Abstract
The double-stranded RNA-activated protein kinase R (PKR) is an important component of antiviral defense. PKR participates in different signaling pathways in response to various stimuli to regulate translation via phosphorylation of the eukaryotic initiation factor 2α, and transcription via activating NF-κB and IRF-1, to induce pro-inflammatory cytokines. Here we show PKR regulates interleukin-10 induction in response to double-stranded RNA, bacterial lipopolysaccaride, and Sendai virus infection. Using chemical inhibitors, dominant negative constructs, and genetic knockouts, we demonstrate that the PKR-mediated interleukin-10 induction engages JNK and NF-κB. Together, our data demonstrate the role of PKR in regulating an anti-inflammatory cytokine. The findings have significance in antiviral as well as broader innate immune responses.
Protein kinase R (PKR)3 is a ubiquitously expressed serine/threonine protein kinase induced by type I interferons (IFNs) and activated by double-stranded RNA (dsRNA), polyanionic molecules, and protein activators (1). It is an important component of the innate immune system to suppress viral infection. The best-characterized cellular substrate of PKR is the eukaryotic initiation factor 2α. Through this mechanism the kinase inhibits translation and primes the cell for apoptosis (2, 3). PKR also induces pro-inflammatory cytokines, including interleukin (IL)-12 and IL-6, in response to dsRNA, tumor necrosis factor α (TNFα), lipopolysaccaride (LPS), and IL-1 in part through activation of NF-κB (4–7). Recently, PKR was demonstrated to be involved in mycobacterium-induced IL-10 in human monocytes (8).
IL-10 is a class II α-helical cytokine, and the founding member of a family of structurally related cytokines, including IL-19, IL-20, IL-22, IL-24, and IL-26 (9). IL-10 is expressed by monocytes, macrophages, T-cells, B-cells, as well as some tumor cells (10–12). IL-10 inhibits production of various pro-inflammatory cytokines including IL-1, IL-6, IL-12, and TNFα (13–17). IL-10 also inhibits the T-cell activation capacity of monocytes or macrophages by down-regulating expression of MHC class II antigen and several co-stimulatory molecules such as CD86 and CD80 (18, 19). Potent inducers of IL-10 include LPS, mycobacteria, microbial lipoproteins, a number of viruses, and cytokines, such as TNFα (20–24). Several transcription factors including Sp1, NF-κB, c-Maf, IRF-1, and c-Jun can regulate IL-10 induction (25–29). The action of IL-10 is mediated by binding to the IL-10 heterodimeric receptor that belongs to the class II cytokine receptor family (16, 30). Receptor binding leads to activation of Jak1 and Tyk2 kinase followed by activation of STAT3. Although the mechanisms of IL-10 induction in response to LPS or other cytokines have been explored, the mechanism of IL-10 induction in response to viral infection has not been elucidated. Here we identify a novel role for PKR in IL-10 induction in response to dsRNA and viral infection. Furthermore, we characterize the mechanisms regulating PKR-dependent IL-10 induction to show the kinase functions via both NF-κB and JNK activation.
EXPERIMENTAL PROCEDURES
Reagents and Plasmids—The chemical inhibitors SP600125, PD98059, SB203580, and BAY-11-7085 were purchased from Calbiochem. The effective concentration of each chemical inhibitor was deduced empirically for each cell line (supplemental Fig. S1, A and B) and was used within the concentration range commonly reported in the literature. LPS (from Escherichia coli serotype 0111:B4), pIC, and 2-aminopurine were purchased from Sigma. The neutralizing antibody for IL-10 (JES5–2A5), and flow cytometry antibodies were from BD Pharmingen. The neutralizing antibody for mouse IFNβ (RMMB-1) was from PBL Interferon Source, and the control IgG antibody (sc-2026) from Santa Cruz Biotechnology. PKR (D-20) and JNK1 antibodies were purchased from Santa Cruz Biotechnology, and phospho-JNK antibody was purchased from Cell Signaling. The pCMVeGFP-blast and pCMVeIκB-SR constructs were kindly given by Dr. J. Didonato (Cleveland Clinic Foundation). Dr. Ze'ev Ronai (The Ruttenberg Cancer Center) provided the PEF61/DN-ATF2 plasmid. The pGL2-IL-10 luc –1538/+65-IL-10 promoter construct was obtained from Dr. David Mosser (University of Maryland) and subsequently cloned into the XhoI and HindIII sites of pGL3 basic vector (Promega). Two NF-κB binding sites in the IL-10 promoter were mutated by site-directed mutagenesis using the QuikChange® kit (Stratagene). Mutant primer pairs were 5′-CTGAGGTAGTAGGAGAAGTAAATACTGAAGGGAAGGTCCAGAC-3′ with 5′-GTCTGGACCTTCCCTTCAGTATTTACTTCTCCTACTACCTCAG-3′ for the NF-κB site at –861/–851, and 5′-ACCTTTGCCAGGAAGGCAAAACTGAGCCTTCAGTATAAAAG-3′ with 5′-CTTTTATACTGAAGGCTCAGTTTTGCCTTCCTGGCAAAGGT-3′ for the NF-κB site at –59/–39. Dominant negative c-Jun was obtained by PCR amplification of human c-Jun (nucleotides 123–331) and insertion into the HindIII and BamHI sites of pCMV-10 (Sigma). Oligonucleotide containing NF-κB consensus sequence was purchased from Santa Cruz Biotechnology (sc-2505). An RNA interference (RNAi) construct targeting PKR was prepared using a previously validated small interfering RNA sequence cloned into pcDNA6.2-GW/EmGFP-miR (Invitrogen). A control vector pcDNA6.2-GW/EmGFP-miR-neg (Invitrogen) expressing a microRNA against LacZ was used as a control in RNAi experiments. C57Bl6 wild-type (wt) mice were obtained from Jackson Laboratory.
Cell Culture—Cells were isolated from wt and pkr knock-out (pkr-ko) (31) isogenic C57Bl6 mice, then subsequently immortalized by transformation with oncogenic retrovirus (expressing ras and myc) as described previously (32). The derived cell lines were adherent and appear flattened without processes (supplemental Fig. S2). The cells did not express the granulocyte marker Gr-1 (data not shown), but did express the macrophage lineage markers Mac-1 and F4/80 as determined by FACS analysis. Additional FACS analysis showed the cells phagocytosed propidium iodide (PI)-labeled Staphylococcus aureus (Pansorbin, Calbiochem) (Fig. 4A). Based on the cell morphology, expression of archetypal surface markers, and the functional assay, the cell lines were characterized as macrophages and designated as spleen-derived macrophages (SMs). SM cells were maintained in Dulbecco's modified Eagle's medium in the presence of 10% fetal bovine serum. SM cells stably expressing the IκB super-repressor (IκB-SR) or, as a control, GFP were generated by infection with lentivirus constructs (pCMVeIκB-SR-blast or pCMVeGFP-blast, respectively), and were maintained in the presence of 10 μg/ml blastocidin (Invitrogen). SM cells (1 × 105) were transiently transfected for RNAi experiments with 0.5 μg of plasmid DNA using Nucleofector solution L and the Nucleofector II device set at X-001 (Amaxa).
