Abstract
We report a role for CA repeats in the 3′-untranslated region (3′-UTR) in regulating CD154 expression. Human CD154 is encoded by an unstable mRNA; this instability is conferred in cis by a portion of its 3′-UTR that includes a polypyrimidine-rich region and CA dinucleotide repeat. We demonstrate similar instability activity with the murine CD154 3′-UTR. This instability element mapped solely to a conserved 100-base CU-rich region alone, which we call a CU-rich response element. Surprisingly, the CA dinucleotide-rich region also regulated reporter expression but at the level of translation. This activity was associated with poly(A) tail shortening and regulated by heterogeneous nuclear ribonucleoprotein L levels. We conclude that the CD154 3′-UTR contains dual cis-acting elements, one of which defines a novel function for exonic CA dinucleotide repeats. These findings suggest a mechanism for the association of 3′-UTR CA-rich response element polymorphisms with CD154 overexpression and the subsequent risk of autoimmune disease.
Post-transcriptional regulation plays a critical role in immune homeostasis at the level of the intact animal. Mutations that perturb post-transcriptional control of tumor necrosis factor (TNF)4 gene expression through the 3′-UTR AU-rich element (ARE) exhibited severe joint and bowel inflammation due to TNF overexpression (1, 2). These studies and others established that the 3′-UTR ARE regulates TNF expression at the level of nuclear export, mRNA stability and translation (1, 3-5).
CD154 (CD40 ligand) is a member of the TNF gene family; it is transiently expressed by activated T lymphocytes to play a central, nonredundant role in humoral and cellular immunity (6-9). Humans with mutations of the CD154 gene experience a severe immunodeficiency characterized by high levels of IgM and the absence of IgG (6, 8). These studies and others underscore the central role of CD154 interacting with CD40 on B cells in mediating Ig class switching. Like TNF and interleukin-2, CD154 mRNA initially exhibits rapid (t½ 20-30 min) decay in activated human T cells (10-12). After prolonged (>24 h) activation, CD154, but not cytokine, mRNA exhibits increased stability (11). A distinct pattern of post-transcriptional regulation is also supported by the ability of LFA-3-CD2 interactions to selectively increase CD154 mRNA stability (13). These studies suggested that the post-transcriptional regulation of CD154 involves cis-trans pathways other than ARE-dependent gene expression.
This hypothesis was supported by chimeric reporter gene studies that mapped a cis-acting instability element to a ∼330-nucleotide (nt) region in the human CD154 3′-UTR that lacked an ARE (14). This region contained a polypyrimidine (CU)-rich region juxtaposed to an extended CA dinucleotide repeat. Changes in CD154 mRNA turnover correlated with the cytoplasmic levels of p55 and p25 proteins that directly contacted cytidines or uridines in this region (10). The p55 protein was purified and sequenced and found to be polypyrimidine tract-binding protein (PTB), also known as hnRNP I (14). Overexpression of PTB increased the cytoplasmic stability of reporter mRNA containing the CD154 3′-UTR instability element (14). The p25 protein derived from a splice isoform of PTB missing exons 3-10 that resulted in an in frame deletion was referred to as PTB-T (14). Overexpression of PTB-T decreased luciferase mRNA stability when this 330-nt instability sequence in the human CD154 3′-UTR was present (14). These data indicated that the competition of specific PTB proteins for binding to CU-rich sequences in this region modulated CD154 mRNA decay (14). With prolonged T cell activation, cytoplasmic PTB levels increased relative to PTB-T (10, 14), consistent with the increased stability of CD154 mRNA seen at later time points (11).
Studies of the increased CD154 mRNA stability seen in activated (24 h) human T cells as well as the D1.1 mutant of the Jurkat cell line demonstrated that PTB proteins from D1.1 cell cytoplasm interacted at three separate sites within this region (11, 15). Deletions that eliminated PTB binding to the CD154 3′-UTR decreased the stability of CD154 mRNA seen in D1.1 cells. Subsequent work suggested that the interaction of PTB and nucleolin with this region of the CD154 3′-UTR correlated with the increased mRNA stability (16). Thus, the stabilization of CD154 mRNA that occurs with prolonged T cell activation involves the differential effects of PTB splice isoforms as well as the interaction of PTB with other trans-acting factors.
Despite these insights, it was still unclear if the dispersed 3′-UTR CU-rich elements in the human CD154 3′-UTR alone conferred the increased CD154 mRNA turnover seen early in T cell activation as well as with chimeric reporter genes (10-12, 14). This is particularly important due to the presence of other 3′-UTR elements (polycytidines, CA repeats) in this region that are associated with post-transcriptional gene regulation (17-21). The conserved murine CD154 3′-UTR has advantages for this analysis, given the condensed (180 nt) nature of the CU- and CA-rich regions (Fig. 1). Using transient transfection of chimeric reporter constructs, we demonstrated that the conserved 100-base CU-rich region in the murine CD154 3′-UTR functions as a cis-acting cytoplasmic instability element.
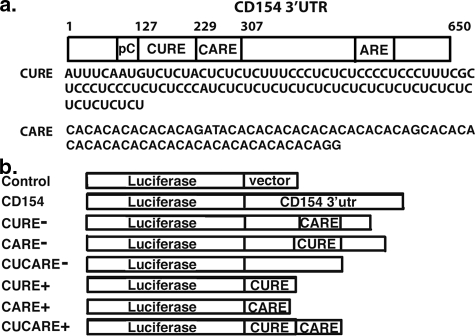
FIGURE 1.
Diagram of murine CD154 mRNA 3′-UTR. a, locations of CUREs, CAREs, polycytidines (pC), and ARE sequences are shown, numbered in reference to translational stop site. CURE and CARE nucleotide sequences are shown immediately below. b, design of pcDNA 3.1- or pTRE-based luciferase reporter plasmids and abbreviations.
This analysis of the CU-rich instability element (CURE) was confounded by the discovery of a second cis-acting element in the CA-rich region that regulates translation. Immediately 3′ to the CURE is a series of 7, 10, and 17 CA dinucleotide repeats. CA dinucleotide repeats are the most common simple repeat in the mammalian genome (22), but their functional role in gene expression has only recently been explored. The polymorphic nature of intronic CA repeats have been shown to regulate splicing efficiency and gene expression (18-21). In contrast, there is little understanding of the role of the exonic CA repeats, which are less frequent. Our studies demonstrate that 3′-UTR CA-rich repeats regulate gene expression at the level of translation. Recent work indicates that the CA repeats in the CD154 3′-UTR are polymorphic and influence both gene expression and autoimmune disease risk (23, 24). Our study not only defines the role of hnRNP L in regulating the function of exonic CA repeats but also provides a molecular mechanism of how CA repeat polymorphisms influence CD154 expression to increase the risk of rheumatoid arthritis (RA) and systemic lupus erythematosus (SLE).
EXPERIMENTAL PROCEDURES
Plasmids and Antisera—The pcDNA 3.1/LUC and tetracycline-responsive vector pTRE-LUC utilize the bovine growth hormone and β-globin polyadenylation signal sequences, respectively, and have been previously described (14). The murine CD154 3′-UTR corresponding to nt 23-650 relative to the translational stop site was amplified from total cellular RNA from B6 splenocytes activated with PMA (10 ng/ml; Sigma) plus ionomycin (0.5 μm) and cloned into TOPO 2.1 (Invitrogen). Sequencing confirmed identity with murine CD154 (Gen-Bank™ accession number 560692). Deletion of the CURE (nt 127-228) or the CARE (nt 229-306) from the CD154 3′-UTR corresponds to the CURE- and CARE-, respectively, whereas the CURE and CARE (nt 127-306) deletion is referred to as CUCARE-. All were generated by QuikChange (Stratagene) deletion from TOPO 2.1, as were the polycytidine and ARE mutations in the context of the CU/CARE-, and confirmed by sequencing. These sequences were released by EcoRI and cloned into the XbaI site in pcDNA 3.1 firefly luciferase (pcDNA3.1/LUC) vector (14). The CUCARE+ reporter was amplified and cloned into TOPO 2.1 and then released and ligated into pcDNA 3.1 luciferase, as described above. The CURE (CURE+) and CARE (CARE+) reporters were generated by QuikChange from plasmids containing the CUCARE+ in TOPO 2.1 and cloned into pcDNA 3.1 luciferase as described above. The pTRE-Luc vectors were generated by cloning inserts from the TOPO 2.1 vectors into the EcoRV site downstream of the luciferase coding region. The CARE was cloned downstream of the polyadenylation signal sequence in the BamHI site of pGL3-promoter vector (Promega).
