Abstract
Expression of the genes for resistance to heavy metals and metalloids is transcriptionally regulated by the toxic ions themselves. Members of the ArsR/SmtB family of small metalloregulatory proteins respond to transition metals, heavy metals, and metalloids, including As(III), Sb(III), Cd(II), Pb(II), Zn(II), Co(II), and Ni(II). These homodimeric repressors bind to DNA in the absence of inducing metal(loid) ion and dissociate from the DNA when inducer is bound. The regulatory sites are often three- or four-coordinate metal binding sites composed of cysteine thiolates. Surprisingly, in two different As(III)-responsive regulators, the metalloid binding sites were in different locations in the repressor, and the Cd(II) binding sites were in two different locations in two Cd(II)-responsive regulators. We hypothesize that ArsR/SmtB repressors have a common backbone structure, that of a winged helix DNA-binding protein, but have considerable plasticity in the location of inducer binding sites. Here we show that an As(III)-responsive member of the family, CgArsR1 from Corynebacterium glutamicum, binds As(III) to a cysteine triad composed of Cys15, Cys16, and Cys55. This binding site is clearly unrelated to the binding sites of other characterized ArsR/SmtB family members. This is consistent with our hypothesis that metal(loid) binding sites in DNA binding proteins evolve convergently in response to persistent environmental pressures.
Arsenic, a toxic metalloid, is currently and has always been ranked first on the Superfund List of Hazardous Substances (available on the World Wide Web), in part because of its environmental ubiquity. As a consequence, nearly all organisms have genes that confer resistance to arsenic. Environmental arsenic is sensed by members of the ArsR/SmtB family of metalloregulatory proteins (1-3). These winged helix repressor proteins specifically bind to arsenic and other toxic metals. Consequently, they control expression of genes involved in arsenic biotransformation and efflux. For example, the ArsR repressor encoded by Escherichia coli plasmid R773 binds to the promoter region of its respective ars operon in the absence of As(III) or Sb(III) (4). This homodimeric repressor has the sequence Cys32-Val-Cys34-Asp-Leu-Cys37 in the DNA binding domain of each monomer (5, 6). The three sulfur thiolates of the cysteine residues form a very specific three-coordinate binding site for the trivalent metalloids As(III) and Sb(III). Binding of metalloids to R773 ArsR is presumed to induce a conformational change, leading to dissociation from the DNA and hence derepression. The Staphylococcus aureus CadC is a Cd(II)/Pb(II)/Zn(II)-responsive member of the ArsR/SmtB family that has four cysteine residues in the inducer binding domain (7). Of these four cysteine residues, two come from one subunit, whereas the other two come from the other subunit of the homodimer (8, 9). The position of this metal binding site in CadC is congruent to that of the R773 ArsR but is formed between two monomers. CadC also has a second type of metal binding site (DXHX10HX2E) for Zn(II) at the dimer interface that is not a regulatory site. This site, however, is identical to the regulatory Zn(II) site of SmtB from Synechococcus PCC 7942 (10). Another member of the ArsR/SmtB family is the ArsR from Acidithiobacillus ferrooxidans (AfArsR),3 which has three cysteine residues (Cys95, Cys96, and Cys102) at the dimer interface rather than in or near the DNA binding domain (11). These three cysteine residues form a three-coordinate or S3 binding site for trivalent metalloids (12). Although both the As(III) binding site of AfArsR and the Zn(II) binding site of SmtB are at the C-terminal dimerization domain, the former is formed of three cysteine residues within a single subunit (two sites per dimer), whereas the latter is formed by four residues, two residues from one monomer and two from the other. Thus, metal(loid) binding sites appear to arise by convergent evolution, even in homologous proteins.
In the present study, we focused on two CgArsRs, which regulate expression of the ars operons of Corynebacterium glutamicum, and their arsenic binding properties. C. glutamicum is a Gram-positive nonpathogenic rod-shaped soil bacterium with high intrinsic resistance to arsenic. Analysis of the arsenic resistance mechanisms in C. glutamicum revealed the presence of two inducible ars operons (ars1 and ars2) that are located distant from each other in the chromosome (13). Expression of both operons is induced by arsenite. Their repressors, CgArsR1 and CgArsR2, which are 66% identical, both have three cysteine residues, two N-terminal residues (Cys15 and Cys16) that have no counterpart in any of the other characterized members of the ArsR/SmtB family and a third cysteine residue (Cys55) that is near but not within the putative DNA binding domain of the regulatory proteins. The positions of these cysteine residues in the two CgArsRs resemble those of CadC but appear to have arisen independently. Homology modeling of CgArsR1 suggests two As(III) binding sites, each of which is formed by Cys15 and Cys16 from one subunit and Cys55 from the other subunit. The results of in vivo and in vitro studies support this hypothesis. Thus, CgArsR1 provides a fresh instance of convergent and parallel evolution of metal(loid) binding sites built on the framework of a common DNA-binding protein.
MATERIALS AND METHODS
Strains, Media, and Reagents—Escherichia coli cells were grown in LB medium at 37 °C. C. glutamicum was grown in Tryptone soya broth (or Tryptone soya broth with 2% agar added) at 30 °C (14). Media were supplemented with 100 μg/ml ampicillin, 25 μg/ml kanamycin, 12.5 μg/ml tetracycline, or 25 μg/ml chloramphenicol (8 μg/ml for C. glutamicum), as required. E. coli strain Top 10 (Invitrogen) was used for plasmid construction and mutagenesis, strain BL21(DE3) (Novagen, Madison, WI), was used for protein expression analysis and strain S17-1 (13) as the donor in conjugation assays. Bacterial growth was monitored as A600 nm. All reagents were obtained from commercial sources.