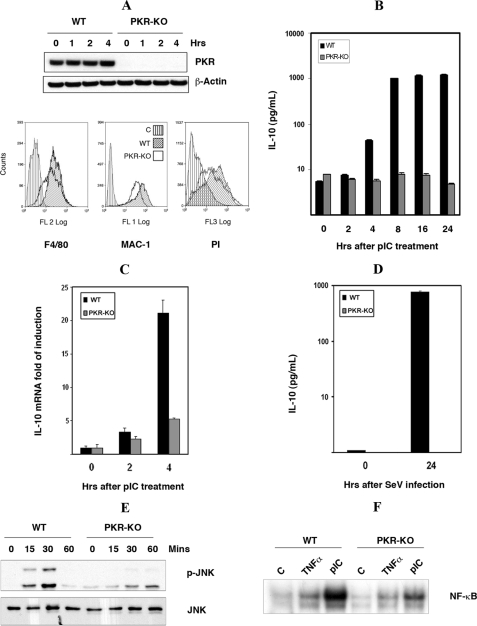
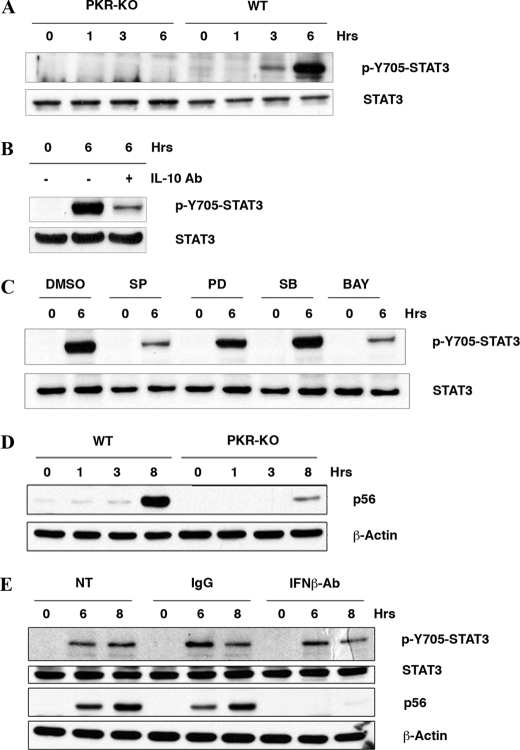
FIGURE 4.
The induction of IL-10 by dsRNA is PKR-dependent. A, characterization of wt and pkr-ko SM cells, by Western blot analysis with an antibody to PKR (upper panel), flow cytometric analysis with antibodies detecting macrophage markers (F4/80 and MAC-1), and phagocytic assay measuring uptake of PI (lower panel). Only the wt SM control cell line is shown in each graph, as the pkr-ko control was equivalent. The control (C) in each graph was either without antibody or incubated at low temperature to inhibit phagocytosis. B, induction of IL-10 by dsRNA was measured in wt and pkr-ko SM cells treated with pIC (50 μg/ml) for the indicated time periods and assessed by ELISA for IL-10 protein levels. C, to assess the requirement for PKR in an alternative cell system, wt and pkr-ko primary BMM were treated with pIC (50 μg/ml) for indicated times, total RNA was extracted, and IL-10 mRNA was measured by real-time quantitative PCR. D, to validate the dsRNA response with a biologically pertinent stimulus, wt and pkr-ko SM cells were infected with Sendai virus (SeV) at a concentration of 80 HAU for 24 h and the level of IL-10 protein in the cell supernatants measured by ELISA. The experiment was done in triplicate. In keeping with the previous data showing a role for JNK and NF-κB in dsRNA-mediated IL-10 induction: E, PKR-dependent activation of JNK was measured in wt and pkr-ko SM cells treated with pIC (50 μg/ml) for indicated times, by Western blot using the phospho-JNK (p-JNK) antibody. Total JNK level was measured to verify equal loading. F, PKR-dependent activation of NF-κB was measured by EMSA in cell lysates from wt and pkr-ko SM cells, either untreated (C), treated with pIC (50 μg/ml), or as a control TNFα (25 ng/ml) for 30 min.
Bone marrow-derived macrophages (BMM) were prepared from the femurs of a C57Bl6 wt or pkr-ko mice. Bone marrow cells were differentiated in culture for 7–10 days in Dulbecco's modified Eagle's medium supplemented with 15% fetal bovine serum, 20% L-cell conditioned medium (as the source of murine macrophage colony-stimulating factor), with penicillin and streptomycin (10 units and 10 μg/ml, respectively) to produce macrophages.
Cell Lysis and Immunoblotting—Prior to lysis, cells were washed twice in cold phosphate-buffered saline (PBS). Cell extracts were prepared by lysing the cells with lysis buffer (50 mm Tris-HCl, pH 7.4, 150 mm NaCl, 50 mm NaF, 10 mm β-glycerophosphate, 0.1 mm EDTA, 10% glycerol, 1% Triton X-100) supplemented with protease/phosphatase inhibitors (1 mm phenylmethylsulfonyl fluoride, 1 mm sodium orthovanadate, 2 μg/ml each of leupeptin, aprotinin, and pepstatin) followed by incubation on ice for 20 min. Lysates were subjected to centrifugation at 14,000 × g for 20 min, and the supernatant collected and protein quantified by Bradford assays. 30 μg of cell lysates were fractionated onto a 10% SDS-PAGE gel, and subsequently transferred to a polyvinylidene difluoride membrane (0.45 μm), and probed with their respective antibodies according to the manufacturer's recommendations.
ELISA—IL-10 secretion by stimulated macrophages was measured using the mouse IL-10 ELISA set (BD Pharmingen) according to the manufacturer's protocol. Cells (3 × 105) were seeded per well of a 12-well plate. The following day, cells were subjected to treatment as indicated.
