Abstract
Voltage-gated K+ channels are dynamic macromolecular machines that open and close in response to changes in membrane potential. These multisubunit membrane-embedded proteins are responsible for governing neuronal excitability, maintaining cardiac rhythmicity, and regulating epithelial electrolyte homeostasis. High resolution crystal structures have provided snapshots of K+ channels caught in different states with incriminating molecular detail. Nonetheless, the connection between these static images and the specific trajectories of K+ channel movements is still being resolved by biochemical experimentation. Electrophysiological recordings in the presence of chemical modifying reagents have been a staple in ion channel structure/function studies during both the pre- and post-crystal structure eras. Small molecule tethering agents (chemoselective electrophiles linked to ligands) have proven to be particularly useful tools for defining the architecture and motions of K+ channels. This Minireview examines the synthesis and utilization of chemical tethering agents to probe and manipulate the assembly, structure, function, and molecular movements of voltage-gated K+ channel protein complexes.
Modular Architecture = Current Diversity
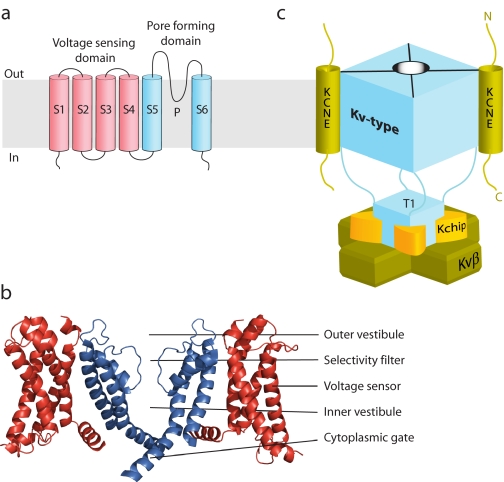
Voltage-gated K+ channels (Kv-type) function as multiprotein complexes made up of ion-conducting α-subunits and regulatory β-subunits. The α-subunits have a voltage-sensing domain (S1–S4) and a pore-forming domain (S5-P-S6), which tetramerizes to form a K+ conduction aqueous pore (Fig. 1a) (1). Most Kv channels are closed at negative resting membrane potentials but open upon membrane depolarization. Voltage dependence results from the tight linkage between voltage sensor movement from the resting to the active state and the cytoplasmic gate of the pore domain (Fig. 1b). Kv α-subunits often assemble with members of the KCNE family of type I transmembrane peptides (KCNE1–KCNE5 (E1–E5)2) to form complexes with strikingly different gating properties and pharmacological sensitivities (2). Co-assembly with some KCNE β-subunits shifts the equilibrium of the voltage sensors to favor the active state (3, 4), enabling Kv channels to be open and operational in non-excitable cells (5, 6). In addition to forming complexes with membrane-embedded β-subunits, a slew of water-soluble regulatory proteins can dock to the cytoplasmic portions of Kv channels to diversify K+ conduction (Fig. 1c) (7–10).
FIGURE 1.
Topological, structural, and schematic model of Kv channel α- and β-subunits. a, topology diagram of a Kv channel. The voltage-sensing domain is shown in red, and the pore-forming domain in blue. b, high resolution structure of the Kv2.1 paddle chimera channel (Protein Data Bank code 2R9R). For clarity, the front and back subunits have been removed. c, schematic model of a Kv-type macromolecular complex. Kv channels that assemble with membrane-embedded KCNE β-subunits do so with a 4:2 stoichiometry. Water-soluble regulatory subunits (Kv channel-interacting proteins (Kchip) and Kvβ) dock onto the N-terminal cytoplasmic tetramerization domain (T1) with a 4:4 stoichiometry. The fourth Kv channel-interacting protein and cytoplasmic C-terminal docking proteins are not visible or shown.