Construction of Strains and Plasmids and Mutagenesis—For CgArsR1 and CgArsR2 expression in E. coli, plasmids pETArsR1 and pETArsR2 were constructed; in both plasmids, the arsR genes were under control of the T7 promoter and fused to a C-terminal His tag. Cysteine mutants were constructed with these plasmids.
For in vivo reporter gene expression, a C. glutamicum host strain lacking the two chromosomal ars operons, including arsR1 and arsR2, was constructed. This strain, termed 2Δars, was made for incorporation of wild type arsR1 and seven mutants in single copy at the original position on the chromosome based on the use of the mobilizable plasmid pK18mob (15). A bifunctional plasmid containing the original regulatory region from the main operator/promoter site of the ars1 operon (o/p) fused in the arsB direction to the egfp2 reporter gene were used for further in vivo analysis. The methods for strain and plasmid construction and for construction of mutants are given in the supplemental materials.
Purification of Wild Type and Mutant CgArsRs—E. coli BL21(DE3) bearing pETarsR1, pETarsR2, or the seven arsR1 mutants were cultured and induced with 0.3 mm isopropyl β-d-1-thiogalactopyranoside. Cells were harvested, washed once with a buffer consisting of 50 mm MOPS-KOH, pH 7.5, containing 20% (v/v) glycerol, 0.5 m NaCl, 20 mm imidazole, and 10 mm 2-mercaptoethanol (buffer A) and suspended in 10 ml of buffer A/g of cells. The cells were lysed with a French press at 20,000 p.s.i. with the addition of diisopropyl fluorophosphate (2.5 μl/g) and further centrifugation (150,000 × g for 1 h). The supernatant was loaded onto a Ni(II)-nitrilotriacetic acid column preequilibrated with buffer A at a flow rate of 0.5 ml/min. The column was washed with 150 ml of buffer A, followed by elution with 60 ml of buffer A containing 0.2 m imidazole. The eluted protein was concentrated with a 10 kDa cut-off Amicon Ultra-centrifugal filter (Millipore Corp., Billerica, MA) and further purified by gel filtration using Superdex 75 (Amersham Biosciences) in a 45 × 1.5-cm column with a total bed volume of 80 ml. The protein was eluted with a buffer consisting of 50 mm MOPS-KOH, pH 7.5, 0.5 m NaCl, 20% (v/v) glycerol, 0.2 mm EDTA, and 2 mm dithiothreitol at a flow rate of 0.3 ml/min. CgArsR1 was identified by SDS-PAGE (16). Fractions containing wild type and mutant proteins were concentrated by ultra-filtration centrifugation. Protein concentrations were estimated by the method of Bradford (17) or by determining the A280 nm using a calculated extinction coefficient of 1615 cm-1 (18).
Cross-linking Assays—Dibromobimane (bBBr) (Invitrogen) cross-linking was performed as described (8). CgArsR1 was reduced in the presence of 2 mm tris(2-carboxyethyl) phosphine on ice for 30 min. Reductants were removed by dialysis prior to the assay. Purified CgArsR1 (30 μm, except for C15S, which was 100 μm) was incubated with a 3-10-fold molar excess of bBBr. Samples were analyzed by SDS-PAGE using 16% gels. The gels were visualized under UV365 nm light and then stained with Coomassie Blue.
X-ray Absorption Spectroscopy (XAS)—Purified wild type and mutant CgArsR1 at 4 mm concentration were incubated on ice for 90 min with 2-3 mm sodium arsenite in a buffer consisting of 50 mm MOPS-KOH, pH 7.5, and 0.5 m NaCl. Glycerol, 30% (v/v) final concentration, was added, and arsenic-loaded protein was injected into Lucite cells covered with Kapton tape. Sample cells were flash-frozen in liquid nitrogen. XAS data were collected at the Stanford Synchrotron Radiation Laboratory on beamline 9-3 in a manner previously described (12). Data reduction and analysis was performed using the EXAF-SPAK software suite (available on the World Wide Web). Prior to data reduction and averaging, each fluorescence channel of each scan was examined for spectral anomalies. Published XAS spectra represent the average of five scans. Data reduction involved fitting the pre-edge spectral region with a second order polynomial and the EXAFS region with a cubic spline constructed of three regions. Following spline subtraction, EXAFS data were converted into k space using an E0 at 11,885 eV. The k3 weighted EXAFS was truncated at 13.0 Å-1 (due to monochromator glitches at higher k), Fourier-transformed, and simulated. The final published fitting results are from analysis of the raw unfiltered data. Data were fit using single scattering amplitude and phase functions generated using the Feff 7.2 for theoretical model generation (19). Single scattering Feff models were calculated for oxygen, sulfur, and arsenic coordination to simulate arsenic-nearest neighbor ligand scattering. Values for the scale factor (Sc) and E0, obtained previously from fitting crystallographically characterized arsenic models compounds, were equal to 0.98 and -10, respectively (20). During the fitting analysis, the scale factor and E0 were fixed. The coordination number was fixed but manually varied at half-integer values. Values for the bond length and Debye-Waller factor were allowed to vary freely. The lowest mean square deviation between data and fit, corrected for the number of degrees of freedom (F′), was used to judge best fit EXAFS simulations and to justify the addition of supplemental interactions within the simulation.