Chromatin Immunoprecipitation (ChIP) Assay—ChIP assays were conducted using the EZ-ChIP Assay kit (Upstate Biotechnology) following the manufacturer's protocol. Cells were washed twice with ice-cold PBS and were scraped off the plates. The cells were recovered by centrifugation at 700 × g at 4 °C and lysed with 1 ml of SDS lysis buffer containing protease inhibitors. Chromatin was sheared by sonication using the sonic dismembrator 500 from Fisher Scientific. The power output was set at the microtip level (4.5) at 15% cycle efficiency for 20 cycles of 10-s pulses to generate sheared chromatin size between 1 kb and 200 bp. Sheared chromatin (100 μl, equivalent to 2 × 106 cells) was used for each immunoprecipitation with respective antibodies. The antibodies used were p50 (SC-7178X) and p65 (SC-109X) from Santa Cruz Biotechnology, and the primers used to amplify the IL-10 promoter surrounding the –861/–851 site were: Fwd: 5′-AATGAAAGGCAATAGGGGACTC-3′ and Rev: 5′-TGTTGAAGGATGGAGATGTTAG-3′. Primers to amplify the IL-10 promoter region surrounding the –59/–39 site were: Fwd: 5′-GACCAGTTCTTTAGCGCTTACAAT-3′ and Rev: 5′-GTCTCTAAATGTAGACCTCCTGTTC-3′. Control primers to amplify the promoter region of α-amylase were: Fwd: 5′-TCAGTTGTAATTCTCCTTGTACGG-3′ and Rev: 5′-CATTCCTTGGCAATATCAACC-3′. PCR conditions were: 94 °C for 3 min, 94 °C for 30 s, 58 °C for 30 s, 72 °C for 40 s, for 35 cycles. The PCR products were resolved through a 1.5% agarose gel to confirm the specificity of amplification.
FACS Analysis—Cells (106/sample) were incubated with an antibody to block Fc receptors (clone 2.4G2, BD Pharmingen) before addition of antibodies against Mac-1 and Gr-1 (clones M1/70 and RB6–8C5, respectively) conjugated to fluorescein isothiocyanate or phycoerythrin diluted in FACS buffer (PBS + 2% FCS + 10 mm NaN3). Cells were washed with FACS buffer before overnight fixation in FACS buffer containing 1% paraformaldehyde. Following resuspension in FACS buffer, flow cytometric analysis was conducted on a FACScan flow cytometer (Becton Dickinson) and data analyzed using FlowJo software (TreeStar Inc).
Phagocytosis assays were conducted by incubating SM cells (1 × 106), suspended in 100 μl of PBS with 2% FCS, 10 μl of Pansorbin (5% w/v)(Calbiochem), and 10 μl of PI-labeled Pansorbin (5% w/v) at 37 °C, or as a control on ice, for 40 min in the dark. The reaction was stopped by adding 1 ml of cold PBS and 100 μl of trypan blue, then rinsed three times with cold PBS with 2% FCS, and centrifugation at 1000 × g for 5 min. Cells taking up the PI label were discriminated by FACS analysis.
Gene Expression Analysis—Total RNA was extracted using Trizol (Invitrogen) and 1 μg of total RNA was reverse-transcribed using oligo(dT) primer and Superscript II reverse transcriptase (Invitrogen). Reverse-transcripted samples were diluted 3-fold, and 2 μl of sample were used for real-time quantitative PCR with SYBR green (Applied Biosystems); the PCR was run on an i-Cycler iQ (Bio-Rad) machine. IL-10 mRNA expression level was normalized relative to GAPDH mRNA. The primers used for IL-10 were: Fwd: 5′-GGTTGCCAAGCCTTATCGGA-3′ and Rev: 5′-ACCTGCTCCACTGCCTTGCT-3′ (33). The primers for GAPDH were: Fwd: 5′-TTCACCACCATGGAGAAGGC-3′ and Rev: 5′-GGCATGGACTGTGGTCATGA-3′.
EMSA—Following treatment with TNF-α, IL-1α (R & D Systems), or pIC, cells were harvested in ice-cold PBS and pelleted at 1,000 × g at room temperature for 2 min. The cell pellet was resuspended on ice in whole-cell extract lysis buffer composed of 25 mm HEPES (pH 7.7); 0.3 m NaCl; 1.5 mm MgCl2; 0.2 mm EDTA; 0.5% Triton X-100 (v/v), 3 mm dithiothreitol; the phosphatase inhibitors 30 mm β-glycerophosphate, 50 mm NaF, 1 mm sodium orthovanadate (Na3VO4), and 20 mm p-nitrophenyl phosphate (Calbiochem); and the protease inhibitors aprotinin, leupeptin, bestatin, and pepstatin (all at 10 mg/ml), 100 mm N-tosyl-l-phenylalanine chloromethyl ketone, and 1 mm phenylmethylsulfonyl fluoride (Sigma). Lysates were rotated in the cold for 45 min and centrifuged at 13,000 × g for 15 min. Whole cell extract (10 μg) was incubated with 5 μg of polyoligonucleotides (dI-dC) and 20,000 cpm (0.2 ng) of the labeled oligonucleotide probe corresponding to the NF-κB-binding site. After incubation for 30 min on ice, the bound and free DNAs were fractionated on 5% native polyacrylamide gels.
Transfection—RAW264.7 cells were transiently transfected using the Lipofectamine transfection and PLUS reagents (Invitrogen). For transfection with luciferase reporter plasmids, 1.6–2 × 105 RAW264.7 cells were plated per well of a 12-well plate. The following day, cells were washed with PBS and transfected with 0.4 μg of reporter plasmid, 0.5 μg of co-transfected plasmid, and 0.1 μg of pRL-null vector (Promega) per well, per the manufacturers instructions. After 24 h, cells were either treated with pIC (50 μg) or left untreated for 16 h. Luciferase assays were performed using the dual luciferase reporter assay system as described by the manufacturer (Promega). All firefly luciferase values were normalized to Renilla luciferase levels.
RESULTS
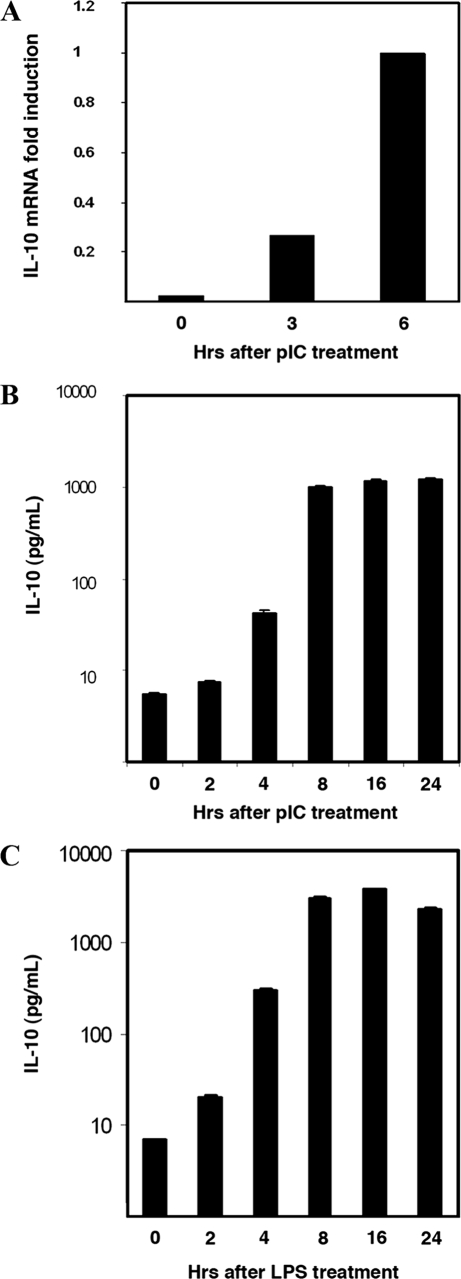
IL-10 Is Induced in Response to dsRNA—A number of viruses have been found to be potent inducers of IL-10 (20–24). To explore this phenomenon, spleen-derived macrophage cells (SMs) were treated with pIC to simulate dsRNA intermediates of viral replication. RNA from pIC-treated cells was used for real-time quantitative PCR. IL-10 mRNA was found to be highly induced (Fig. 1A). Appropriately, IL-10 protein was also induced in cells, with levels in the culture medium peaking at 8 h (Fig. 1B). As LPS has previously been shown to induce IL-10, this treatment was used to confirm that the SM cell line responds appropriately (Fig. 1C). The data show the reported induction of IL-10 by viral infection is, at least in part, via a dsRNA response.