Modification of Cysteines in K+ Channels
Chemical tinkering with ion channels started in the 1960s when Karlin and Winnik (11) discovered that native extracellular cysteine residues on the acetylcholine receptor were modifiable with thiol-specific reagents and that ion channel modification could be monitored using electrical recordings. The cloning of K+ channels in the 1990s provided the opportunity to pick apart the structure and function of K+ channels with the site-directed introduction of non-native and removal of pesky native cysteines. Cysteine-modifying reagents commonly used for ion channels include methanethiosulfonates (MTS) and maleimides, which rapidly modify water-exposed cysteines on living cells and in isolated patches of membrane (12). The membrane-impermeant versions, MTSET and MTSES, have been invaluable for probing the nooks and crannies of K+ channels (13–15). Cysteine cross-linking of adjacent K+ channel subunits has also placed distance constraints between the voltage sensors and the pore-forming domain (16). These zero-length tethers (disulfide bonds) were used to covalently tie up the four subunits of the tetramerization domain (T1), demonstrating that the K+ ions must flow around this hanging cytoplasmic domain shared by the Shaker-type Kv channels (Fig. 1c) (17). Other biophysical probes have been attached to thiol-specific electrophiles to expand the study of K+ channel structure/function beyond electrophysiological and biochemical experimentation (18, 19). Most of the structural and functional details garnered from earlier chemical modification studies were satisfyingly confirmed by the first K+ channel crystal structure (20). Now aided by the many high resolution blueprints of K+ channels, the next generation of chemical tethering agents is calibrated to target, label, and modulate specific K+ channel complexes.
Tethered Blockers as “Tape Measures”
Tethered blockers are chemical reagents consisting of an ion channel inhibitor or pore blocker linked to a cysteine-modifying group (21) (although the repertoire of protein-modifying groups is expanding (22)). These bifunctional molecules typically act as affinity labeling reagents, binding first to the ion channel and then reacting with the target cysteine. Tethered blockers with a high affinity ligand greatly accelerate the kinetics of modification by increasing the local concentration of the cysteine-modifying agent near the channel (23). Once the channel is modified, the tethered blocker conversely creates a localized concentration of inhibitor near the cysteine modification site. Because the effective molarity of the inhibitor is directly dependent on the tether length (24), an ideal length tether can raise the effective concentration into the tens of millimolar, affording potent inhibition. If the tether is too short, the inhibitor cannot reach its binding site; if it is too long, the effective concentration decreases because the inhibitor samples more three-dimensional space. In practice, however, overly long tethers only modestly reduce the inhibitor's effective concentration, and thus, the magnitude of channel inhibition is usually unaffected (23).
Blaustein et al. (21) exploited the “Goldilocks scenario” of tether length to measure the distances from the extracellular ends of the K+ channel voltage sensor to the external tetraethylammonium blocking site at the entrance of the pore (Fig. 2a). By synthesizing a panel of quaternary ammoniums (QA) linked to maleimides with varying length polyglycine tethers (Fig. 2b), they could systematically find a tether length that was “just right” to block the channel. The KcsA crystal structure (20) was then used to calibrate the distances measured with the different length tethered blockers. These blockers, in combination with electrophysiology, revealed that the top of the S3 segment is the farthest away from the center of the Shaker K+ channel, followed closely by the S1 segment, and that the extended S3-S4 loop is the closest. Assuming that the shortest tether that demonstrated any irreversible block reflected the extended length of the tether, the approximate distances of 30 Å for S3 and S1 and 18 Å for the S3-S4 loop were measured. The recent paddle chimera channel structure (25) indicates that these tethered blocker-measured distances are systematically shorter than the distances in the crystal structure. If the tethers are recalibrated using 50% block (instead of the first sign of inhibition) as the benchmark for extended tether length, then the tethered blockers would yield similar distances for the voltage sensor in the active state. However, this hindsighted recalibration assumes that the extracellular ends of the voltage sensor move no closer to the blocking site in the resting state. Nonetheless, these molecular tape measures provide a straightforward approach to determine the radial arrangement of helices around the central conduction pore of K+ channel complexes.
