardt, D. P., Sakai, L. Y., and Timpl, R. (1996) FEBS Lett. 384 247-250 [DOI] [PubMed] [Google Scholar]
- 7.Sakai, L. Y., Keene, D. R., and Engvall, E. (1986) J. Cell Biol. 103 2499-2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kielty, C. M., Wess, T. J., Haston, L., Ashworth, J. L., Sherratt, M. J., and Shuttleworth, C. A. (2002) J. Muscle Res. Cell Motil. 23 581-596 [PubMed] [Google Scholar]
- 9.Gibson, M. A., and Cleary, E. G. (1987) Immunol. Cell Biol. 65 345-356 [DOI] [PubMed] [Google Scholar]
- 10.Trask, B. C., Broekelmann, T. J., and Mecham, R. P. (2001) Biochemistry 40 4372-4380 [DOI] [PubMed] [Google Scholar]
- 11.Brown-Augsburger, P., Broekelmann, T., Mecham, L., Mercer, R., Gibson, M. A., Cleary, E. G., Abrams, W. R., Rosenbloom, J., and Mecham, R. P. (1994) J. Biol. Chem. 269 28443-28449 [PubMed] [Google Scholar]
- 12.Finnis, M. L., and Gibson, M. A. (1997) J. Biol. Chem. 272 22817-22823 [DOI] [PubMed] [Google Scholar]
- 13.Werneck, C. C., Trask, B. C., Broekelmann, T. J., Trask, T. M., Ritty, T. M., Segade, F., and Mecham, R. P. (2004) J. Biol. Chem. 279 23045-23051 [DOI] [PubMed] [Google Scholar]
- 14.Trask, T. M., Crippes Trask, B., Ritty, T. M., Abrams, W. R., Rosenbloom, J., and Mecham, R. P. (2000) J. Biol. Chem. 275 24400-24406 [DOI] [PubMed] [Google Scholar]
- 15.Trask, B. C., Trask, T. M., Broekelmann, T., and Mecham, R. P. (2000) Mol. Biol. Cell 11 1499-1507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reinboth, B., Hanssen, E., Cleary, E. G., and Gibson, M. A. (2002) J. Biol. Chem. 277 3950-3957 [DOI] [PubMed] [Google Scholar]
- 17.Miyamoto, A., Lau, R., Hein, P. W., Shipley, J. M., and Weinmaster, G. (2006) J. Biol. Chem. 281 10089-10097 [DOI] [PubMed] [Google Scholar]
- 18.Hanssen, E., Hew, F. H., Moore, E., and Gibson, M. A. (2004) J. Biol. Chem. 279 29185-29194 [DOI] [PubMed] [Google Scholar]
- 19.Gibson, M. A., Finnis, M. L., Kumaratilake, J. S., and Cleary, E. G. (1998) J. Histochem. Cytochem. 46 871-885 [DOI] [PubMed] [Google Scholar]
- 20.Segade, F., and Mecham, R. P. (2005) Biochim. Biophys. Acta 1731 215-224 [DOI] [PubMed] [Google Scholar]
- 21.Segade, F., Suganuma, N., Mychaleckhj, J. C., and Mecham, R. P. (2007) Int. J. Biochem. Cell Biol. 39 2303-2313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Segade, F., Trask, B. C., Broekelmann, T., Pierce, R. A., and Mecham, R. P. (2002) J. Biol. Chem. 277 11050-11057 [DOI] [PubMed] [Google Scholar]
- 23.Chen, Y., Faraco, J., Yin, W., Germiller, J., Francke, U., and Bonadio, J. (1993) J. Biol. Chem. 268 27381-27389 [PubMed] [Google Scholar]
- 24.Brown-Augsburger, P. B., Broekelmann, T., Rosenbloom, J., and Mecham, R. P. (1996) Biochem. J. 318 149-155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramirez, F., and Dietz, H. C. (2007) J. Cell. Physiol. 213 326-330 [DOI] [PubMed] [Google Scholar]
- 26.Carta, L., Pereira, L., Arteaga-Soli, E., Lee-Arteaga, S. Y., Lenart, B., Starcher, B., Merkel, C. A., Sukoyan, M., Kerkis, A., Hazeki, N., Keene, D. R., Sakai, L. Y., and Ramirez, F. (2006) J. Biol. Chem. 281 8016-8023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu, Y. J., Xu, F. H., Shen, H., Liu, Y. Z., Deng, H. Y., Zhao, L. J., Huang, Q. Y., Dvornyk, V., Conway, T., Davies, K. M., Li, J. L., Recker, R. R., and Deng, H. W. (2004) J. Clin. Endocrinol. Metab. 89 875-882 [DOI] [PubMed] [Google Scholar]