FIGURE 1.
IL-10 is induced by dsRNA. Levels of IL-10 were measured in SM cells treated with the dsRNA mimic pIC (50 μg/ml) for the indicated time periods. A, total RNA was extracted and analyzed by real-time quantitative PCR to measure the induction of the IL-10 transcript. B, the cell supernatants were analyzed by ELISA to measure induction of IL-10 protein. C, SM cells were treated with LPS (10 ng/ml) for the indicated time periods, and the cell supernatants were analyzed by ELISA for IL-10 levels.
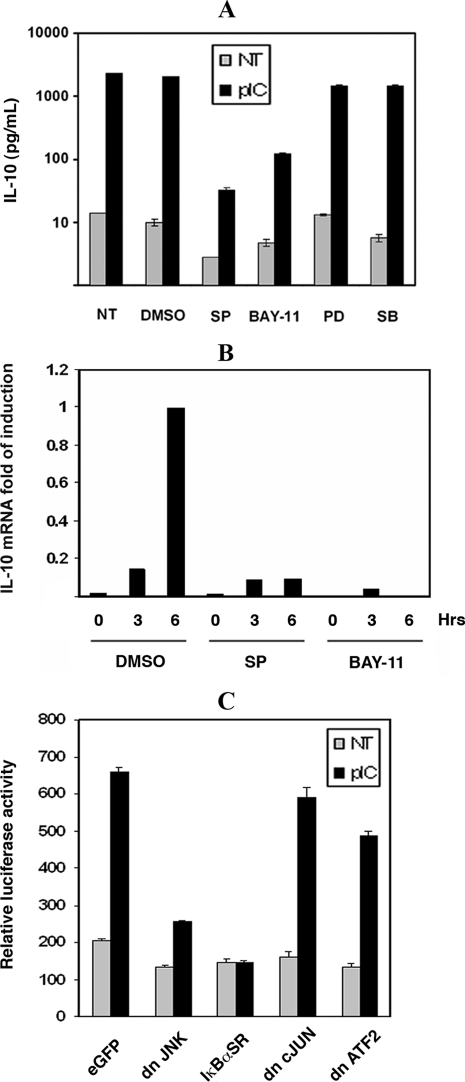
JNK and NF-κB Regulate IL-10 Induction—To identify the essential factors that mediate IL-10 induction, signaling pathways regulated by dsRNA were explored. Members of the MAP kinase family with NF-κB have been reported to regulate IL-10 induction in macrophages in response to LPS (8, 26, 34). To assess a role for these proteins in the dsRNA-mediated induction of IL-10, SM cells were treated with the chemical inhibitors, SP600125, PD98059, SB203580, or BAY-11-7085, to gauge the involvement of JNK, Erk1/2, p38, and NF-κB (respectively). Induction of both IL-10 mRNA and protein was significantly blocked in response to pIC only in the presence of the JNK and IKK inhibitor (Fig. 2, A and B).
FIGURE 2.
dsRNA-mediated IL-10 induction requires JNK and NF-κB. A, identity of signal transduction proteins that regulate the induction of IL-10 was investigated by a 1-h pretreatment of SM cells with the inhibitors SP600125 (SP; 10 μm), PD98059 (PD; 20 μm), SB203580 (SB; 4 μm), or BAY-11-7085 (BAY-11; 5 μm), or as a control the solvent (DMSO), or untreated (NT), prior to stimulation with pIC (50 μg/ml) for 16 h. IL-10 levels were then measured by ELISA. B, IL-10 mRNA levels were measured by real-time quantitative PCR after 1 h of pretreatment of SM cells with SP600125 (10 μm) and BAY-11-7085 (5 μm) followed by stimulation with pIC (50 μg/ml) for 3 and 6 h. C, dsRNA-mediated induction of IL-10 was assessed using a firefly luciferase reporter construct directed by the murine IL-10 promoter fragment –1538 to +65 (pGL3B-IL-10-LUC). The reporter construct was transiently cotransfected into RAW264.7 cells with constructs expressing; GFP (eGFP), dominant negative (dn) JNK1, c-Jun, and ATF2, or the IκBα super-repressor (dn JNK, IκBαSR, dn cJUN, dn ATF2, respectively). Following transfection, untreated controls (NT) were compared with cells treated with pIC (50 μg/ml) for 16 h. IL-10 promoter activity was measured by luciferase assay. The experiment was done in triplicate.
The role of NF-κB and JNK in pIC-regulated IL-10 induction was further examined by promoter reporter assay. Dominant negative JNK1 or the IκBα super-repressor (IκBα-SR) was transfected into the murine macrophage cell line, RAW 264.7, expressing the murine IL-10 promoter (–1538/+65) as a luciferase reporter construct, and then treated with pIC. As was shown for the endogenous IL-10 mRNA and protein, the IL-10 promoter was induced more than 3-fold in response to pIC (Fig. 2C). The promoter induction was significantly reduced by dominant negative JNK1 and IκBα-SR constructs. In contrast, the dominant negative c-Jun and ATF2 constructs did not alter the activation of the IL-10 promoter.