Short hairpin RNA cDNA (HuSH plasmids) targeting hnRNP L were purchased from Origene; their activity was confirmed in HeLa cells by immunoblotting. Generation of pcDNA3.1-hnRNP L resulted from release of hnRNP L from pFASTBac hnRNP L (generously provided by Dr. Albert Bindereif) through EcoRI and XhoI digestion and cloning into EcoRI and XhoI sites in pcDNA 3.1. Mouse anti-green fluorescent protein antibody and goat anti-TIA (T-cell intracellular antigen) antiserum were obtained from Invitrogen and Santa Cruz Biotechnology, respectively. Human serum containing antibodies that react with Ge-1, a component of the cytoplasmic RNA processing body, was previously described (25). Fluorescein isothiocyanate- or rhodamine-conjugated, species-specific donkey anti-human, anti-mouse, and anti-goat IgG antisera were obtained from Jackson ImmunoResearch Laboratories.
Transient Transfection Assay of Reporter Gene Activity—Except for small interfering RNA and hnRNP L overexpression studies, 5 × 104 HeLa cells were transfected with 50 ng of luciferase vectors, 1.5 μl of Lipofectamine (Invitrogen), and 4 μl of PLUS (Invitrogen) in 0.5 ml of RPMI for 3.5 h at 37 °C 5% CO2, after which 0.5 ml of RPMI plus 20% fetal calf serum was added. After 20 h, cells were lysed, and luciferase activity was determined by luminometry. Individual experiments were analyzed for 3′-UTR-specific effects by dividing the mean luciferase activity from triplicate transfections of pcDNA3.1/LUC- or pTRE-LUC-based expression plasmids by that obtained from cells transfected with the corresponding control vector, which was assigned a value of 100%. Identical results were obtained when we utilized cotransfection of Renilla luciferase vectors to control for changes in transfection efficiency (data not shown). In short hairpin RNA and hnRNP L overexpression experiments, cells were transiently transfected at day -2 with either 500 ng of HuSH 303 or HuSH L79 or 250 ng of empty pcDNA 3.1 control vector or pcDNA 3.1 hnRNP L, followed by a repeat cotransfection of these plasmids along with the corresponding luciferase reporters at day 0 with 40-60% transfection efficiency of the effector vectors. Separate cultures received equivalent amounts of the corresponding control vector (empty or containing an irrelevant RNA sequence) on days -2 and 0. Transient transfection of human peripheral blood mononuclear cells was performed using AMAXA nucleofection. After being transiently transfected overnight, luciferase activity was measured. Additional cultures were activated with PMA/ionomycin for 4 h prior to analysis of luciferase activity.
RNA Analysis by Quantitative RT-PCR—Cytoplasmic RNA was extracted as previously described (26). Nuclei were pelleted and resuspended in Solution 1 (10 mm Tris, 150 mm NaCl, 1.5 mm MgCl2, and 0.65% Nonidet P-40) and spun through a 30% sucrose cushion, resuspended in Solution 1, and spun through a 30% sucrose cushion again. Nuclear RNA was then extracted as previously described (27). Poly(A) RNA was purified using Oligotex beads (Qiagen). Isolation of a polysome-enriched fraction was performed as previously described (28). Cells were washed three times with 1× phosphate-buffered saline and resuspended in 3.5 ml of Buffer A (10 mm Tris-HCl, pH 7.6, 1 mm KAc, 1.5 mm MgAc, 2 mm dithiothreitol, 1 mg/ml pepstatin A, 1 mg/ml leupeptin, 1 mm pefabloc). The inclusion of EDTA (10 mm) in the homogenization buffer was used to disrupt polysomes prior to purification (29). Cells were lysed by 20 strokes with a Teflon pestle homogenizer at 1,500 rpm and centrifuged at 12,000 × g for 10 min to pellet the nuclei, followed by layering of the supernatant onto a 30% sucrose gradient in buffer A that was centrifuged at 4 °C for 5 h at 36,000 rpm. RNA was extracted from polysomes and S130, and RT-PCR analysis was performed as described (14, 27). For studies of mRNA stability, Tet-Off HeLa cells (Clontech) were purchased and carried according to the manufacturer's instructions and transiently transfected as described above, allowed to recover overnight, and then treated with doxycycline (1 μg/ml) to shut off transcription for specified times. Analysis of the effects of priming on gene expression utilized cytoplasmic RNA that was digested with TurboDNase I (Ambion) and then reverse transcribed with either oligo(dT), random hexamers, or a luciferase-specific primer (5′-TTTGGCGGTTGTTACTTGAC-3′) and Superscript II RT (Invitrogen). Reverse transcriptions were analyzed for luciferase transcripts using primers for luciferase, GAPDH, and H4 histone RNA: luciferase, 5′-GGTGGCTCCCGCTGAATTGG-3′ (upper) and 5′-CCGTCATCGTCTTTCCGTGC-3′ (lower); GAPDH, 5′-ACCACCTTCTTGATGTCATC-3′ (upper) and 5′-CAAGGCTGTGGGCAAGGTCA-3′ (lower); H4 histone, 5′-CAACATTCAGGGCATCACCAA-3′ (upper) and 5′-CCCGAATCACATTCTCCAAGAA-3′ (lower).
Oligo(dT) and random hexamer (RH) priming of reverse transcriptions were analyzed for GAPDH and histone H4 transcripts to control for input RNA. Real time PCR was performed using a Bio-Rad iCycler and IQ SYBR Green Supermix (Bio-Rad product number 170-8882). The luciferase/GAPDH or H4 transcript ratio was calculated for each sample, where Ct = threshold cycle and ΔCt = luciferase Ct - GAPDH (oligo(dT)-primed RT) or H4 (RH-primed RT) Ct. ΔΔCt =ΔCt1 - ΔCt2, where ΔCt1 is CD154 and ΔCt2 is control with the -fold difference = 2-ΔΔCt.
In these experiments, the percentage inhibition of CD154 3′UTR-dependent luciferase expression seen with each vector and priming method was calculated and then divided by the inhibition seen with the empty control vector, which was assigned a value of 100%. In some instances (Fig. 4), data are presented where the ΔCt obtained with oligo(dT) for a given transfection is subtracted from that obtained with random hexamer (ΔCtRH - ΔCtdT) or luciferase-specific priming. Student's t test was performed, and p values were determined using Excel.
FIGURE 4.
The presence of the 3′-UTR-CARE is associated with differential priming of reverse transcription by oligo(dT) and random hexamers. a, total cytoplasmic RNA extracts from transient transfections of HeLa cells with specified pcDNA 3.1-based vectors were analyzed by quantitative PCR using luciferase primers following reverse transcription with either oligo(dT) or random hexamers. Poly(A) purification was omitted. The 3′-UTR CARE and the CUCARE+, but not the CURE alone, affected the measurable amount of input luciferase RNA as a function of priming with increased levels seen with random hexamers. Data are shown as a percentage of input RNA in control transfection. b, summary of effect of CARE on input luciferase mRNA measured by quantitative PCR following priming shown as random hexamers relative to oligo(dT) (n = 8).c, cytoplasmic RNA was analyzed by quantitative PCR following priming of reverse transcription with oligo(dT), RHs, or a luciferase-specific primer. Data are presented as a ratio of the effect of reverse transcription primer or luciferase-specific primer on apparent input RNA relative to that seen with oligo(dT). The 3′-UTR CARE increased the ratio of input luciferase mRNA with both luciferase-specific priming and random hexamers by 4-fold relative to that seen with oligo(dT). In contrast, little or no effect was seen on luciferase mRNA from cells transfected with control and 3′-UTR-CURE-containing plasmids.