DNA Footprinting—DNA footprinting was performed as described (12). Complementary DNA primers in which one was labeled at the 5′-end with WellRED D3 fluorescence dye (Integrated DNA Technologies, Coralville, IA), as indicated, were used to amplify the C. glutamicum ars1 o/p) region. Two pairs of primers were used, one for determining the arsRB direction, and the other for the arsBR direction. After optimization, a high molar excess of CgArsR1 was added at room temperature for 30 min. As a negative control, 5 μm bovine serum albumin was used in place of CgArsR. The samples were loaded into 96-well plates and assayed with a CEQ 2000XL DNA sequencer (Beckman Coulter, Fullerton, CA). The data were analyzed with CEQ 2000 fragment analysis software.
DNA Mobility Shift Assays—DNA mobility shift assays were performed as described (21). Two probes corresponding to the arsR-arsB intergenic region from operon ars1 (igR1B1) and ars2 (igR2B2) were PCR-amplified from C. glutamicum ATCC13032 chromosomal DNA.
DNA Binding by Fluorescence Anisotropy—DNA binding studies by fluorescence anisotropy were performed as described (12) using a Photon Technology International spectrofluorimeter fitted with polarizers in the L format. Complementary 30-mer oligonucleotides, one of which was labeled at the 5′-end with carboxyfluorescein, were synthesized (Integrated DNA Technologies, Inc.) based on the DNA footprinting results. Wild type or mutant CgArsR1s were titrated with 0.4 ml of 30 nm fluorescein-labeled ars1 o/p DNA. Changes in anisotropy were calculated after each addition using the supplied Felix32 software. To measure DNA dissociation of CgArsR1 upon binding to As(III)/Sb(III), 30 nm carboxyfluorescein-labeled DNA was saturated with 10 μm wild type or mutant CgArsR1. CgArsR1 was treated with methyl methanethiosulfonate to modify the cysteine thiolates to -S-S-CH3. Dissociation of CgArsR1 was induced by the addition of varying amounts of As(III) or Sb(III).
RESULTS
CgArsR1 and CgArsR2 Have Atypical Metalloid Binding Sites—A structure-based multiple alignment of CgArsR1 and CgArsR2 with other ArsR homologues and CadC was performed using the structure of CadC (9) (Fig. 1A). CgArsR1 and CgArsR2 are 66% identical to each other and exhibit high similarity to other members of the ArsR/SmtB family. Notably, both CgArsR1 and CgArsR2 lack the typical Cys32-Cys34-Cys37 three-coordinate As(III) binding site of the well characterized R773 ArsR. They also lack the Cys95-Cys96-Cys105 As(III) binding site of AfArsR (12). CadC binds Cd(II) in a four-coordinate binding site composed of four cysteine residues. CadC has a second, nonregulatory four-coordinate Zn(II) binding site composed of residues Asp101 and His103 from one monomer and His114′ and Glu117′ from the other monomer. The two CgArsRs lack the acidic and histidine residues of those sites as well. How do these ArsRs respond to environmental As(III), and where are the metalloid binding sites? Although they lack the binding sites identified in homologous metalloregulators, they have three cysteine residues, Cys15, Cys16, and Cys55 that, from the alignment, do not correspond to metal(loid) ligands of homologues. From the structure-based alignment with CadC, a homology model of CgArsR1 was constructed (Fig. 1B). Like CadC, CgArsR1 has a putative helix-turn-helix DNA binding domain consisting of α4 and α5. In CadC, the Cd(II) binding domain is composed of Cys58 and Cys60 in α4 of one subunit and Cys7 and Cys11 from the N-terminal region of the other subunit. Although CgArsR1 has no corresponding cysteine residues, Cys55 is proposed to be located just before α4, and Cys15 and Cys16 are in α1. Either Cys15 or Cys16 can be positioned close enough to Cys55 to form a two-coordinate complex with As(III). Since the side chains of Cys15 or Cys16 would face in opposite directions, the α1 helix would have to be unraveled from the N-terminal end to allow them to form a three-coordinate complex with Cys55 (Fig. 1C). Our working hypothesis, therefore, is that these three cysteine residues form a three-coordinate As(III) binding site, where Cys15 and Cys16 come from one subunit and the Cys55 comes from the other. The experiments below were designed to test that hypothesis.
FIGURE 1.
Multiple alignment of CgArsR homologues and homology modeling. A, representative ArsR homologues (accession numbers in parentheses) are from plasmid pI258 CadC (P20047), Synechocystis sp. PCC 7942 SmtB (P30340), plasmid R773 ArsR (P15905), C. glutamicum CgArsR1 (YP_225794) and CgArsR2 (NP_599514), A. ferrooxidans AfArsR (AAF69241), and Mycobacterium tuberculosis H37Rv CmtR (NP_216510). The structure-based multiple alignment was calculated using 3DCoffee (Pubmed ID 15201059). B, modeling of As(III)-free CgArsR was performed with SWISS-MODEL (Pubmed ID 12824332) (26), using the crystal structure of CadC (9) as template. The two monomers are colored in purple (subnunit a) and green (subunit b), respectively. The cysteines are shown in ball-and-stick models. The DNA binding domain is α4-turn-α5. The model was drawn using PyMOL (27). The side chain sulfur atoms of Cys15, Cys16, and Cys55 are shown as yellow spheres. C, binding is proposed to occur in two steps: 1) As(III) (blue sphere) binds first to the thiolates of Cys55 of one subunit and either Cys15 or Cys16 (shown here for only Cys15) in α1 of the other subunit, forming a low affinity S2O site, and 2) the end of α1 unravels to allow the adjacent cysteine residue to become the third ligand to As(III), forming a high affinity S3 site. The model of the As(III)-bound form was built by manually adjusting the N termini so that the geometry of As(III) coordination derived from the EXAFS results.