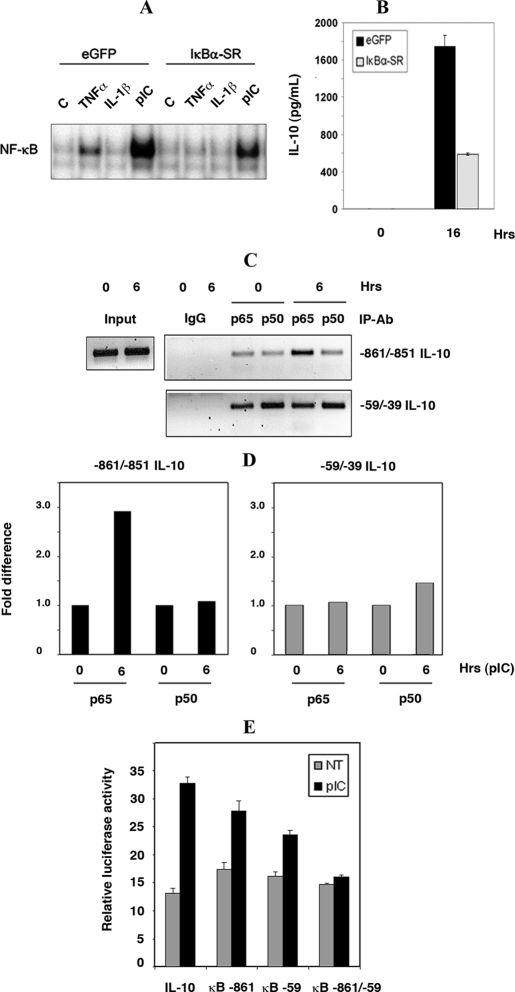
NF-κB Regulates the IL-10 Promoter via a Novel Distal Binding Site—The involvement of NF-κB in dsRNA-mediated IL-10 induction was verified using an electrophoretic mobility shift assay with lysates of SM cells expressing the IκBα-SR. Treatment of SM cells with pIC showed significant NF-κB DNA binding activity (Fig. 3A). Control treatments compared TNFα and IL-1β treatments. Appropriately, NF-κB DNA binding activity was significantly reduced in cells expressing IκBα-SR. Accordingly, the induction of IL-10 was reduced 3-fold in response to pIC in cells expressing IκBα-SR compared with control GFP-expressing cells (Fig. 3B).
FIGURE 3.
dsRNA-mediated NF-κB-dependent regulation of IL-10 at a novel distal site on the gene promoter. A, direct activation of NF-κB was measured by EMSA in whole cell lysates (20 μg) from SM cells stably expressing IκBα-SR or GFP. Cells were untreated (C), or stimulated with pIC (50 μg/ml), and as controls TNFα (25 ng/ml), or IL-1β (5 ng/ml) for 30 min. B, role of NF-κB in dsRNA induction of IL-10 was verified by comparing IL-10 protein levels in supernatants from SM cells either stably expressing the IκBα-super-repressor (IκBα-SR) or, as a control, GFP (eGFP), either untreated or treated with pIC (50 μg/ml) for 16 h. C, binding of NF-κB to specific elements within the IL-10 promoter was measured by ChIP analysis. Protein complexes were immunoprecipitated with anti-p65, or anti-p50 antibodies from SM cell lysates either untreated or stimulated with pIC (50 μg/ml) for 6 h. Two regions of the IL-10 promoter, surrounding regions –59/–39 and –861/–851, were amplified from DNA obtained from the ChIP. As a positive control amplification of regions surrounding the α-amylase promoter is shown using input DNA before ChIP (Input). The specificity of the ChIP assay was tested using rabbit IgG. D, the difference in NF-κB binding to the proximal (–59/–39) or distal (–861/–851) sites in the IL-10 promoter was measured by quantitative PCR. Measures were normalized to the zero time point for each site. E, the functionality of each NF-κB binding site within the murine IL-10 promoter was tested by transient transfection of RAW264.7 cells with pGL3B-IL-10-LUC that were either wild-type (IL-10), mutated at the distal site (κB –861), or proximal site (κB –59), and at both sites (κB –861/–59).
To verify the binding of NF-κB specifically to the IL-10 promoter, a ChIP assay was performed. SM cells were stimulated with pIC for 6 h and ChIP performed using anti-p65 and p50 antibodies. Analysis of the murine IL-10 promoter sequence (using TESS software) identified a putative NF-κB binding site between the nucleotides –861/–851 (relative to the transcription start site). This site, with a second previously reported site at –59/–39 within the IL-10 promoter (26), was interrogated. Fig. 3C confirms that NF-κB associates with the murine IL-10 promoter and identifies the distal, –861/–851, region as the pertinent regulatory motif mediating the response to pIC. Increased binding to the distal NF-κB site in the IL-10 promoter was confirmed by quantitative PCR (Fig. 3D). Although the relevance of either NF-κB site can only be accurately assessed in context of the cellular chromatin, we confirmed the functionality of the distal site in an exogenous promoter context using an IL-10 promoter (–1538/+65) reporter assay in which the proximal and distal sites were mutated, separately and together, to prevent NF-κB binding (Fig. 3E). The data demonstrate that JNK and NF-κB are key regulators of the dsRNA-mediated induction of IL-10 in SM and RAW 264.7 cells.
dsRNA-mediated IL-10 Induction Is PKR-dependent—As PKR is implicated in signaling pathways triggered by dsRNA, as well as LPS in macrophage cells, and is reported to activate both JNK and NF-κB (4, 35), a role in IL-10 induction was investigated. The response to pIC was compared between wt and pkr-ko SM cells (characterized in Fig. 4A and supplemental Fig. S2) by measuring IL-10 levels in the culture supernatant with an ELISA. Remarkably, pIC-mediated IL-10 induction was entirely lost in the pkr-ko cells (Fig. 4B). Treatment of wt SMs with a chemical inhibitor of PKR, 2-aminopurine, or RNAi targeting PKR and then treatment with LPS (10 ng/ml) verified that the difference observed between the wt and pkr-ko cell lines was entirely due to the genetic deletion of pkr (supplemental Fig. S3).
PKR dependence of the dsRNA-mediated IL-10 induction was also measured in primary BMM isolated from wt or pkr-null mice. Supporting the observation using SMs, the induction of IL-10 mRNA was found to be 4-fold higher in the wt BMM compared with the pkr-ko BMM after 4 h of pIC treatment (Fig. 4C). Interestingly, this defect was largely rectified in the BMM at later time points (data not shown).
To confirm the importance of PKR in the IL-10 response in a biologically relevant stress response, wt and pkr-ko SM cells were infected with Sendai virus, and the levels of IL-10 in cell supernatants were measured by ELISA. As shown for pIC, IL-10 was highly induced by Sendai virus only in the wt SM cells (Fig. 4D).
The preceding data predict that PKR mediates the induction of IL-10 via activation of JNK and NF-κB. To confirm this, we examined the activation status of JNK and NF-κBinwtand pkr-ko SM cells. Consistent with the previous observations, JNK activation was decreased in pkr-ko SM cells, as measured by the levels of the phosphorylated protein detected by Western analysis (Fig. 4E). Similarly, NF-κB activity was severely impaired in the pkr-ko compared with the wt cells treated with pIC, measured by EMSA (Fig. 4F). The data show that PKR is critical in the dsRNA signaling pathway leading to IL-10 production in SM cells, and is upstream to JNK and NF-κB.