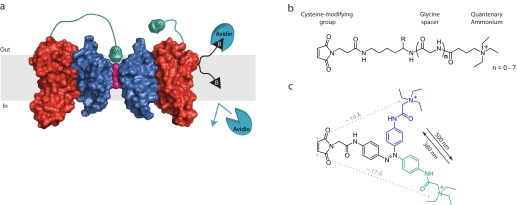
FIGURE 2.
Tethered blockers and inhibitors of K+ channel function. a, Kv channel modified with QA-tethered blockers and a biotin depth charge. The linker on the tethered blocker on the left voltage sensor is long enough to allow the QA to reach its binding site and block conduction, whereas the tethered blocker on the right voltage sensor is too short, and the conductance of the channel is unaffected. Also shown is a membrane-embedded residue that is biotinylated (black triangle) with a linker that permits avidin binding from the extracellular side, but not from the intracellular milieu. The potassium ions are shown in pink, the pore-forming domain in blue, and the voltage-sensing domain in red, and only two Kv subunits are shown for clarity. b, chemical structures of tethered blockers. The effective tether length can be adjusted by incorporating glycine residues Gly(n) using standard solid-phase peptide synthesis: Gly(0), 21 Å; and Gly(7), 45 Å. c, a photoisomerizable azobenzene linker that changes its effective length by ∼7 Å upon exposure to different wavelengths of light.
Measuring Voltage Sensor Movement
The voltage-induced movements within K+ channels are critical for electrical excitability, and thus, the movement of the voltage sensor has been intensely studied (13, 18, 26). There has been much debate surrounding the magnitude of voltage sensor displacement (in particular, the S4 helix of the paddle motif) upon depolarization (27). Two sets of molecular tape measures have been employed to measure the distance transversed by the voltage sensor from the resting to active state. The first set of reagents, biotins linked to maleimides, act as depth charges, measuring the membrane depths of transmembrane residues by obliterating channel function upon avidin binding (Fig. 2a) (28). Because membrane-buried cysteines are refractory to maleimide modification, the channels must be first labeled in the quasi-aqueous environment of detergent micelles and then reconstituted into a lipid bilayer. Thus, purified bacterial channels are ideal for this tethering approach. The maleimidobiotins were first calibrated on the rigid KvAP pore domain (29), which demonstrated that the effective linker length of the probes faithfully mirrors the average membrane thickness of ∼30 Å. This set of calibrated reagents was then used to examine the motions of the four KvAP voltage sensor helices. Two of these helices (S1 and S2) do not change their position perpendicular to the membrane (z axis) because their avidin accessibility is similar to that of the static pore domain. In contrast, the voltage sensor paddle domain (S3b and S4 helices) shows varied accessibility. The biotinylated S3b helix has increased one-sided reactivity to extracellularly applied avidin, implying that the outer leaflet of the membrane near S3 may be substantially dimpled to permit avidin binding. On the other hand, several residues in the S4 helix are accessible to avidin from both sides of the membrane with a linker as short as 10 Å. Assuming that the channel's proteomembranous environment around S4 is a constant barrier to avidin regardless of the position of the voltage sensor, this double-sided accessibility implies that the S4 helix undergoes a 10–15-Å transition in the z axis from the resting to active state. This distance is significantly larger than what was predicted by fluorescence-based approaches (30, 31). However, the recent higher resolution paddle chimera structure (25) hints that the S4 helix will need to drop back into the membrane a significant amount to achieve the total charge movement measured with gating currents (26), supporting the distance measured by the maleimidobiotin depth charges.