- 28.Morris, B. J. (2005) Hypertension 46 1256-1258 [DOI] [PubMed] [Google Scholar]
- 29.Pausova, Z., Gaudet, D., Gossard, F., Bernard, M., Kaldunski, M. L., Jomphe, M., Tremblay, J., Hudson, T. J., Bouchard, G., Kotchen, T. A., Cowley, A. W., and Hamet, P. (2005) Hypertension 46 1280-1285 [DOI] [PubMed] [Google Scholar]
- 30.Kozel, B. A., Wachi, H., Davis, E. C., and Mecham, R. P. (2003) J. Biol. Chem. 278 18491-18498 [DOI] [PubMed] [Google Scholar]
- 31.Davis, E. C. (1993) Lab. Invest. 68 89-99 [PubMed] [Google Scholar]
- 32.Wagenseil, J. E., Nerurkar, N. L., Knutsen, R. H., Okamoto, R. J., Li, D. Y., and Mecham, R. P. (2005) Am. J. Physiol. Heart Circ. Physiol. 289 H1209-H1217 [DOI] [PubMed] [Google Scholar]
- 33.Starcher, B., Green, M., and Scott, M. (1995) Respiration 62 252-257 [DOI] [PubMed] [Google Scholar]
- 34.Broekelmann, T. J., Kozel, B. A., Ishibashi, H., Werneck, C. C., Keeley, F. W., Zhang, L., and Mecham, R. P. (2005) J. Biol. Chem. 280 40939-40947 [DOI] [PubMed] [Google Scholar]
- 35.Segade, F., Broekelmann, T. J., Pierce, R. A., and Mecham, R. P. (2000) Matrix Biol. 19 671-682 [DOI] [PubMed] [Google Scholar]
- 36.Werneck, C. C., Vicente, C. P., Weinberg, J. S., Shifren, A., Pierce, R. A., Broekelmann, T. J., Tollefsen, D. M., and Mecham, R. P. (2008) Blood 111 4137-4144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chaudhry, S. S., Cain, S. A., Morgan, A., Dallas, S. L., Shuttleworth, C. A., and Kielty, C. M. (2007) J. Cell Biol. 176 355-367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gregory, K. E., Ono, R. N., Charbonneau, N. L., Kuo, C. L., Keene, D. R., Bachinger, H. P., and Sakai, L. Y. (2005) J. Biol. Chem. 280 27970-27980 [DOI] [PubMed] [Google Scholar]
- 39.Isogai, Z., Ono, R. N., Ushiro, S., Keene, D. R., Chen, Y., Mazzieri, R., Charbonneau, N. L., Reinhardt, D. P., Rifkin, D. B., and Sakai, L. Y. (2003) J. Biol. Chem. 278 2750-2757 [DOI] [PubMed] [Google Scholar]
- 40.Dallas, S. L., Keene, D. R., Bruder, S. P., Saharinen, J., Sakai, L. Y., Mundy, G. R., and Bonewald, L. F. (2000) J. Bone Miner. Res. 15 68-81 [DOI] [PubMed] [Google Scholar]
- 41.Karonen, T., Jeskanen, L., and Keski-Oja, J. (1997) Br. J. Dermatol. 137 51-58 [PubMed] [Google Scholar]
- 42.Gibson, M. A., Hughes, J. L., Fanning, J. C., and Cleary, E. G. (1986) J. Biol. Chem. 261 11429-11436 [PubMed] [Google Scholar]
- 43.Gibson, M. A., Kumaratilake, J. S., and Cleary, E. G. (1989) J. Biol. Chem. 264 4590-4598 [PubMed] [Google Scholar]