Activation of STAT3 in Response to dsRNA Is PKR-dependent—A consequence of IL-10 induction is the subsequent activation of STAT3 (36). To measure the downstream effect of IL-10 induction following pIC treatment, and to corroborate a role for PKR in this pathway, we examined the activation of STAT3 in response to pIC in wt and pkr-ko SM cells. Remarkably, STAT3 activation, measured by phosphorylation of the tyrosine residue at position 705, was exclusively observed in the wt and not in the pkr-ko SM cells on pIC treatment (Fig. 5A). Appropriately, a neutralizing antibody to IL-10 ablated STAT3 activation (Fig. 5B). As predicted from our previous results, JNK and NF-κB inhibitors (SP600125 and BAY-11-7085) significantly decreased pIC-mediated STAT3 activation (Fig. 5C). Furthermore, STAT3 phosphorylation was unaffected by inhibitors of Erk1/2 and p38 MAP kinases.
FIGURE 5.
PKR activates STAT3 via production of IL-10 independent of IFN. A, STAT3 activation in wt and pkr-ko SM cells treated with pIC was measured by Western blot analysis using a phospho-Y705-STAT3 antibody. Total STAT3 level was measured to verify equal loading. B, STAT3 activation was attenuated in wt SM incubated with an IL-10 neutralizing antibody (IL-10 Ab, 20 μg/ml) for 1 h prior to stimulation with pIC for 6 h. C, role for JNK and NF-κB in dsRNA-mediated, IL-10 induction and ensuing STAT3 activation was corroborated by pretreating wt SM cells with DMSO, or the inhibitors; SP600125 (SP; 10 μm), PD98059 (PD; 20 μm), SB203580 (SB; 4 μm), and BAY-11-7085 (BAY; 5 μm) for 1 h prior to stimulation with pIC (50 μg/ml) for 6 h, followed by Western blot analysis using a phospho-Y705-STAT3 and total STAT3 antibodies. D, induction of the IFN-inducible protein p56 was measured by Western blot of lysates from wt and pkr-ko SM cells treated with pIC (50 μg/ml) for indicated times. β-Actin levels were measured to verify equal loading. E, the role of the IFN response in IL-10 signaling was assessed by incubating wt SM cells with control IgG or an IFNβ neutralizing antibody (12.5 μg) for 1 h before stimulation with pIC for 6 and 8 h. Cell lysates were probed by Western blot to measure STAT3 activation using the phospho-Y705 STAT3 and total STAT3 antibodies. Suppression of the IFN response was gauged by probing for levels of p56.
LPS-mediated induction of IL-10 has been reported to be dependent upon induction of IFNβ (37). PKR has been shown to regulate induction of IFNβ, via NF-κB and IRF1 in mouse embryonic fibroblasts (38). More recent reports, however, have emphasized the role of TLR3 and RIG-I in production of type I IFNs in response to viral infection (39, 40). We sought to estimate the contribution of PKR by monitoring induction of p56, a well-established IFN-inducible gene product, using wt and pkr-ko SM cells. Induction of p56 was markedly reduced in the absence of PKR (Fig. 5D), implying PKR is required for full induction of IFN. To gauge the requirement of IFNs in the dsRNA-mediated induction of IL-10, SM cells were treated with a neutralizing antibody to IFNβ or, as a control, IgG antibody, prior to pIC. This treatment did not markedly reduce the production of IL-10 (data not shown) and did not affect STAT3 phosphorylation (Fig. 5E). Appropriately, the IFN-dependent amplification of p56 was blocked. Thus PKR-mediated IL-10 induction and STAT3 activation are largely independent of IFNβ production in SM cells.
DISCUSSION
Previous reports have indicated a role for PKR in the induction of pro-inflammatory cytokines, including TNFα, IL-6, and IL-12 p40, in response to different mitogenic or stress stimuli (4, 7, 41, 42). Recently, PKR was implicated in the up-regulation of the anti-inflammatory cytokine IL-10, likely by NF-κB and Erk1/2, in response to mycobacterial infection (8). Here we have shown for the first time that IL-10 is expressed in response to dsRNA (Fig. 1, A–C), and that this induction is mediated by PKR (Fig. 4, B and C). Moreover, induction of IL-10 in response to Sendai virus infection was also PKR-dependent (Fig. 4D). Using different inhibitors we demonstrated that the PKR-mediated induction of IL-10 in response to pIC involves activation of JNK1 and NF-κB (Fig. 2, A–C). JNK1 has been accredited with regulating the expression of pro-inflammatory cytokines and chemokines. This role as a positive regulator of IL-10 induction in response to dsRNA is a novel function for the MAP kinase. NF-κB has previously been demonstrated to induce IL-10, with a recent report showing binding of the p50 subunit to the –59/–39 region of the mouse IL-10 promoter in response to LPS (26). However, we did not detect binding to this region in response to pIC. Instead, we identified a novel NF-κB binding site at the –861/–851 region of the IL-10 promoter (Fig. 3, C–E). Intriguingly, this distal NF-κB binding site appears to be conserved in the human IL-10 promoter sequence (supplemental Fig. S4). Consequently, the same transcription factor may engage the IL-10 promoter at different sites with different stimuli. Our data also suggest different transcription factors are recruited to the IL-10 promoter under different stimuli. Contrary to the proposed mechanisms in mycobacterial infected human monocytes, Erk1/2 inhibitors did not affect pIC-mediated IL-10 induction (Fig. 2, A and C). Similarly, although c-Jun has been shown to induce IL-10 in Th2 cells in response to PMA and to bind to the human IL-10 promoter, in our assays the presence of dominant negative c-Jun or its heterodimeric partner ATF2 had no effect upon IL-10 induction in response to pIC (Fig. 2C) (29). In keeping with a PKR-dependent role for IL-10 induction in response to dsRNA, ensuing protein responses were shown to be impaired. IL-10-dependent STAT3 phosphorylation was lost in the PKR-null SM (Fig. 5, A and B). PKR had previously been described to activate STAT3 in response to platelet-derived growth factor (43). Our data identify a mechanism by which PKR might affect STAT3 activation, whereby PKR-mediated activation of JNK and NF-κB induces IL-10, which then binds to its receptor to activate Jak1 and Tyk2, with the resultant binding and phosphorylation of STAT3. Following its activation, STAT3 forms homo- or heterodimers with other STAT members to regulate transcription. Hence, it would be interesting to look at the downstream targets of STAT3 upon dsRNA treatment.