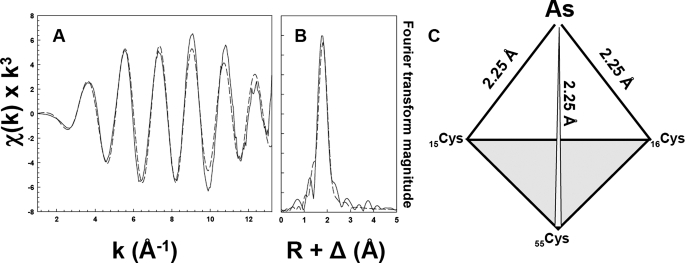
The As(III) Binding Site in CgArsR1 Is Composed of Three Sulfur Thiolates—Binding of As(III) and the geometry of the binding site was analyzed by XAS. Near edge x-ray absorption fine structure analysis indicates that arsenic in purified wild type CgArsR1 is stably bound as As(III) (data not shown). The observed value for the first inflection point energy is 11,866 eV, consistent with the observed values for As(III) binding sites (20). Simulation analysis of the EXAFS data for CgArsR1 was best fit with a single nearest neighbor ligand environment constructed with only three sulfur atoms (Fig. 2, A and B). The average bond length of arsenic-sulfur was calculated to be 2.25 Å. Attempts to simulate these data with a mixed sulfur and oxygen nearest neighbor environment were unsuccessful. These results are consistent with an As(III) binding site composed of the sulfur thiolates of Cys15, Cys16, and Cys55 (Fig. 2C).
FIGURE 2.
EXAFS and Fourier transforms of CgArsR1 XAS data. EXAFS spectra of CgArsR1 with bound As(III) (A) and the corresponding Fourier transform (B) are shown as dashed lines, with the simulations of EXAFS and FT data shown as solid lines. A schematic representation of the putative coordination environment in CgArsR1 is shown in C. Sample preparation was as described under “Materials and Methods.”
Contribution of CgArsR1 Cysteine Residues to Metalloregulation in Vivo—To examine whether Cys15, Cys16, and Cys55 are involved in As(III) sensing in vivo, the double ars operon-deleted 2Δars was constructed. In an additional step, wild type and mutant copies of the arsR1 gene were integrated into the chromosome of C. glutamicum 2Δars at the same position of the original ars1 operon, creating strains encoding wild type ArsR1 (2Δarswt) and variants C15S, C16S, C55S, C15S/C16S, C15S/C55S, C16S/C55S, or C15S/C16S/C55S. These strains were used as host for the bifunctional mobilizable plasmid pECGFPars, which contains the original ars1 o/p regulatory region fused in the arsB direction to the reporter gene egfp2 (a variant of gfp) that has been commonly used for expression analysis in corynebacteria.
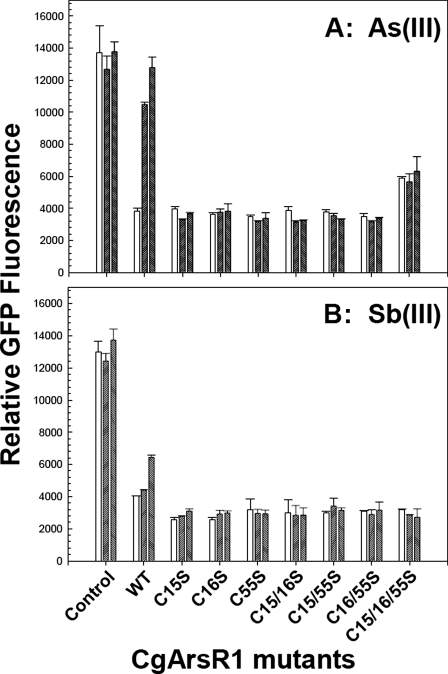
Cells of C. glutamicum lacking the two ars operons had constitutive GFP expression (Fig. 3). Cells with wild type arsR1 expressed egfp2 at repressed levels in the absence of metalloid, and the addition of either As(III) or Sb(III) induced expression. As(III) induced well at 10 μm. Sb(III) is not taken up very well by C. glutamicum and is therefore not as effective as an inducer as As(III). Even so, Sb(III) induced more at 30 μm than at 10 μm. All of the C. glutamicum strains containing the single (C15S, C16S, and C55S), double (C15S/C16S, C15S/C55S, and C16S/C55S), or triple (C15S/C16S/C55S) arsR1 mutants repressed the egfp2 expression, demonstrating that the mutant ArsR1s were capable of binding to the ars1 o/p. In contrast, no induction was observed with any of the mutants, suggesting that they do not dissociate from the DNA in the presence of As(III) (Fig. 3A) or Sb(III) (Fig. 3B). These results are consistent with the hypothesis that each of the three cysteine residues contributes to the inducer binding site.
FIGURE 3.
In vivo regulation of egfp2(gfp) expression from the arsB promoter using CgArsR1 and the seven Cys mutants in the presence or absence of As(III) or Sb(III). Construction of C. glutamicum 2Δars, further integration of arsR1 derivatives, mobilization of the reporter gene (gfp), sample preparation, and fluorescence measurements were performed as described under “Materials and Methods.” Strains C. glutamicum 2Δars (Control), 2Δars containing the arsR1 integrated (2Δarswt; WT), and the single (C15S, C16S, and C55S), double (C15S/C16S, C15S/C55S, and C16S/C55S), or triple (C15S/C16S/C55S) arsR1 mutant integrated were analyzed in the absence of arsenite (left bars) or the presence of 10 μM (middle bars) or 30 μm (right bars) As(III) (A) or in the absence (left bars) or presence of 10 μm (middle bars) or 30 μm (right bars) Sb(III) (B).