Whereas there has been much ado about voltage sensor movement perpendicular to the membrane, there is also evidence that the paddle motif moves inward, toward the ion conduction pore, during depolarization (32). The previously described QA-tethered blockers (21) are ideal yardsticks for this movement because they measure distances emanating from the center of the channel. To measure the radial movements of the Shaker K+ channel paddle motif, the kinetics of tethered block were compared using two depolarization protocols that position the voltage sensor in primarily the active or resting state (33). For S3b residues, the kinetics of tethered block are state-independent, indicating that these residues do not change their exposure to the extracellular milieu or their distance from the ion conduction pore. This lack of radial movement is not due to the extended S3-S4 loop in Shaker because shortening it to mimic the KvAP paddle does not reveal any shrouded S3b movements. In contrast, the kinetics of tethered block for S4 are state-dependent, and an inward movement of 2–5 Å was calculated using a polymer statistical approach. Combining these results with the depth charge measurements provides an overall depolarization trajectory for the S4 helix: an ∼10–15-Å upward movement resulting in the top of the S4 helix moving 2–5 Å closer to the ion conduction pathway. The lack of accessibility changes for the S3b helix by either tethering approach does not imply that this helix remains static during the transition from the resting to active state and leaves open the possibility for further experimentation and refinement of voltage sensor movements. One uncharted protein movement for tethering approaches is rotational, whether it is helical rotation or the entire voltage-sensing domain rotating around the pore-forming unit. Synthesis of tethering agents that report on rotational movements would be a valuable tool in pinning down the molecular gyrations of ion channels.
Controlling Neuronal Firing
The ability of tethered blockers to report on K+ channel architecture suggested that this approach could be used to control neuronal K+ channel function. Inspired by Lester's photo-chromatic activators of the acetylcholine receptor (34), Kramer and co-workers (35) designed a K+ channel tethered blocker with a light-activable linker. This photoswitchable reagent, MAL-AZO-QA, is composed of a maleimide linked to a quaternary amine through an azobenzene spacer (Fig. 2c). In visible light, the azobenzene adopts an extended trans-configuration, whereas irradiation with 380-nm light causes the azobenzene to adopt a condensed cis-conformation, shortening the linker by ∼7 Å. Modification of Shaker K+ channels (Q422C) with MAL-AZO-QA creates a synthetic photoisomerizable azobenzene-regulated K+ (or SPARK) channel that is turned “on” or “off” with a particular wavelength of light. SPARK channels “open” upon exposure to 380-nm light because the shorter linker prevents the tethered blocker from reaching the ion conduction pathway. Exposing the unblocked SPARK channel to 500-nm light provides the necessary linker length to block conductance and turn the channel off. To control neuronal excitability, cultured hippocampal neurons were transfected with SPARK channels that are open at resting membrane potentials. Irradiation (390 nm) of these “SPARKed” neurons silences spontaneous action potentials, whereas exposure to 500-nm light restores activity. Opening SPARK channels silences neuronal activity by hyperpolarizing the cell, thereby inhibiting the firing of action potentials. To promote action potential firing, neurons were transfected with a SPARK channel that lacked K+ selectivity (D-SPARK) (35). “Opening” of D-SPARK with long-wave UV light permits cations to flow into the cell, causing depolarization and triggering a burst of action potentials. Action potential firing could be shut off by “closing” the D-SPARK channels with visible light. Although first developed for K+ channels, this approach appears to be generalizable because ligand-gated channels have also been “SPARed” (36).
Assessing Assembled Complexes
Many Kv channels co-assemble with KCNE β-subunits to form membrane-embedded complexes in various tissues (2). Although assembly with KCNE subunits is required for proper physiological function of the Kv α-subunit, the stoichiometry of K+ channel·KCNE complexes has been a long-standing debate: two, four, and a combination of two and four KCNE subunits per complex have been reported (37–39). In addition, the realization that multiple KCNE subunits are expressed in the same tissue (40, 41) has raised the possibility that two different KCNE subunits could assemble with the same K+ channel.