In this study we do not unequivocally identify the primary receptor regulating the response to pIC. PKR could directly mediate this response, as activation of PKR by dsRNA and viral infection is well established. Alternatively, another dsRNA receptor may be engaged that subsequently signals via PKR. Toll-like receptor 3 (TLR3), for instance, could be the primary receptor. In accord with this, PKR has been shown to be involved in TLR3 signaling (5). It is worth noting, however, that earlier reports have shown that TLR3 is dispensable in Sendai virus-induced gene expression (44). Conversely, the data here show that PKR is required for the induction of IL-10 by this virus (Fig. 4D). It has been proposed that the TLR3-independent response to Sendai virus is principally mediated by another key dsRNA receptor, RIG-I (45). However, RIG-I has not been ascribed an important role in myeloid cells, as used in this study (40, 46–49). This is sustained by data here showing the RIG-I responsive protein p56 was not induced in the SM cells in response to pIC (Fig. 5D). A recent report identified a critical role for IFN in the induction of IL-10 in BMM in response to LPS (37). PKR has previously been shown to regulate the production of IFNβ in mouse embryonic fibroblasts (31, 38, 50). The data here identify an impairment of the IFN-induced p56 in pkr-ko SM (Fig. 5D), implying PKR also regulates IFNβ production in macrophages. However, in contrast to the signaling events triggered by LPS in BMMs, experiments here with neutralizing antibodies to IFNβ showed pIC-mediated IL-10 induction is independent of IFN (Fig. 5E). This may reflect a different regulation mechanism in the response to pIC as opposed to LPS. Correspondingly, Toshchakov et al. (51) showed that TLR2, unlike TLR4 agonists, induce pro-inflammatory genes independent of IFNβ induction. Consequently, PKR may participate differently in signaling pathways triggered by TLR4 versus TLR3 or other dsRNA receptors (52). Alternatively, PKR could be responding directly to pIC to induce IL-10. Another possibility is that the independence from IFN signaling shown here may reflect the cell lines being investigated. Previous data have shown macrophages and myeloid dendritic cells, but not the key IFN-producing plasmacytoid dendritic cells, producing high levels of IL-10 in response to TLR9 signaling (53). Furthermore, IL-10 suppresses proinflammatory cytokines produced by plasmacytoid dendritic cells in trans. Consequently, uncoupling of IL-10 production from the IFN response, as shown here, is likely to be pivotal in the regulation of the inflammatory response.
Macrophage activation in response to different pathogens needs to be tightly regulated to prevent overproduction of pro-inflammatory cytokines that can result in tissue damage or autoimmune disorders. Earlier reports have described a role for PKR in the production of pro-inflammatory cytokines. Our findings demonstrate a novel role for PKR in the regulation of the anti-inflammatory cytokine IL-10. This response is demonstrated to be mediated through activation of JNK and NF-κB. Therefore, PKR may act as a regulator of innate immunity by maintaining the balance between pro- and anti-inflammatory cytokines in response to viral infection.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grants R01 AI034039 and P01 CA062220. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1–S4.
Footnotes
The abbreviations used are: PKR, protein kinase R; IFN, interferon; dsRNA, double-stranded RNA; IL, interleukin; TNFα, tumor necrosis factor α; LPS, lipopolysaccaride; RNAi, RNA interference; wt, wild-type; pkr-ko, pkr knockout; SM, spleen-derived macrophages; IκBα-SR, IκBα super-repressor; BMM, bone marrow-derived macrophages; PBS, phosphate-buffered saline; ChIP, chromatin immunoprecipitation; PI, propidium iodide; TLR, toll-like receptor; GFP, green fluorescent protein; FACS, fluorescent-activated cell sorting; FCS, fetal calf serum; EMSA, electrophoretic mobility shift assay; ELISA, enzyme-linked immunosorbent assay; JNK, c-Jun N-terminal kinase.
References
- 1.Williams, B. R., and Sadler, A. J. (2006) UCSD-Nature Molecule Pages (2006) doi: 10.1038/mp.a000792.01 [DOI]
- 2.Scheuner, D., Patel, R., Wang, F., Lee, K., Kumar, K., Wu, J., Nilsson, A., Karin, M., and Kaufman, R. J. (2006) J. Biol. Chem. 281 21458–21468 [DOI] [PubMed] [Google Scholar]
- 3.Hsu, L. C., Park, J. M., Zhang, K., Luo, J. L., Maeda, S., Kaufman, R. J., Eckmann, L., Guiney, D. G., and Karin, M. (2004) Nature 428 341–345 [DOI] [PubMed] [Google Scholar]
- 4.Zamanian-Daryoush, M., Mogensen, T. H., DiDonato, J. A., and Williams, B. R. (2000) Mol. Cell. Biol. 20 1278–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jiang, Z., Zamanian-Daryoush, M., Nie, H., Silva, A. M., Williams, B. R., and Li, X. (2003) J. Biol. Chem. 278 16713–16719 [DOI] [PubMed] [Google Scholar]
- 6.Gil, J., Garcia, M. A., Gomez-Puertas, P., Guerra, S., Rullas, J., Nakano, H., Alcami, J., and Esteban, M. (2004) Mol. Cell. Biol. 24 4502–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Meusel, T. R., Kehoe, K. E., and Imani, F. (2002) J. Immunol. 168 6429–6435 [DOI] [PubMed] [Google Scholar]
- 8.Cheung, B. K., Lee, D. C., Li, J. C., Lau, Y. L., and Lau, A. S. (2005) J. Immunol. 175 7218–7225 [DOI] [PubMed] [Google Scholar]
- 9.Fickenscher, H., Hor, S., Kupers, H., Knappe, A., Wittmann, S., and Sticht, H. (2002) Trends Immunol. 23 89–96 [DOI] [PubMed] [Google Scholar]
- 10.de Waal Malefyt, R., Yssel, H., Roncarolo, M. G., Spits, H., and de Vries, J. E. (1992) Curr. Opin. Immunol. 4 314–320 [DOI] [PubMed] [Google Scholar]
- 11.Gastl, G. A., Abrams, J. S., Nanus, D. M., Oosterkamp, R., Silver, J., Liu, F., Chen, M., Albino, A. P., and Bander, N. H. (1993) Int. J. Cancer 55 96–101 [DOI] [PubMed] [Google Scholar]
- 12.Dummer, W., Bastian, B. C., Ernst, N., Schanzle, C., Schwaaf, A., and Brocker, E. B. (1996) Int. J. Cancer 66 607–610 [DOI] [PubMed] [Google Scholar]
- 13.D'Andrea, A., Aste-Amezaga, M., Valiante, N. M., Ma, X., Kubin, M., and Trinchieri, G. (1993) J. Exp. Med. 178 1041–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Waal Malefyt, R., Figdor, C. G., Huijbens, R., Mohan-Peterson, S., Bennett, B., Culpepper, J., Dang, W., Zurawski, G., and de Vries, J. E. (1993) J. Immunol. 151 6370–6381 [PubMed] [Google Scholar]