DNA Binding Properties of CgArsR1 and CgArsR2 and Contribution of Cysteine Residues to Metalloregulation in Vitro—To examine the DNA binding properties of CgArsRs and the contribution of cysteine residues to metalloid-induced dissociation from the DNA, protein-DNA interactions were analyzed in several ways. First, binding was examined by DNA mobility shift assays. In the two C. glutamicum ars operons, the gene organization is different from the R773 arsRDABC in that the corresponding arsRs genes are expressed divergently from the other ars genes. The ability of CgArsR1 and CgArsR2 to bind to their own ars intergenic regulatory DNA (in cis) and also their ability to bind to the other ars intergenic DNA (in trans) were determined. In both cases, bands corresponding to the amplified intergenic regions from either ars1 (igR1B1) (Fig. 4) or ars2 (igR2B2) (data not shown), respectively, were retarded by either CgArsR1 or CgArsR2. These probes were 150 bp (igR1B1) and 180 bp (igR2B2) and contained a portion of the arsR gene as well as the sequence past -70. As shown below, this probe has two sequences that footprint with CgArsR1, but only the upstream sequence binds with low affinity and should not affect DNA mobility in this assay. Instead, the slight differences in mobility can be attributed to increasing concentrations of repressor. Either repressor bound to either promoter site, despite the fact that they have different palindromic sequences in their intergenic regions. Since both repressors appeared to have similar binding abilities, subsequent biochemical analysis was performed with only CgArsR1.
FIGURE 4.
Electrophoretic mobility shift assays of CgArsR-DNA binding. The igR1B1 PCR-amplified band (150 bp) was assayed in the presence (+) or absence (-) of CgArsR1 or CgArsR2 at the indicated concentrations of protein. In each case, a retarded band was observed upon PAGE using a 10% nondenaturating gel when the repressor protein was present.
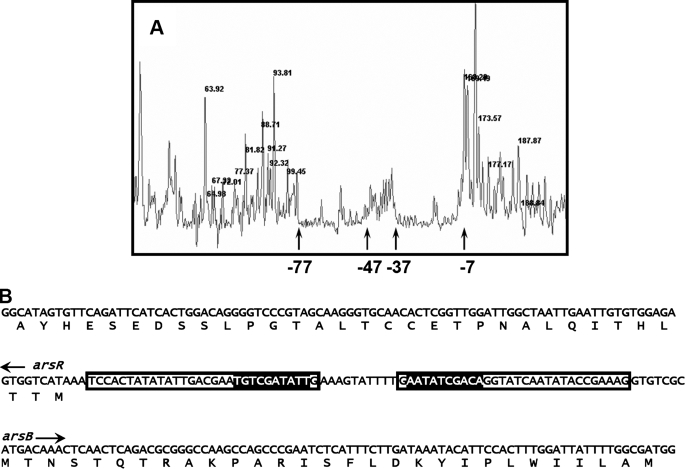
Second, to examine DNA binding properties in more detail, the sequence in the intergenic region between the arsR1 and arsB1 genes to which CgArsR1 binds was determined by DNase I footprinting (Fig. 5). CgArsR1 were observed to bind to two regions of 30 base pairs, from -7 to -37 bp and -47 to -77, each numbered relative to the start of arsB.
FIGURE 5.
DNA footprinting. A, DNase I footprinting was performed as described under “Materials and Methods” using WellRED D3 dye-labeled double-stranded DNA. The sizes of major fragments in nucleotides are shown at the top of the peaks. The sequences bound to CgArsR1 are shown between the arrows. The positions -7, -37, -47, and -77 indicate nucleotides upstream of the start of the arsB gene. B, the sequence of the double-stranded DNA is shown with the binding sequences boxed. The highlighted sequences indicate a palindromic repeat in the intervening region between the arsR and arsB genes of the ars1 operon. The start sites of the arsR and arsB genes are indicated.
DNA binding was further analyzed by fluorescence anisotropy (Fig. 6) Double-stranded oligonucleotides of 30 bp, each corresponding to the -7 to -37 and -47 to -77 sequences, were labeled with carboxyfluorescein for anisotropy measurements. The free probes tumble rapidly in solution and hence have relatively low optical anisotropy. Binding of CgArsR1 and hence the emitted light is partially polarized. Dissociation of the repressor from the DNA reverses the increase in polarization. CgArsR1 bound strongly to the -7 to -37 probe (Fig. 6A). Despite the footprinting results, CgArsR1 bound only weakly to the -47 to -77 probe. It is possible that binding of the repressor to the -7 to -37 region cooperatively increases binding to the -47 to -77 region. Wild type CgArsR1 bound to the -7 to -37 probe with an apparent affinity of 0.15 μm. Each single, double, and triple cysteine mutant bound to the -7 to -37 probe, although with reduced affinity ranging from 0.5 to 2 μm. When the cysteine residues in wild type CgArsR1 were chemically modified with methyl methanethiosulfonate, the affinity of the modified repressor was similar to that of the cysteine mutants. These results demonstrate that none of the cysteine residues is required for binding to ars1 o/p DNA.
FIGURE 6.