Immunoprecipitation Analysis—HeLa cells were transiently transfected as specified and cultured overnight, or human peripheral blood mononuclear cells were activated for 24 h with PMA (10 ng/ml) and ionomycin (1 μm). Cytoplasmic and nuclear extractions were performed as previously described (10, 14) with the addition of Protector RNase inhibitor (Roche Applied Science). Immunoprecipitations from extracts were performed in immunoprecipitation buffer (10 mm Tris, pH 7.6, 1.5 mm MgCl2, 0.05% Triton X-100, 150 mm NaCl, 2 μm pefabloc, 2 ng/ml each leupeptin and pepstatin A, 4 units/ml RNase inhibitor from Roche Applied Science) in parallel with 4D11 (anti-RNP L) and BB7 (anti-PTB) antibodies as well as a mouse IgG isotype control bound to protein A-Sepharose beads (Amersham Biosciences). Beads were washed six times with 10 volumes of immunoprecipitation buffer, and 5% of packed beads were boiled in SDS-PAGE loading buffer and resolved by 12% SDS-PAGE and immunoblotted. To study the RNA tethering of PTB and hnRNP L, RNase A digestion was performed prior to immunoprecipitation, as previously described (30). Similar results were obtained with the incubation of beads with RNase A (1 unit/ml, 37 °C, 30 min; Roche Applied Science) at 37 °C treatment following immunoprecipitation. The remaining beads were digested with proteinase K (Roche Applied Science) and extracted with phenol/chloroform. Following DNase I digestion, the presence of human CD154 or luciferase mRNA in each precipitation was measured by oligo(dT)-based reverse transcription and quantitative PCR using primer specific for luciferase (described above) or human CD154, GAPDH (described above), interleukin-2, or γ-interferon primers as listed below: CD154, 5′-TTGCGGGCAACAATCCATTCACTT (upper) and 5′-GTGGGCTTAACCGCTGTGCTGTATT (lower); interleukin-2, 5′-AACCTCAACTCCTGCCACA (upper) and 5′-CCTGGTCACTTTGGGATTCT (lower); γ-interferon, 5′-GGCAAGGCTATGTGATTAC (upper) and 5′-TTTCCATTTGGGTACAGTCA (lower).
Analysis of Poly(A) Tail Length—Poly(A) tail length was measured by a ligase-mediated polyadenylation tail assay (LM-PAT) assay (31), in which the 3′-end of the poly(A) tail was hybridized to a primer containing oligo(dT)16 and a GC “anchor” sequence, 5′-GCGAGCTCCGCGGCCGCT16-3′ (Operon). Target mRNA (100 ng) was incubated with phosphorylated oligo(dT)16 (Roche Applied Science) at 42 °C in the presence of 10 units of T4 DNA ligase (Invitrogen) saturating the poly(A) tail, thereby creating an oligo(dT) copy of the poly(A) tail. At 42 °C, the 3′-end of the poly(A) tail remained largely unpaired due to unfavorable hybridization conditions. The oligo(dT)-GC anchor sequence was added at a 10-fold molar excess, and the temperature was reduced to 12 °C, enabling selective hybridization to the unpaired 3′-ends. Reverse transcription was performed (Superscript II reverse transcriptase; Invitrogen), followed by PCR using a primer corresponding to the GC-rich sequence in the oligo(dT) anchor along with a primer specific for the mRNA to be analyzed. Primers used for LM-PAT assay were as follows: luciferase, GCCATCTGTTGTTTGCC; mCD154, CTGTCTACAGCACTGTCGGG; mTNF, CACCTGGCCTCTCTACCTTG. Primers are designed so that the PCR product encodes a restriction enzyme site: MnlI, AseI, and HphI, respectively. Products are resolved by agarose gel electrophoresis, and the identity of the visualized band was confirmed by restriction enzyme digestion.
Immunohistochemistry—Hep-2 cells were transfected with a eukaryotic expression plasmid encoding green fluorescent protein-hnRNP L using the Effectene transfection system (Qiagen). To induce stress granule formation, Hep-2 cells were exposed to sodium arsenite (0.5 mm) for 1 h at 37 °C. For immunofluorescent staining, Hep-2 cells were fixed with 2% paraformaldehyde in PBS and permeabilized by treatment with 100% methanol. Cells were stained with primary and secondary antisera as previously described (25).
RESULTS
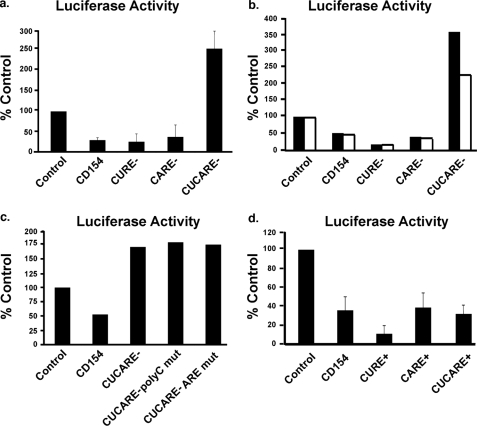
The Murine CD154 3′-UTR Has Two cis-Acting Elements That Independently Regulate Reporter Gene Expression—The murine CD154 3′-UTR exhibits highly conserved CU- and CA-rich elements as well as polycytidine (pC) and ARE (Fig. 1). Relative to its human counter-part, the murine CURE and CARE are condensed (∼180 nt). Transient transfection of HeLa cells with chimeric reporter constructs demonstrated that the conserved murine CD154 3′-UTR functioned to reduce luciferase expression (Fig. 2a). The magnitude of this effect was equivalent to that seen with the human CD154 3′-UTR (14). Prior studies established that human CD154 mRNA levels and stability were regulated by the interaction of PTB proteins with canonical binding sites in a region in the CU-rich portion of the 3′-UTR (11, 14). Thus, deletion of these PTB binding sites contained in the CURE of the murine CD154 3′-UTR (CURE-) was predicted to eliminate its ability to inhibit reporter gene expression (Fig. 1). Unexpectedly, little or no decrease in inhibition was seen with the CURE-CD154 3′-UTR (Fig. 2a). Deletion of the CARE (CARE-) from the 3′-UTR also retained inhibitory activity. Excision of both CURE and CARE (CUCA-) increased luciferase activity that approached or exceeded that seen with controls. Similar effects were seen with transient transfection of human peripheral blood mononuclear cells under basal and short term activation conditions (Fig. 2b). These data suggest that CURE and CARE both regulated reporter gene expression.
FIGURE 2.
The murine CD154 3′-UTR has two cis-acting elements that regulate reporter gene expression cytoplasmic poly(A)+ mRNA accumulation. a, murine CD154 3′-UTR limits pcDNA 3.1 reporter gene expression in transiently transfected HeLa cells. Deletion of both CURE and CARE (CUCARE-) was required to eliminate inhibitory effect of CD154 3′-UTR on luciferase gene expression. Data shown are an average of four experiments, with error bars representing the S.E. (n = 4). b, the CARE and CURE function to regulate reporter gene expression in human peripheral blood mononuclear cells under both resting and activated conditions. Transient transfection with the specified reporter vectors was performed, and luciferase activity was measured under basal (black bars) or activated (open bars) conditions, where phorbol 12-myristate 13-acetate (10 ng/ml) and ionomycin (1 μm) were added for the last 4 h. c, mutations of the polycytidine or ARE in the context of the CUCARE-deletions had no effect on gene expression. HeLa cells were transiently transfected with pcDNA 3.1-based plasmids containing the control, CD154 3′-UTR, or CUCARE-, mutation. The CUCARE-polyCmut and the CUCARE-AREmut additionally contained deletions of the polycytidine and ARE sequences, respectively. Error bars are not visible at this scale in this representative experiment (n = 4). d, the CURE and CARE alone are each sufficient to limit reporter gene expression effect on luciferase expression, whereas combination of the CURE and CARE (CUCARE+) did not enhance their activity (n = 4).