- 15.Gruber, M. F., Williams, C. C., and Gerrard, T. L. (1994) J. Immunol. 152 1354–1361 [PubMed] [Google Scholar]
- 16.Moore, K. W., de Waal Malefyt, R., Coffman, R. L., and O'Garra, A. (2001) Annu. Rev. Immunol. 19 683–765 [DOI] [PubMed] [Google Scholar]
- 17.Smale, S. T., and Fisher, A. G. (2002) Annu. Rev. Immunol. 20 427–462 [DOI] [PubMed] [Google Scholar]
- 18.de Waal Malefyt, R., Haanen, J., Spits, H., Roncarolo, M. G., te Velde, A., Figdor, C., Johnson, K., Kastelein, R., Yssel, H., and de Vries, J. E. (1991) J. Exp. Med. 174 915–924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ding, L., Linsley, P. S., Huang, L. Y., Germain, R. N., and Shevach, E. M. (1993) J. Immunol. 151 1224–1234 [PubMed] [Google Scholar]
- 20.Schols, D., and De Clercq, E. (1996) J. Virol. 70 4953–4960 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Giambartolomei, G. H., Dennis, V. A., Lasater, B. L., and Philipp, M. T. (1999) Infect. Immun. 67 140–147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.van Furth, A. M., Verhard-Seijmonsbergen, E. M., Langermans, J. A., van Dissel, J. T., and van Furth, R. (1999) Infect. Immun. 67 3714–3718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Libraty, D. H., Airan, L. E., Uyemura, K., Jullien, D., Spellberg, B., Rea, T. H., and Modlin, R. L. (1997) J. Clin. Investig. 99 336–341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Daftarian, P. M., Kumar, A., Kryworuchko, M., and Diaz-Mitoma, F. (1996) J. Immunol. 157 12–20 [PubMed] [Google Scholar]
- 25.Brightbill, H. D., Plevy, S. E., Modlin, R. L., and Smale, S. T. (2000) J. Immunol. 164 1940–1951 [DOI] [PubMed] [Google Scholar]
- 26.Cao, S., Zhang, X., Edwards, J. P., and Mosser, D. M. (2006) J. Biol. Chem. 281 26041–26050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao, S., Liu, J., Song, L., and Ma, X. (2005) J. Immunol. 174 3484–3492 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ziegler-Heitbrock, L., Lotzerich, M., Schaefer, A., Werner, T., Frankenberger, M., and Benkhart, E. (2003) J. Immunol. 171 285–290 [DOI] [PubMed] [Google Scholar]
- 29.Wang, Z. Y., Sato, H., Kusam, S., Sehra, S., Toney, L. M., and Dent, A. L. (2005) J. Immunol. 174 2098–2105 [DOI] [PubMed] [Google Scholar]
- 30.Williams, L. M., Ricchetti, G., Sarma, U., Smallie, T., and Foxwell, B. M. (2004) Immunology 113 281–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang, Y. L., Reis, L. F., Pavlovic, J., Aguzzi, A., Schafer, R., Kumar, A., Williams, B. R., Aguet, M., and Weissmann, C. (1995) EMBO J. 14 6095–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grabstein, K. H., Hess, B., Weisser, K. E., Clark, L., Goodwin, R., and Overell, R. W. (1992) J. Immunol. 149 1524–1530 [PubMed] [Google Scholar]
- 33.Wang, X., and Seed, B. (2003) Nucleic Acids Res. 31 e154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lucas, M., Zhang, X., Prasanna, V., and Mosser, D. M. (2005) J. Immunol. 175 469–477 [DOI] [PubMed] [Google Scholar]
- 35.Goh, K. C., deVeer, M. J., and Williams, B. R. (2000) EMBO J. 19 4292–4297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Finbloom, D. S., and Winestock, K. D. (1995) J. Immunol. 155 1079–1090 [PubMed] [Google Scholar]
- 37.Chang, E. Y., Guo, B., Doyle, S. E., and Cheng, G. (2007) J. Immunol. 178 6705–6709 [DOI] [PubMed] [Google Scholar]
- 38.Kumar, A., Yang, Y. L., Flati, V., Der, S., Kadereit, S., Deb, A., Haque, J., Reis, L., Weissmann, C., and Williams, B. R. (1997) EMBO J. 16 406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Alexopoulou, L., Holt, A. C., Medzhitov, R., and Flavell, R. A. (2001) Nature 413 732–738 [DOI] [PubMed] [Google Scholar]
- 40.Kato, H., Sato, S., Yoneyama, M., Yamamoto, M., Uematsu, S., Matsui, K., Tsujimura, T., Takeda, K., Fujita, T., Takeuchi, O., and Akira, S. (2005) Immunity 23 19–28 [DOI] [PubMed] [Google Scholar]
- 41.Kumar, A., Haque, J., Lacoste, J., Hiscott, J., and Williams, B. R. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 6288–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paludan, S. R. (2001) J. Virol. 75 8008–8015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Deb, A., Zamanian-Daryoush, M., Xu, Z., Kadereit, S., and Williams, B. R. (2001) EMBO J. 20 2487–2496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lopez, C. B., Moltedo, B., Alexopoulou, L., Bonifaz, L., Flavell, R. A., and Moran, T. M. (2004) J. Immunol. 173 6882–6889 [DOI] [PubMed] [Google Scholar]
- 45.Li, K., Chen, Z., Kato, N., Gale, M., Jr., and Lemon, S. M. (2005) J. Biol. Chem. 280 16739–16747 [DOI] [PubMed] [Google Scholar]
- 46.Elco, C. P., Guenther, J. M., Williams, B. R., and Sen, G. C. (2005) J. Virol. 79 3920–3929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Diebold, S. S., Montoya, M., Unger, H., Alexopoulou, L., Roy, P., Haswell, L. E., Al-Shamkhani, A., Flavell, R., Borrow, P., and Reis e Sousa, C. (2003) Nature 424 324–328 [DOI] [PubMed] [Google Scholar]
- 48.Kato, H., Takeuchi, O., Sato, S., Yoneyama, M., Yamamoto, M., Matsui, K., Uematsu, S., Jung, A., Kawai, T., Ishii, K. J., Yamaguchi, O., Otsu, K., Tsujimura, T., Koh, C. S., Reis e Sousa, C., Matsuura, Y., Fujita, T., and Akira, S. (2006) Nature 441 101–105 [DOI] [PubMed] [Google Scholar]
- 49.Yoneyama, M., Kikuchi, M., Natsukawa, T., Shinobu, N., Imaizumi, T., Miyagishi, M., Taira, K., Akira, S., and Fujita, T. (2004) Nat. Immunol. 5 730–737 [DOI] [PubMed] [Google Scholar]
- 50.Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S., and Reis e Sousa, C. (2004) Science 303 1529–1531 [DOI] [PubMed] [Google Scholar]
- 51.Toshchakov, V., Jones, B. W., Perera, P. Y., Thomas, K., Cody, M. J., Zhang, S., Williams, B. R., Major, J., Hamilton, T. A., Fenton, M. J., and Vogel, S. N. (2002) Nat. Immunol. 3 392–398 [DOI] [PubMed] [Google Scholar]
- 52.Asakura, Y., Fujiwara, Y., Kato, N., Sato, Y., and Komatsu, T. (2007) Am. J. Hematol. 82 640–642 [DOI] [PubMed] [Google Scholar]
- 53.Boonstra, A., Rajsbaum, R., Holman, M., Marques, R., Asselin-Paturel, C., Pereira, J. P., Bates, E. E., Akira, S., Vieira, P., Liu, Y. J., Trinchieri, G., and O'Garra, A. (2006) J. Immunol. 177 7551–7558 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.