- 44.Chen, E., Larson, J. D., and Ekker, S. C. (2006) Blood 107 4364-4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Judge, D. P., and Dietz, H. C. (2005) Lancet 366 1965-1976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pereira, L., Lee, S. Y., Gayraud, B., Andrikopoulos, K., Shapiro, S. D., Bunton, T., Biery, N. J., Dietz, H. C., Sakai, L. Y., and Ramirez, F. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 3819-3823 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cohn, R. D., van Erp, C., Habashi, J. P., Soleimani, A. A., Klein, E. C., Lisi, M. T., Gamradt, M., ap Rhys, C. M., Holm, T. M., Loeys, B. L., Ramirez, F., Judge, D. P., Ward, C. W., and Dietz, H. C. (2007) Nat. Med. 13 204-210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ng, C. M., Cheng, A., Myers, L. A., Martinez-Murillo, F., Jie, C., Bedja, D., Gabrielson, K. L., Hausladen, J. M., Mecham, R. P., Judge, D. P., and Dietz, H. C. (2004) J. Clin. Invest. 114 1586-1592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Arteaga-Solis, E., Gayraud, B., Lee, S. Y., Shum, L., Sakai, L., and Ramirez, F. (2001) J. Cell Biol. 154 275-281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Neptune, E. R., Frischmeyer, P. A., Arking, D. E., Myers, L., Bunton, T. E., Gayraud, B., Ramirez, F., Sakai, L. Y., and Dietz, H. C. (2003) Nat. Genet. 33 407-411 [DOI] [PubMed] [Google Scholar]
- 51.Zacchigna, L., Vecchione, C., Notte, A., Cordenonsi, M., Dupont, S., Maretto, S., Cifelli, G., Ferrari, A., Maffei, A., Fabbro, C., Braghetta, P., Marino, G., Selvetella, G., Aretini, A., Colonnese, C., Bettarini, U., Russo, G., Soligo, S., Adorno, M., Bonaldo, P., Volpin, D., Piccolo, S., Lembo, G., and Bressan, G. M. (2006) Cell 124 929-942 [DOI] [PubMed] [Google Scholar]
- 52.Jensen, S. A., Reinhardt, D. P., Gibson, M. A., and Weiss, A. S. (2001) J. Biol. Chem. 276 39661-39666 [DOI] [PubMed] [Google Scholar]
- 53.Pogue, R., and Lyons, K. (2006) Curr. Top. Dev. Biol. 76 1-48 [DOI] [PubMed] [Google Scholar]
- 54.Lee, S. J. (2007) PLoS ONE 2 e789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Choy, L., and Derynck, R. (2003) J. Biol. Chem. 278 9609-9619 [DOI] [PubMed] [Google Scholar]
- 56.Blakytny, R., Ludlow, A., Martin, G. E., Ireland, G., Lund, L. R., Ferguson, M. W., and Brunner, G. (2004) J. Cell. Physiol. 199 67-76 [DOI] [PubMed] [Google Scholar]
- 57.Brunner, G., and Blakytny, R. (2004) Thromb. Haemost. 92 253-261 [DOI] [PubMed] [Google Scholar]
- 58.Agah, A., Kyriakides, T. R., Lawler, J., and Bornstein, P. (2002) Am. J. Pathol. 161 831-839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Puolakkainen, P. A., Bradshaw, A. D., Brekken, R. A., Reed, M. J., Kyriakides, T., Funk, S. E., Gooden, M. D., Vernon, R. B., Wight, T. N., Bornstein, P., and Sage, E. H. (2005) J. Histochem. Cytochem. 53 571-581 [DOI] [PubMed] [Google Scholar]
- 60.Bradshaw, A. D., and Sage, E. H. (2001) J. Clin. Invest. 107 1049-1054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kyriakides, T. R., Rojnuckarin, P., Reidy, M. A., Hankenson, K. D., Papayannopoulou, T., Kaushansky, K., and Bornstein, P. (2003) Blood 101 3915-3923 [DOI] [PubMed] [Google Scholar]
- 62.Kyriakides, T. R., Zhu, Y. H., Smith, L. T., Bain, S. D., Yang, Z., Lin, M. T., Danielson, K. G., Iozzo, R. V., LaMarca, M., McKinney, C. E., Ginns, E. I., and Bornstein, P. (1998) J. Cell Biol. 140 419-430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Speakman, J. R. (2004) J. Nutr. 134 2090S-2105S [DOI] [PubMed] [Google Scholar]