Mutations of other candidate cis-acting elements (the retained AURE, poly(C) sequence) in the context of CURE and CARE deletion (CUCA-) had no effect on reporter gene expression (Fig. 2c). Insertion of the CURE (CARE+) or CARE (CARE+) alone in the 3′-UTR of reporter genes demonstrated that each cis-acting element was equipotent in reducing luciferase activity (Fig. 2d). Thus, either the CURE or the CARE is sufficient to transfer their effect when present in the 3′-UTR of heterologous transcripts. However, combining the CURE and CARE (CUCA+) did not further enhance their effects on luciferase expression (Fig. 2d).
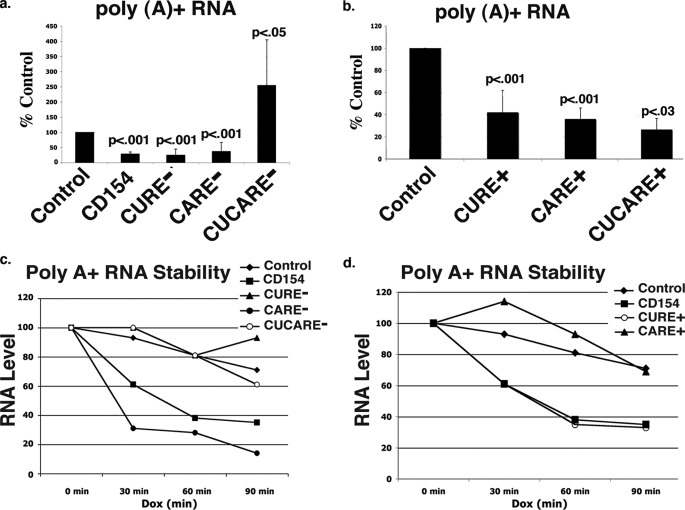
The CURE and CARE in the Murine CD154 3′-UTR Regulate Reporter Gene Expression through Different Pathways—Using transient transfection of plasmid reporters into HeLa cells, we examined the effect of the murine CD154 3′-UTR on cytoplasmic poly(A) mRNA levels, using oligo(dT) priming and RT-PCR (Fig. 3). Consistent with the luciferase activity data, the presence of the murine CD154 3′-UTR markedly reduced steady state levels of luciferase mRNA; deletion of the CURE or CARE alone had no effect on this inhibition (Fig. 3a). Mutation of both the CARE and CURE increased mRNA levels relative to controls. The presence of either the CURE or CARE alone reduced cytoplasmic levels of poly(A) luciferase mRNA (Fig. 3b). Combination of the CURE and CARE (CUCARE+) resulted in similar changes to that of the CURE and CARE alone.
FIGURE 3.
The CURE and CARE represent separate cis-acting elements that differentially regulate the inhibitory activity of the CD154 3′-UTR. a, the murine CD154 3′-UTR limits pcDNA 3.1 reporter gene expression in transiently transfected HeLa cells at the level of cytoplasmic poly(A) mRNA as measured by oligo(dT)-primed RT-PCR. Deletion of both CURE and CARE (CUCARE-) was required to eliminate inhibitory effect of CD154 3′-UTR on poly(A) mRNA (n = 6). p values are shown relative to that seen with luciferase controls. b, the CURE and CARE alone are sufficient to limit cytoplasmic poly(A) mRNA accumulation (n = 6). c and d, pTRE-based plasmids were transiently transfected into HeLa Tet-Off cells with transcriptional inhibition triggered by the addition of doxycycline (1 μg/ml). Cytoplasmic RNA was extracted at the time of doxycycline addition and specified times thereafter with poly(A) RNA purified and analyzed by oligo(dT)-primed RT-PCR. The level of input RNA measured at time 0 for each vector is assigned a value of 100%, although both the CURE- and CARE-vectors reduced mRNA levels. Increased mRNA decay rates are only seen with reporters (c, CD154, CARE-, and CURE+) containing the CURE in their 3′-UTR. Data shown are representative of three experiments each.
Previous studies with tetracycline-repressible reporters demonstrated that a region containing both the CURE and CARE in the human CD154 3′-UTR increased cytoplasmic mRNA decay (14). Using this approach, the murine CD154 3′-UTR increased the rate of decay of cytoplasmic poly(A) mRNA (Fig. 3, c and d, and Table 1). Deletion of both the CURE and CARE (CUCA-) increased the stability of poly(A) luciferase mRNA, consistent with its effects on luciferase activity and mRNA accumulation. In contrast, mutation of the CARE (CARE-) still resulted in rapid mRNA turnover, suggesting a role of the retained CURE. In contrast, deletion of the CURE (CURE-) increased cytoplasmic poly(A) mRNA stability (Fig. 3c and Table 1).
TABLE 1.
Cytoplasmic poly(A) mRNA decay summary
t½ was calculated from the decay curve in each experiment performed as described in Fig. 3, utilizing transcient transfections of specified vectors into HeLa cells. Data shown represent mean ± S.E. from a specified number of experiments.
| Vector | t½ | No. of experiments |
|---|---|---|
| min | ||
| Control | >90 | 9 |
| CD154 | 48 ± 9 | 6 |
| CURE− | >90 | 2 |
| CARE− | 53 ± 20 | 3 |
| CU/CARE− | >90 | 4 |
| CURE+ | 42 ± 2.5 | 2 |
| CARE+ | >90 | |
| CU/CARE+ | 70 ± 8 | 2 |
Examination of the activity of the isolated 3′-UTR CURE and CARE was performed (Fig. 3d). The CURE alone (CURE+) increased mRNA decay, establishing that it alone confers cytoplasmic mRNA instability. When combined with the stability of the CURE-reporter, these data indicate that the instability associated with the CD154 3′-UTR derives solely from the CURE. In contrast, the CARE element alone (CARE+) had no effect on the decay of poly(A)+ mRNA, despite decreasing luciferase RNA levels. Thus, the CARE reduced luciferase activity and poly(A)+ mRNA levels independent of cytoplasmic decay, both in isolation as well as in the context of the CD154 3′-UTR (CURE-). The combination of the CURE and the CARE (CUCARE+) in the 3′-UTR resulted in cytoplasmic mRNA stability that was intermediate between that of the CURE and CARE alone, suggesting their interaction (Table 1).
The 3′-UTR CARE Regulates mRNA Poly(A) Tail Length in the Nucleus and Cytoplasm—The 3′-UTR CARE reduced cytoplasmic poly(A) mRNA levels without increasing mRNA decay, prompting examination of its function as a transcriptional silencer when placed downstream of the reporter gene. No effect was seen (data not shown). Since measurements of reporter mRNA by quantitative RT-PCR relied on the use of cytoplasmic poly(A) mRNA, we tested if these results were influenced by an effect of the 3′-UTR CARE on poly(A) tail length. Total cytoplasmic RNA was analyzed by RT-PCR in which the effect of priming with either oligo(dT) or random hexamers was compared (Fig. 4a). The 3′-UTR CURE reduced luciferase mRNA levels independent of priming, consistent with its effects on cytoplasmic mRNA decay. In contrast, random hexamer priming increased >3-fold the apparent levels of input CARE+ luciferase RNA relative to that seen with oligo(dT) (Fig. 4b). The CUCARE+ vector showed an intermediate result to both the CURE and CARE, suggesting that it also reduced poly(A) tail length. Similar results (4-fold increases) were seen in separate transfections in which luciferase-specific priming of reverse transcription and random hexamers were compared, indicating that this effect was not unique to random hexamer priming (Fig. 4c).
Since oligo(dT)-dependent priming of RT-PCR was selectively affected by the 3′-UTR CARE, we tested if poly(A) tail length was affected using the LM-PAT assay (31). The 3′-UTR CARE reduced the poly(A) tail to <50 adenylates, relative to the control and 3′-UTR CURE reporter mRNA, where poly(A) tails as long as 150 bases were seen (Fig. 5a). The effect of the 3′-UTR CARE on poly(A) tail length was evident in the CUCARE+. This result was consistent with the difference seen between random hexamer and oligo(dT) priming (Fig. 4a).