CgArsR1 binding to DNA assayed by fluorescence anisotropy. DNA binding assays were performed as described under “Materials and Methods.” A, fluorescently labeled double-stranded DNA was titrated with the indicated concentrations of wild type CgArsR1 (•), methyl methanethiosulfonate-modified wild type CgArsR1 (▴), C15S (○), C16S (▾), C55S(▵), C15S/C16S (▪), C15S/C55S (□), C16S/C55S (⋄), C15S/C16S/C55S (♦), and wild type CgArsR1 preincubated with 0.3 mm sodium arsenite (▿). B, double-stranded DNA with bound wild type CgArsR1 was titrated with the indicated concentrations of sodium arsenite without GSH in the assay buffer (•) and with 2 mm GSH (○), C16S (▾), or methyl methanethiosulfonate-modified wild type CgArsR1 (▵) titrated with the indicated concentrations of sodium arsenite. C, double-stranded DNA with bound wild type CgArsR1 (•), CgArsR1 with 2 mm GSH (○), or C16S (▾) titrated with the indicated concentrations of potassium antimonyl tartrate.
The effect of binding of As(III) (Fig. 6B) or Sb(III) (Fig. 6C) on dissociation of the repressor from the ars1 o/p DNA was assayed by fluorescent anisotropy. Wild type CgArsR1 exhibited half-maximal dissociation at 0.15 mm As(III). The concentration of Sb(III) required for half-maximal dissociation was ∼10 μm. For unexplained reasons, the addition of reduced glutathione increased the apparent affinity for As(III) but not Sb(III). In contrast to the in vivo results (Fig. 3), Sb(III) was more effective in vitro than As(III). This may represent relatively poor uptake of Sb(III) in vivo compared with As(III). However, none of the single, double, or triple mutants dissociated from the DNA with concentration of As(III) or Sb(III) as high as 1 mm. These data indicate that each of the three cysteine residues is necessary for dissociation from the DNA and hence derepression.
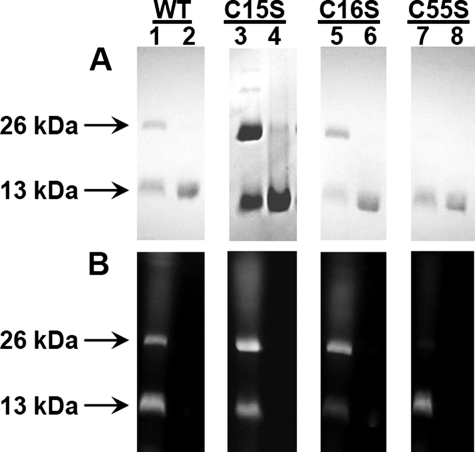
Cys15 and Cys16 from One Subunit Are in Proximity of Cys55 in the Other Subunit—In CadC, the Cd(II) binding site is composed of two cysteine residues from the N-terminal region of one subunit and two cysteine residues from α4 of the DNA binding domain of the other subunit of the homodimer (9). In the homology model of CgArsR1 built on the CadC structure, the three-coordinate As(III) binding site is proposed to consist of Cys15 and Cys16 from one subunit and Cys55 from the other (Fig. 1C). A testable prediction of that model is that the thiolates of either Cys15 or Cys16 should be close enough to the thiolate of Cys55 to cross-link with a bifunctional thiol reagent, such as bBBr, which forms a fluorescent adduct following reaction with two thiolates that are within 3-6 Å of each other (22). If so, then the homodimer would not dissociate upon SDS-PAGE following bBBr treatment. Purified wild type CgArsR1 migrated as a monomer on SDS-PAGE (Fig. 7A, lane 2). Following treatment with bBBr, a portion of the protein migrated as a fluorescent dimer (Fig. 7, A and B, lanes 1). The band corresponding to the monomer was also fluorescent, which could result from bimane adduct formation between Cys15 and Cys16 from the same monomer or from formation of bimane adducts between cysteine thiolates and solvent molecules. The single C15S and C16S mutants similarly formed fluorescent dimers (Fig. 7B, lanes 3 and 5). The triple C15S/C16S/C55S mutant did not dimerize or become fluorescent (data not shown). These results indicate that both Cys15 and Cys16 from one subunit are within 3-6 Å of the thiolate of Cys55 in the other subunit (but not necessarily simultaneously, as suggested in Fig. 1C). These results are consistent with the hypothesis that the CgArsR1 As(III) binding site is composed of two cysteine residues from one subunit and one cysteine residue from the other.
FIGURE 7.
Reaction of CgArsR1 with dibromobimane. Wild type CgArsR1 (lanes 1 and 2) and single mutants C15S (lanes 3 and 4), C16S (lanes 5 and 6), and C55S (lane 7 and 8) were analyzed by SDS-PAGE on 16% polyacrylamide gels with (lanes 1, 3, 5, and 7) or without (lanes 2, 4, 6, and 8) reaction with dibromobimane. The gels were stained with Coomassie Blue (A) and visualized on a transilluminator for fluorescence (B). The positions of the 13-kDa monomer and 26-kDa dimer are indicated by the arrows.