FIGURE 5.
The presence of the 3′-UTR-CARE is associated with a shortened poly(A) tail but increased cytoplasmic mRNA stability. a, the presence of the 3′-UTR-CARE is associated with increased deadenylation of reporter gene as well as native CD154 mRNA. Cytoplasmic RNA extracts from transiently transfected HeLa cells with specified pcDNA 3.1-based plasmids were assayed for poly(A) tail length. Cytoplasmic luciferase mRNA from transient transfected HeLa cells containing the 3′-UTR CARE has a markedly shortened (<50-nt) poly(A) tail relative to control or 3′-UTR CURE as measured by the LM-PAT assay. The arrow indicates the predicted length of fully deadenylated fragments. Note the extended poly(A) tail length (∼150 A bases) from transfections with control and CURE constructs. Presence of CURE markedly reduced luciferase mRNA while not affecting poly(A) length; the data shown in this gel have twice as much PCR product in this lane loaded to make this product visible by EtBr staining. The presence of the CURE and CARE (CUCA+) yielded a short poly(A) tail as well. b, the 3′-UTR CARE results in decreased luciferase mRNA poly(A) tail length in nuclear, cytoplasmic, and polysomal fractions. Right-hand arrows show the predicted fragments following MnlI restriction enzyme digestion, establishing the identity of the amplified product. c, the presence of the CARE in the 3′-UTR resulted in increased mRNA stability, whether measured by RT-PCR following oligo(dT)- or RH-dependent priming.
Furthermore, the effect of the 3′-UTR CARE was apparent in both nuclear and cytoplasmic extracts (Fig. 5b). Since the nuclei had been purified by spinning through a sucrose cushion twice prior to RNA extraction, these data suggest that the effect on poly(A) tail length by the 3′-UTR CARE is either transduced in the nucleus or that the cytoplasmic mRNA that are being measured copurify with the nucleus.
The 3′-UTR CARE Increases Cytoplasmic mRNA Stability in Association with Polysomal Loading of Hypoadenylated mRNA—In order to better understand the effect of the 3′-UTR CARE on cytoplasmic mRNA stability, we examined its effects over a prolonged period of time (Fig. 5c). Interestingly, the 3′-UTR CARE increased luciferase mRNA stability relative to controls independent of priming (Fig. 5c). Thus, the 3′-UTR CARE slows decay as well as deadenylation of this hypoadenylated mRNA in the cytoplasm. This latter result supports the model that the 3′-UTR CARE is regulating poly(A) tail length in the nucleus.
Subsequent analysis demonstrated that despite a short poly(A) tail, all of the luciferase mRNA containing the 3′-UTR CARE was found in the polysomal pellet that that had passed through a 30% sucrose gradient (data not shown). The disruption of polysomes by EDTA (10 mm) reduced polysomal levels of luciferase-CARE, GAPDH, and histone mRNA 4-8-fold. Thus, the presence of these mRNA in the pellet was dependent on intact polysomes (data not shown). LM-PAT analysis demonstrated that this efficient loading of luciferase mRNA containing the 3′-UTR CARE onto polysomes occurred despite a shortened poly(A) tail (Fig. 5b, right lanes). These data suggest that the effects of the 3′-UTR CARE on translation occurs despite normal polysomal loading, as reported with other mRNA (32, 33).
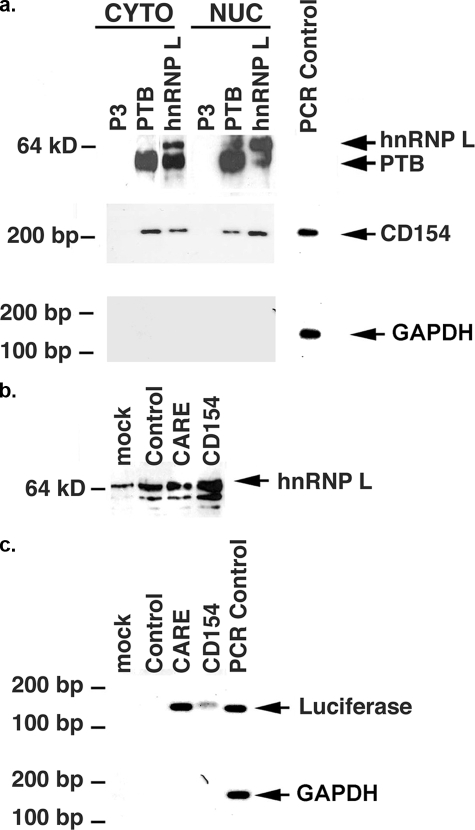
hnRNP L Binds CD154 mRNA through the 3′-UTR CARE to Regulate Poly(A) Tail Length—The in vitro binding of PTB proteins to the human CD154 3′-UTR was previously shown (10, 14, 15). Similar approaches have shown that the RNA-binding protein hnRNP L exhibits remarkable binding specificity and affinity for CA repeats (18-20). Immunoprecipitation of cytoplasmic and nuclear extracts from activated human peripheral blood mononuclear cells was performed (Fig. 6a). Immunoblotting demonstrated that PTB and hnRNP L coprecipitate. This coprecipitation was eliminated by RNase A digestion, indicating that the PTB-hnRNP L interaction is RNA-dependent (data not shown). RNA extraction of these immunoprecipitates followed by RT-PCR demonstrated that both PTB and hnRNP L bind native CD154 mRNA in vivo. No GAPDH, interleukin-2, or γ-interferon RNA was found in these immunoprecipitates, establishing the selectivity of PTB and hnRNP L with CD154 mRNA (Fig. 6a; data not shown). The in vivo interaction of hnRNP L with the murine CD154 3′-UTR and CARE was examined (Fig. 6, b and c). Following transient transfection of HeLa cells with reporter vectors that lacked (control) or contained these sequences, hnRNP L immunoprecipitates were analyzed for the presence of luciferase mRNA. Luciferase mRNA coprecipitated with hnRNP L in a 3′-UTR CARE-dependent manner. The specificity of this RNA-protein interaction was shown by the inability of hnRNP L to immunoprecipitate either GAPDH or luciferase mRNA that lacked a 3′-UTR CARE.
FIGURE 6.
hnRNP L interacts with CD154 mRNA in a 3′-UTR CARE-dependent manner. a, PTB and hnRNP L were immunoprecipitated from cytosolic and nuclear extracts from activated (PMA/ionomycin, 24 h) T cells. PTB (BB7 MoAb), hnRNP L (4D11), and P3 (irrelevant isotype control) were used. PTB and hnRNP L were coimmunoprecipitated in both extracts, although hnRNP L in PTB immunoprecipitate is not appreciated with this exposure. Immediately below is shown CD154 mRNA in hnRNP L and PTB immunoprecipitates from both cytoplasm and nucleus. No GAPDH, interleukin-2, or γ-interferon mRNA was detectable in either PTB or hnRNP L immunoprecipitates. b, immunoblot of hnRNP L immunoprecipitate from cytoplasmic extracts from HeLa cells transfected with control, CARE, or CD154 3′-UTR containing luciferase vectors or no vector (mock). c, hnRNP L immunoprecipitates contain luciferase mRNA in a 3′-UTR CARE-dependent manner, since no product is seen in extracts transfected with control vector lacking these sequences. Further specificity is shown by inability to detect GAPDH mRNA in immunoprecipitates.
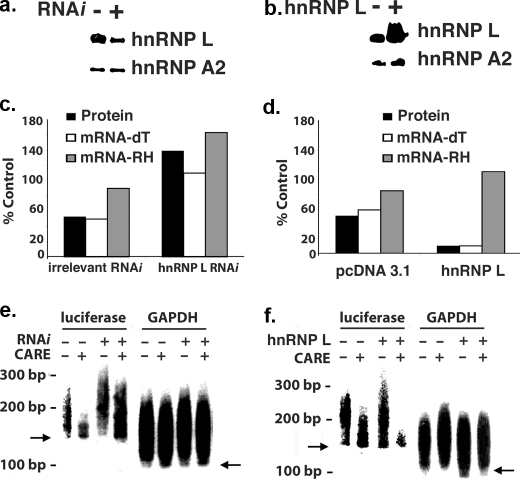
The functional significance of the hnRNP L-3′-UTR CARE interaction was tested. Knockdowns of hnRNP L levels by RNA interference eliminated the inhibitory effects of the 3′-UTR CARE on luciferase expression and poly(A) tail length (Fig. 7). Overexpression of hnRNP L strikingly enhanced the activity of the 3′-UTR CARE, increasing the inhibition of luciferase activity from ∼50% to 90% (Fig. 7). This enhanced suppression was accompanied by increased luciferase mRNA deadenylation, as measured by oligo(dT) priming of RT-PCR or LM-PAT assay. We conclude that hnRNP L binds the 3′-UTR CARE in vivo to regulate nuclear poly(A) tail length.
FIGURE 7.
Modulation of hnRNP L levels alters the ability of the 3′-UTR CARE to regulate poly(A) tail length. a and b, immunoblotting showing modulation of nuclear hnRNP L levels by RNA interference (a) or overexpression (b). Experiments utilized transient transfection of HeLa cells with irrelevant (HuSH 303) and hnRNP L-specific (HuSH L79) short hairpin RNA plasmids (Origene) and pcDNA 3.1-hnRNP L and empty vector control on days -2 and 0 with 40-60% efficiency. Immunoblotting for hnRNP A2 is used as a loading control. The pcDNA 3.1 CARE+ plasmid was cotransfected on day 0. c, knockdown of hnRNP L resulted in loss of 3′-UTR CARE effects on luciferase activity relative to irrelevant small interfering RNA. RT-PCR analysis with either oligo(dT) or RH priming shows that hnRNP L knockdown reduces differences seen with these priming methods, indicating restoration of normal poly(A) tail length. d, overexpression of hnRNP L enhanced CARE-dependent inhibition of luciferase activity and the differential between oligo(dT) and random hexamers in detecting luciferase mRNA. e, hnRNP L knockdown is associated with increased poly(A) tail length. Note that treatment with irrelevant small interfering RNA control had little or no effect. No effect on GAPDH mRNA polyadenylation was seen. The arrows indicate predicted size generated by deadenylated mRNA. f, overexpression of hnRNP L increases 3′-UTR CARE-dependent effects on luciferase expression and shortens the poly(A) tail length in a CARE-dependent manner. Note the lack of effect on GAPDH and control luciferase mRNA. (n = 3 for each experiment in this figure.)
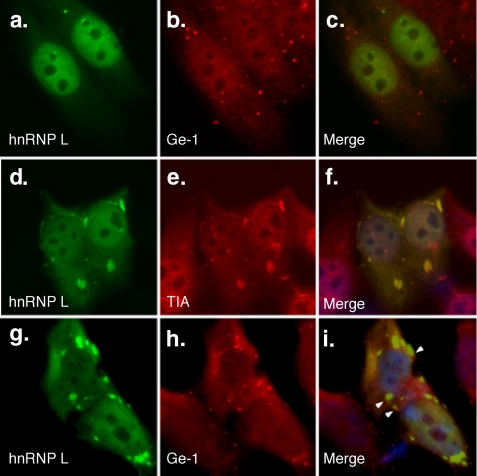
hnRNP L Traffics to Stress Granules but Not Processing Bodies—Cytoplasmic stress granules and mRNA-processing bodies (PBs) are frequently found in close proximity (reviewed in Ref. 34). Stress granules are thought to be sites for the sequestration of translationally inert or incompetent mRNA, whereas PBs mediate mRNA degradation (34). We examined the cytoplasmic trafficking of hnRNP L. In unstressed cells, hnRNP L is predominantly nuclear (Fig. 8a). PBs, identified using antibodies directed against Ge-1, a central component of the PB (25), appear as 5-10 dots in the cytoplasm of these cells (Fig. 8b). After exposure to arsenite-induced stress, hnRNP L localizes to cytoplasmic granules (Fig. 8d) and colocalizes with T-cell intracellular antigen (TIA), a marker for cytoplasmic stress granules (Fig. 8e). Although PBs are found adjacent to hnRNP L-containing stress granules (Fig. 8, g-i), there was no evidence of hnRNP L trafficking to mRNA PBs in either resting or stressed cells.
FIGURE 8.
Trafficking of hnRNP L to stress granules but not processing bodies. After transfection into Hep-2 cells, green fluorescent protein-labeled hnRNP L localized to the nucleus (green; a). Serum containing antibodies directed against mRNA processing body marker Ge-1 stained 5-10 dots in the cytoplasm of Hep-2 cells (red; b). After exposure to arsenite, hnRNP L localized to granules in the cytoplasm (d) and co-localized with T-cell intracellular antigen (TIA), a marker for stress granules (red; e). mRNA-processing bodies (h) localized adjacent to hnRNP L in stress granules (g). Overlap of green and red staining is shown in c, f, and i. 4′,6-Diamidino-2-phenylindole staining in f and i indicate the location of cell nuclei. White arrowheads (i) indicate the location of Ge-1-containing mRNA processing bodies adjacent to hnRNP L-containing stress granules.
DISCUSSION
Post-transcriptional Regulation of CD154 Expression by Adjacent cis-Acting Elements—Previous studies mapped the mRNA instability activity in the human CD154 3′-UTR to a 330-nt region containing CURE and CARE (14, 15). The cis- acting instability element in this region was never precisely identified. Using the conserved mouse CD154 3′-UTR, we demonstrate that the CURE (nt 127-228) alone functions as a cytoplasmic mRNA instability element. Surprisingly, the region immediately 3′ to the CURE, the CARE (nt 229-306), functioned as a separate cis-acting element that independently regulated reporter gene expression. In contrast to the CURE, the 3′-UTR CARE limited reporter gene mRNA poly(A) tail length and translation. Despite this effect on poly(A) tail length, the CARE did not reduce cytoplasmic mRNA levels relative to controls. Moreover, the 3′-UTR CARE enhanced cytoplasmic mRNA stability. Deletion of the CURE and CARE from the 3′-UTR resulted in loss of any inhibitory activity. Thus, CD154 expression is regulated by its 3′-UTR at the levels of mRNA stability and translation, as seen with TNF (1, 3, 4). In contrast to the ability of the TNF 3′-UTR ARE to regulate both mRNA stability and translation (1), these activities segregate to separate cis-acting elements in the CD154 3′-UTR.
Elements in the 3′-UTR have been shown to regulate splicing, poly(A) site selection, nuclear export, cytoplasmic mRNA stability, and translation (1, 3-5, 17, 35-37). CD154 plays a critical, nonredundant role in immunity (6-10). Hence, it is not surprising that a unique pattern of gene regulation has evolved. We now demonstrate that the murine CD154 3′-UTR regulates poly(A) tail length and does so through an exonic CA dinucleotide repeat. The 3′-UTR CARE conferred similar poly(A) shortening to the same degree in both nuclear and cytoplasmic mRNA extracts. The effect on nuclear RNA was seen with washing the nuclear pellet and spinning it through a sucrose cushion twice prior to RNA extraction. These data suggest that the CARE effect is present in the nucleus, although it is possible that this cytoplasmic mRNA is retained in association with the nucleus. Since the 3′-UTR CARE decreased the rate of cytoplasmic deadenylation (Fig. 5c), these data are more consistent with a nuclear effect. If this is eventually confirmed, then consideration of a separate pathway for efficient nuclear export must be considered, since the 3′-UTR CARE did not affect cytoplasmic mRNA levels. This activity parallels that reported for the poly(A)-limiting element, which reduced poly(A) tail length in the nucleus without affecting nuclear export or polysomal loading (38, 39). The observation that the 3′-UTR CARE limits translation without altering polysomal loading is similar to that seen with other systems (33, 34). Clearly, this is an area requiring further study.