DISCUSSION
The ArsR/SmtB family of metalloregulatory proteins is large and diverse, with more than 3000 members identified in archaea, prokaryotic genomes, and plasmids. They respond to a number of transition metals, heavy metals, and metalloids (1-3). Only a few have been examined at the structural level (9, 23, 24), but a striking observation is that, although they all appear to have evolved from a common winged helix repressor, their metal-(loid) binding appears to have evolved independently from each other and occurs in different spatial locations in their respective structures (3, 12, 25) (Fig. 8). These inducer binding sites can be located either near the DNA binding domain, such as found in R773 ArsR or CadC, or near the C-terminal dimer interface, as in SmtB, CmtR, or AfArsR. For softer metals and metalloids, such as As(III), Sb(III), and Cd(II), the binding sites are composed of three or four cysteine residues for As(III) or Cd(II). For harder metals, such as the Zn(II) binding site of SmtB and CadC, carboxylate oxygens and imidazole nitrogens form the sites. Moreover, the binding sites are in some repressors composed of residues within a single subunit (R773 ArsR or AfArsR) or sometimes between subunits of the homodimer (CadC, SmtB, or CmtR). Finally, the metal(loid) binding ligands are frequently located at the beginning or end of helices, where unraveling these helices may induce dissociation of the repressor from the operator/promoter DNA. Ends are preferred, because it would undoubtedly take more energy to break the helix if they were located in the middle.
FIGURE 8.
Location of metal(loid) binding sites in ArsR/SmtB repressors.Metal(loid) binding sites in members of the ArsR/SmtB family of repressor proteins are shown on a surface model of the CadC aporepressor structure either by coloring CadC residues corresponding to each binding site as identified from the structure-based alignment (Fig. 1A) or, in the case of CmtR, by overlaying the two structures. The S3 As(III) binding site of the R773 ArsR (red) formed within each monomer overlaps with the corresponding S4 Cd(II) binding site of CadC (yellow) formed between the N terminus of one subunit and the DNA binding domain of the other subunit. Zn(II) binding sites of CadC and SmtB (cyan) formed between the antiparallel C-terminal α6 helices also overlap. The S3 binding sites of CgArsR1 (green), CmtR (blue), and AfArsR (purple) are at a variety of locations distributed over the surface of the repressor.
However, that reasoning is based on only a small number of examples from the thousands of homologous sequences in data bases. For that reason, we characterized two other members of the ArsR/SmtB family encoded by the two ars operons of C. glutamicum (13). As(III) induces gene expression in cells of C. glutamicum, justifying their designations as As(III)-responsive repressors CgArsR1 and CgArsR2. It was clear from the structure-based alignment that they lack the As(III) binding sites of the other two known As(III)-responsive repressors from plasmid R773, which has an S3 site in the DNA binding site, and A. ferrooxidans, which has an S3 site in the C-terminal dimerization domain.
From the near edge x-ray absorption fine structure results, CgArsR1 also binds As(III), and the EXAFS shows that it is an S3 site. Since CgArsR1 has only three cysteine residues, Cys15, Cys16, and Cys55, these are the logical candidates for the S3 site. Consistent with this idea, mutations in any of the three resulted in loss of As(III) responsiveness in vivo and loss of dissociation from the ars operator/promoter DNA in vitro. The CgArsR1 model suggests that the S3 site is formed intermolecularly between Cys15 and Cys16 in α1 and Cys55, which is located just before α4 of the helix-turn-helix DNA binding domain. This is reminiscent of the S4 Cd(II) binding site of CadC, but none of the cysteine residues in CgArsR1 align with those in CadC. Thus, it is highly likely that the As(III) binding site in CgArsR1 evolved independently of the As(III) binding sites in R773 ArsR or AfArsR or the Cd(II) binding sites of CadC or CmtR.
As discussed above, we have noted that the metal(loid) binding site in each of these repressors has ligands that are located near the start or end of an α helix and have suggested that unraveling of a helix from one end is involved in the conformational change that dissociates the repressor from its cognate DNA sequence. We propose that this is the mechanism of derepression in CgArsR1. Although recognizing that a homology model is not a structure, it predicts that Cys15 and Cys16 are close enough to Cys55 to simultaneously coordinate to As(III), and the bBBr cross-linking results support that idea. How do they form a three-coordinate binding site? Cys15 and Cys16 are a vicinal cysteine pair, which should form one of the strongest types of As(III) binding sites. However, they are predicted to be at the beginning of α1 (Fig. 1C), and their thiolates cannot approach each other closely enough to bind As(III) two-coordinately. We propose that Cys55 and either Cys15 or Cys16 first bind As(III) in a weak complex. This is followed by unraveling the beginning of α1, exposing the third thiolate, which then completes the high affinity As(III) binding site and dissociation of CgArsR1 from the DNA.
In summary, the identification of a third As(III)-responsive repressor in which the inducer binding site is different from that of R773 ArsR, AfArsR, CadC, SmtB, or CmtR supports our hypothesis that the binding sites are the result of independent and recent evolutionary events. Just as animals have a body plan of bilateral symmetry, these repressors are built using a preexisting scaffold of an ancestral winged helix DNA binding protein. Just as animals such as birds, bats, flying squirrels, and flying fish evolved wings convergently, so too have the inducer binding sites of ArsR/SmtB homologues evolved in diverse locations on the surface of the protein in response to environmental pressures by spatial positioning of residues to form three- or four-coordinate metal(loid) binding sites (Fig. 8).
Acknowledgments
A portion of this research was carried out at the Stanford Synchrotron Radiation Laboratory, a national user facility operated by Stanford University on behalf of the United States Department of Energy, Office of Basic Energy Sciences.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI043428 (to B. P. R.), R01 DK068139 (to T. L. S.), and LE040A07, BIO2005-02723, and BIO2008-00519 (to J. A. G. and L. M. M.). The Stanford Synchrotron Radiation Laboratory Structural Molecular Biology Program is supported by the Department of Energy, Office of Biological and Environmental Research, and by the National Institutes of Health, National Center for Research Resources, Biomedical Technology Program. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental “Materials and Methods.”