CARE Function Is Regulated by hnRNP L—CA dinucleotide repeats constitute the most common simple repeat in the mammalian genome (22). These repeats are often polymorphic and typically intronic (18-21). In the human endothelial nitric-oxide synthase gene and murine α2 integrin genes, the length of intronic CA repeats predicted increased splicing efficiency and gene expression, respectively (18-21). Work by the Bindereif laboratory established that hnRNP L specifically bound to the CA repeats with high affinity (20). Longer CA repeats enhanced hnRNP L binding and correlated with increased effects on splicing and gene expression (19, 20).
We confirmed this specificity of hnRNP L binding by demonstrating its interaction with luciferase mRNA in a 3′-UTR CARE-dependent manner. The existence of this interaction as well as its functional importance was confirmed through manipulation of hnRNP L levels in vivo. Overexpression of hnRNP L enhanced CARE-dependent decreases in reporter gene mRNA poly(A) tail length and expression, whereas hnRNP L knockdowns by RNA interference had the opposite effect. The link between decreased mRNA translation and poly(A) tail length is strengthened by their parallel modulation as a function of hnRNP L levels.
Additionally, our studies demonstrate that the hnRNP L-CD154 3′-UTR interaction is maintained in the cytoplasm of normal human T cells. Thus, this interaction might potentially serve similar roles in the post-transcriptional gene regulation of the same mRNA transcript, consistent with the known ability of hnRNP L to shuttle between nucleus and cytoplasm (40). For example, the 3′-UTR CARE-hnRNP L interaction might directly promote nuclear export and cytoplasmic mRNA stability, thereby accounting for these findings in the context of a short poly(A) tail. The trafficking of hnRNP L to stress granules, but not PBs, is consistent with the model that hnRNP L-CARE interactions mediate translational repression without increased mRNA turnover.
CA Dinucleotide Repeats and hnRNP L; a Multifunctional cis-trans Pathway of Gene Regulation—The specificity of hnRNP L binding to CA repeats has been established (20). To date, hnRNP L binding to pre-mRNA and mRNA has been shown to serve multiple roles in post-transcriptional gene regulation. Within introns, the effect of hnRNP L binding on splicing is influenced by the location of the CA repeat (19). Depending on its position relative to the 5′ splice site, splicing inhibition, enhancement, and cryptic splice selection can result (20). Other studies demonstrate that hnRNP L binding to internal exon sequences variably promotes exon silencing, nuclear mRNA stability, and export (41, 42). In contrast to our own findings, the effect of the exonic pre-mRNA processing element on nuclear mRNA stability and export was lost when inserted near the polyadenylation site of the RNA transcript (42). Together with its effects on splicing (20), these data underscore how the site of hnRNP L binding influences function. This functional complexity extends to the cytoplasm, where hnRNP L has been reported to variably influence mRNA stability as well as the function of the hepatitis C virus IRES (43-45). Thus, the context of hnRNP L binding (intron versus exon, proximity to splice or polyadenylation sites) as well as its subcellular location (nucleus, cytoplasm) regulates its downstream effects on gene expression.
Moreover, the length of the CA repeats (and presumably the number of hnRNP L molecules bound) influences the nature of its function. The intronic CA repeats reported to influence splicing are generally longer than the exonic sequences associated with changes in mRNA stability, nuclear export, and translation (18-21, 41-45). In the instance of the murine CD154 3′-UTR, the extended nature of the CARE (7, 10, and 17 CA repeats) is more typical of that seen in intronic CA repeats. The hnRNP L binding consensus sequence was defined as 10 nucleotides, so longer CA repeats probably promote multimeric hnRNP L binding and higher affinity interactions (20, 21). In addition, homodimeric interactions by hnRNP L would be presumably enhanced by the density of binding, which in turn might have quite different consequences for RNA structure and protein-protein interactions.
The Role of the 3′-UTR CARE in CD154 Expression and Disease—CD154 mRNA is rapidly degraded in human T cells following T cell activation (10-13). The mapping of the instability element to the 100-nt CURE in the murine CD154 3′-UTR supports the view that PTB proteins regulate this activity, since this region is rich in canonical PTB binding sites (10, 14). We also identify the role of the hnRNP L-CARE interactions in regulating CD154 expression at the level of mRNA poly(A) tail length and translation. Changes in CD154 mRNA poly(A) tail length and translation would potentially explain the second peak of CD154 expression by activated T cells seen at times (24-48 h) when mRNA levels are decreasing (13, 14).
Although equipotent in isolation, deletion of either the CURE or CARE yielded equivalent effects on luciferase expression to that seen with the CD154 3′-UTR (Figs. 2 and 3), suggesting their functional interaction. This model of interacting cis-acting elements was supported by the finding of enhanced mRNA stability and reduced poly(A) tail length with the CUCARE+ mRNA transcripts relative to CURE alone (Fig. 4 and Table 1). This interaction becomes important in considering the relationships between CD154 CA polymorphisms and the development of SLE and RA. Overexpression of CD154 results in a phenotype suggestive of SLE (46-49), whereas T cells from patients with SLE and RA exhibit enhanced surface expression of CD154 (50, 51). The 3′-UTR CARE is polymorphic in humans, ranging from 20 to 30 CA repeats (23). Studies of allelic variations suggest that 24 CA repeats limited CD154 expression to the greatest degree and conferred protection against the development of RA and SLE (23, 24). Interestingly, CD154 expression by specific CARE polymorphisms correlated inversely with CD154 mRNA stability (24). These observations are consistent with our findings that the CARE regulates translation. Moreover, it suggests the model that minor changes in CA dinucleotide repeat number might alter the interactions between the CARE and the CURE with significant effects on CD154 mRNA translation or turnover. Thus, characterization of the CARE pathway potentially accounts for the genetic linkage of CD154 3′-UTR CA polymorphisms and the development of SLE and RA.