Footnotes
The abbreviations used are: AfArsR, ArsR from A. ferrooxidans; CgArsR, ArsR from C. glutamicum; XAS, x-ray absorption spectroscopy; EXAFS, extended x-ray absorption fine structure; bBBR, dibromobimane; o/p, operator/promoter; MOPS, 4-morpholinepropanesulfonic acid.
References
- 1.Xu, C., and Rosen, B. P. (1999) in Metals and Genetics (Sarkar, B. ed) pp. 5-19, Plenum Press, New York
- 2.Busenlehner, L. S., Pennella, M. A., and Giedroc, D. P. (2003) FEMS Microbiol. Rev. 27 131-143 [DOI] [PubMed] [Google Scholar]
- 3.Campbell, D. R., Chapman, K. E., Waldron, K. J., Tottey, S., Kendall, S., Cavallaro, G., Andreini, C., Hinds, J., Stoker, N. G., Robinson, N. J., and Cavet, J. S. (2007) J. Biol. Chem. 282 32298-32310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu, J., and Rosen, B. P. (1993) J. Biol. Chem. 268 52-58 [PubMed] [Google Scholar]
- 5.San Francisco, M. J., Hope, C. L., Owolabi, J. B., Tisa, L. S., and Rosen, B. P. (1990) Nucleic Acids Res. 18 619-624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shi, W., Wu, J., and Rosen, B. P. (1994) J. Biol. Chem. 269 19826-19829 [PubMed] [Google Scholar]
- 7.Yoon, K. P., Misra, T. K., and Silver, S. (1991) J. Bacteriol. 173 7643-7649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wong, M. D., Lin, Y. F., and Rosen, B. P. (2002) J. Biol. Chem. 277 40930-40936 [DOI] [PubMed] [Google Scholar]
- 9.Ye, J., Kandegedara, A., Martin, P., and Rosen, B. P. (2005) J. Bacteriol. 187 4214-4221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Turner, J. S., Glands, P. D., Samson, A. C., and Robinson, N. J. (1996) Nucleic Acids Res. 24 3714-3721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Butcher, B. G., and Rawlings, D. E. (2002) Microbiology 148 3983-3992 [DOI] [PubMed] [Google Scholar]
- 12.Qin, J., Fu, H. L., Ye, J., Bencze, K. Z., Stemmler, T. L., Rawlings, D. E., and Rosen, B. P. (2007) J. Biol. Chem. 282 34346-34355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ordóñez, E., Letek, M., Valbuena, N., Gil, J. A., and Mateos, L. M. (2005) Appl. Environ. Microbiol. 71 6206-6215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 15.Fernández-González, C., Gil, J. A., Mateos, L. M., Schwarzer, A., Schäfer, A., Kalinowski, J., Puhler, A., and Martin, J. F. (1996) Appl. Microbiol. Biotechnol. 46 554-558 [DOI] [PubMed] [Google Scholar]
- 16.Laemmli, U. K. (1970) Nature 227 680-685 [DOI] [PubMed] [Google Scholar]
- 17.Bradford, M. M. (1976) Anal. Biochem. 72 248-254 [DOI] [PubMed] [Google Scholar]
- 18.Gill, S. C., and von Hippel, P. H. (1989) Anal. Biochem. 182 319-326 [DOI] [PubMed] [Google Scholar]
- 19.Ankudinov, A. L., and Rehr, J. J. (1997) Phys. Rev. B Condens. Matter 56 R1712-R1715 [Google Scholar]
- 20.Ramírez-Solis, A., Mukopadhyay, R., Rosen, B. P., and Stemmler, T. L. (2004) Inorg. Chem. 43 2954-2959 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Letek, M., Valbuena, N., Ramos, A., Ordóñez, E., Gil, J. A., and Mateos, L. M. (2006) J. Bacteriol. 188 409-423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kosower, N. S., Kosower, E. M., Newton, G. L., and Ranney, H. M. (1979) Proc. Natl. Acad. Sci. U. S. A. 76 3382-3386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Cook, W. J., Kar, S. R., Taylor, K. B., and Hall, L. M. (1998) J. Mol. Biol. 275 337-346 [DOI] [PubMed] [Google Scholar]
- 24.Banci, L., Bertini, I., Cantini, F., Ciofi-Baffoni, S., Cavet, J. S., Dennison, C., Graham, A., Harvie, D. R., and Robinson, N. J. (2007) J. Biol. Chem. 282 30181-30188 [DOI] [PubMed] [Google Scholar]
- 25.Harvie, D. R., Andreini, C., Cavallaro, G., Meng, W., Connolly, B. A., Yoshida, K., Fujita, Y., Harwood, C. R., Radford, D. S., Tottey, S., Cavet, J. S., and Robinson, N. J. (2006) Mol. Microbiol. 59 1341-1356 [DOI] [PubMed] [Google Scholar]
- 26.Marti-Renom, M. A., Stuart, A. C., Fiser, A., Sanchez, R., Melo, F., and Sali, A. (2000) Annu. Rev. Biophys. Biomol. Struct. 29 291-325 [DOI] [PubMed] [Google Scholar]
- 27.DeLano, W. L. (2001) The PyMOL User's Manual, DeLano Scientific, San Carlos, CA
- 28.Mateos, L. M., Schafer, A., Kalinowski, J., Martin, J. F., and Puhler, A. (1996) J. Bacteriol. 178 5768-5775 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ramos, A., Honrubia, M. P., Valbuena, N., Vaquera, J., Mateos, L. M., and Gil, J. A. (2003) Microbiology 149 3531-3542 [DOI] [PubMed] [Google Scholar]