This work was supported, in whole or in part, by National Institutes of Health Grant R01AR49834. This work was also supported by a grant from the American College of Rheumatology Research and Education Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: TNF, tumor necrosis factor; UTR, untranslated region; ARE, AU-rich element; nt, nucleotide(s); PTB, polypyrimidine tract-binding protein; hnRNP, heterogeneous nuclear ribonucleoprotein; CURE, CU-rich instability element; RA, rheumatoid arthritis; SLE, systemic lupus erythematosus; CARE, CA-rich instability element; RT, reverse transcription; RH, random hexamer; LM-PAT, ligase-mediated polyadenylation tail; PBS, phosphate-buffered saline; GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
References
- 1.Kontoyiannis, D., Pasparakis, M., Pizarro, T. T., Cominelli, F., and Kollias, G. (1999) Immunity 10 387-398 [DOI] [PubMed] [Google Scholar]
- 2.Taylor, G. A., Carballo, E., Lee, D. M., Lai, W. S., Thompson, M. J., Patel, D. D., Schenkman, D. I., Gilkeson, G. S., Broxmeyer, H. E., Haynes, B. F., and Blackshear, P. J. (1996) Immunity 4 445-454 [DOI] [PubMed] [Google Scholar]
- 3.Carballo, E., Lai, W. S., and Blackshear, P. J. (1998) Science 281 100-104 [DOI] [PubMed] [Google Scholar]
- 4.Piecyk, M., Wax, S., Beck, A. R., Kedersha, N., Gupta, M., Maritim, B., Chen, S., Gueydan, C., Kruys, V., Streuli, M., and Anderson, P. (2000) EMBO J. 19 4154-4163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dumitru, C. D., Ceci, J., Tsatsanis, C., Kontoyiannis, D., Stamatakakis, K., Lin, J. H., Patriotis, C. C., Jenkins, N. A., Copleland, N. G., Kollias, G., and Tsichlis, P. N. (2000) Cell 103 1071-1083 [DOI] [PubMed] [Google Scholar]
- 6.Hollenbaugh, D., Ochs, H. D., Noelle, R. J., Ledbetter, J. A., and Aruffo, A. (1994) Immunol. Rev. 138 23-37 [DOI] [PubMed] [Google Scholar]
- 7.Foy, T. M., Aruffo, A., Bajorath, J., Buhlmann, J. E., and Noelle, R. J. (1996) Annu. Rev. Immunol. 14 591-617 [DOI] [PubMed] [Google Scholar]
- 8.Noelle, R. J. (1996) Immunity 4 415-419 [DOI] [PubMed] [Google Scholar]
- 9.Grewal, I. S., and Flavell, R. A. (1998) Annu. Rev. Immunol. 16 111-135 [DOI] [PubMed] [Google Scholar]
- 10.Rigby, W. F. C., Waugh, M. G., and Hamilton, B. J. (1999) J. Immunol. 163 4199-4206 [PubMed] [Google Scholar]
- 11.Ford, G. S., Barnhart, B., Shone, S., and Covey, L. R. (1999) J. Immunol. 162 4037-4044 [PubMed] [Google Scholar]
- 12.Suarez, A., Mozo, L., Gayo, A., Zamorano, J., and Gutierrez, C. (1997) Eur. J. Immunol. 27 2822-2829 [DOI] [PubMed] [Google Scholar]
- 13.Murakami, K., Ma, W., Fuleihan, R., and Pober, J. S. (1999) J. Immunol. 163 2667-2673 [PubMed] [Google Scholar]
- 14.Hamilton, B. J., Genin, A., Cron, R. Q., and Rigby, W. F. C. (2003) Mol. Cell. Biol. 23 510-525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosinski, P. A., Laughlin, J., Singh, K., and Covey, L. R. (2003) J. Immunol. 170 979-988 [DOI] [PubMed] [Google Scholar]
- 16.Singh, K., Laughlin, J., Kosinski, P. A., and Covey, L. R. (2004) J. Immunol. 173 976-985 [DOI] [PubMed] [Google Scholar]
- 17.Waggoner, S. A., and Liebhaber, S. A. (2003) Exp. Biol. Med. 228 387-395 [DOI] [PubMed] [Google Scholar]
- 18.Hui, J., Reither, G., and Bindereif, A. (2003) RNA 9 931-936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hui, J., Stangl, K., Lane, W. S., and Bindereif, A. (2003) Nat. Struct. Biol. 10 33-37 [DOI] [PubMed] [Google Scholar]
- 20.Hui, J., Hung, L. H., Heiner, M., Schreiner, S., Neumuller, N., Reither, G., Haas, S. A., and Bindereif, A. (2005) EMBO J. 24 1988-1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cheli, Y., and Kunicki, T. J. (2006) Blood 107 4391-4398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mouse Genome Sequencing Consortium (2002) Nature 420 520-562 [DOI] [PubMed] [Google Scholar]
- 23.Citores, M. J., Rua-Figueroa, I., Rodriguez-Gallego, C., Durantez, A., Garcia-Laorden, M. I., Rodriguez-Lozano, C., Rodriguez-Perez, J. C., Vargas, J. A., and Perez-Aciego, P. (2004) Ann. Rheum. Dis. 63 310-317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin-Donaire, T., Losada-Fernandez, I., Perez-Chacon, G., Rua-Figueroa, I., Erausguin, C., Naranjo-Hernandez, A., Rosodo, S., Garcia-Saavedra, A., Citores, M. J., Vargas, J. A., Perez-Aciego, P. (2007) Arth. Res. Ther. 9 R89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yu, J. H., Yang, W. H., Gulick, T., Bloch, K. D., and Bloch, D. B. (2005) RNA 11 1795-1802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gough, N. M. (1988) Anal. Biochem. 173 93-95 [DOI] [PubMed] [Google Scholar]
- 27.Chomczynski, P., and Sacchi, N. (1987) Anal. Biochem. 162 156-159 [DOI] [PubMed] [Google Scholar]
- 28.Brooks, S. A., Connolly, J. E., Diegel, R. J., Fava, R. A., and Rigby, W. F. C. (2002) Arth. Rheum. 46 1362-1370 [DOI] [PubMed] [Google Scholar]
- 29.Nolan, R. D., and Arnstein, H. R. (1969) Eur. J. Biochem. 9 445-450 [DOI] [PubMed] [Google Scholar]
- 30.Lykke-Andersen, J., and Wagner, E. (2005) Genes Dev. 19 351-361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salles, F. J., Richards, W. G., and Strickland, S. (1999) Methods 17 38-45 [DOI] [PubMed] [Google Scholar]
- 32.Petersen, C. P., Bordeleau, M.-E., Pelletier, J., and Sharp, P. A. (2006) Mol. Cell 21 533-542 [DOI] [PubMed] [Google Scholar]
- 33.Nair, A. P., Hirsch, H. H., Colombi, M., and Moroni, C. (1999) Mol. Cell. Biol. 19 889-898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anderson, P., and Kedersha, N. (2006) J. Cell Biol. 172 803-808 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castelo-Branco, P., Furger, A., Wollerton, M., Smith, C., Moreira, A., and Proudfoot, N. (2004) Mol. Cell. Biol. 24 4174-4183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Natalizio, B. J., Muniz, L. C., Arhin, G. K., Wilusz, J., and Lutz, C. S. (2002) J. Biol. Chem. 277 42733-42740 [DOI] [PubMed] [Google Scholar]
- 37.de Moor, C. H., Meijer, H., and Lissenden, S. (2005) Semin. Cell Dev. Biol. 16 49-58 [DOI] [PubMed] [Google Scholar]
- 38.Das Gupta, J., Gu, H., Chernokalskaya, E., Gao, X., and Schoenberg, D. R. (1998) RNA 4 766-776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Peng, J., and Schoenberg, D. R. (2005) RNA 11 1131-1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pinol-Roma, S., Swanson, M. S., Gall, J. G., and Dreyfuss, G. (1989) J. Cell Biol. 109 2575-2587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tong, A., Nguyen, J., and Lynch, K. W. (2005) J. Biol. Chem. 280 38297-38304 [DOI] [PubMed] [Google Scholar]
- 42.Guang, S., Felthauser, A. M., and Mertz, J. E. (2005) Mol. Cell. Biol. 25 6303-6313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shih, S. C., and Claffey, K. P. (1999) J. Biol. Chem. 274 1359-1365 [DOI] [PubMed] [Google Scholar]
- 44.Kim, T. D., Kim, J. S., Kim, J. H., Myung, J., Chae, H. D., Woo, K. C., Jang, S. K., Koh, D. S., and Kim, K. T. (2005) Mol. Cell. Biol. 25 3232-3246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hahm, B., Kim, K. K., Kim, J. H., Kim, T. Y., and Jang, S. K. (1998) J. Virol. 72 8782-8788 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clegg, C. H., Ruffles, J. T., Haugen, H. S., Hoggatt, I. H., Aruffo, A., Durham, S. K., Farr, A. G., and Hollenbaugh, D. (1997) Int. Immunol. 9 1111-1122 [DOI] [PubMed] [Google Scholar]
- 47.Dunn, R. J., Luedecker, C. J., Haugen, H. S., Clegg, C. H., and Farr, A. G. (1997) 45 129-141 [DOI] [PubMed]
- 48.Mehling, A., Loser, K., Varga, G., Metze, D., Luger, T. A., Schwarz, T., Grabbe, S., and Beissert, S. (2001) J. Exp. Med. 194 615-628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Higuchi, T., Aiba, Y., Nomura, T., Matsuda, J., Mochida, K., Suzuki, M., Kikutani, H., Honjo, T., Nishioka, K., and Tsubata, T. (2002) J. Immunol. 168 9-12 [DOI] [PubMed] [Google Scholar]
- 50.Koshy, M., Berger, D., and Crow, M. K. (1996) J. Clin. Invest. 98 826-837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berner, B., Wolf, G., Hummel, K. M., Muller, G. A., and Reuss-Borst, M. A. (2000) Ann. Rheum. Dis. 59 190-195 [DOI] [PMC free article] [PubMed] [Google Scholar]