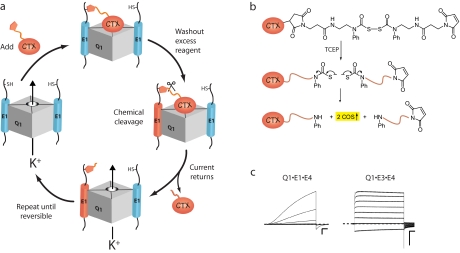
We rationalized that a tethered blocker strategy would allow us to assess the number and composition of KCNE subunits assembled with KCNQ1 (Q1) K+ channels. To determine the stoichiometry of the Q1·E1 complex, we devised an iterative subunit-counting approach that relies on a chemically releasable K+ channel-blocking reagent (Fig. 3, a and b) (42). The extracellularly applied reagent, charybdotoxin (CTX)-Clv, binds to the outer vestibule of the channel with a 1:1 stoichiometry and irreversibly blocks CTX-sensitive Q1 channels (37) by chemically modifying E1 peptides that contain an N-terminal cysteine residue. Irreversible inhibition is relieved by chemically cleaving the linker, washing out the blocking moiety, and leaving behind a permanently modified E1 peptide that cannot participate in subsequent rounds of treatment. A subunit tally is obtained by subjecting the K+ channels to multiple rounds of block and release until block becomes reversible. Success of the counting approach requires that only one E1 subunit is modified per round of treatment and that the nonspecific bimolecular reaction between CTX-Clv and E1 does not appreciably occur. At low nanomolar concentrations, CTX-Clv meets these two absolute requirements (42). Application of CTX-Clv to cells expressing Q1·E1 complexes requires two and only two rounds of treatment before complete reversibility is observed, demonstrating that the stoichiometry of the Q1·E1 complex is 4:2. Moreover, the completeness of each step of the reaction cycle with the Q1·E1 complex rules out any measurable contributions from sub-, supra-, and multiple stoichiometries. The architectural asymmetry of the Q1·E1 complex brings up many questions about its formation in the endoplasmic reticulum, the molecular mechanisms of KCNE modulation, and the drug sensitivity of the complex (2, 43, 44).
FIGURE 3.
Tethering strategies with chemically derivatized scorpion toxins (CTX). a, schematic depiction of the iterative counting strategy used to determine the number of E1 subunits in a Q1·K+ channel complex. b, structure and reductive cleavage of CTX-Clv. Tris(2-carboxyethyl)phosphine (TCEP) cleaves the bis(N-phenylcarbamoyl)disulfane linker, giving rise to two secondary amines and 2 eq of carbonyl sulfide gas. c, families of currents from Q1·E1·E4 and Q1·E3·E4 heteromeric K+ channel complexes. Current traces were revealed by subtracting pretreated currents from the currents elicited after washout of CTX-Mal. Command voltages were –100 to 40 mV (20-mV steps). The dashed line indicates zero current. Scale bars = 1 μA and 0.5 s.
Traditionally, the distinctive opening and closing kinetics of the different Q1·KCNE complexes are used to identify the KCNE subunit in a functioning complex (2). However, this approach is ineffective if a K+ channel co-assembles with two different KCNE subunits to form a heteromeric complex. To determine whether heteromeric Q1·KCNE complexes form, we synthesized a non-cleavable maleimidocharybdotoxin (CTX-Mal) that detects specific KCNE peptides in functioning complexes by irreversible inhibition (45). Using CTX-Mal as a “KCNE sensor,” a single application and washout of this reagent to cells expressing Q1 with two different KCNE subunits (one of which bears an N-terminal cysteine) showed that Q1·E1·E3, Q1·E1·E4, and Q1·E3·E4 heteromeric complexes form, traffic to the cell surface, and are functional. Using mathematical subtraction to visualize the irreversibly blocked current by CTX-Mal, the currents and gating kinetics of the different heteromeric complexes were revealed (Fig. 3c), and a hierarchy of KCNE subunit modulation of Q1 channels was determined: E3 > E1 ≫ E4. Strikingly, Q1·E1·E4 currents are kinetically indistinguishable from cardiac IKs currents, raising the possibility that this heteromeric complex plays a role in cardiac repolarization, a notion bolstered by the discovery that E4 is the most abundant KCNE transcript in heart (40, 41). Although these chemically derivatized peptide toxins were designed to address specific questions about Q1·E1 complex assembly and function, these tools can be readily adapted to study other multisubunit ion channel complexes.
Future Targets
Small molecule tethering reagents offer a unique chemical approach to dissect the structure and function of K+ channel complexes with molecular precision. The abundance of specific, high affinity ligands for K+ and other ion channels makes chemical tethering a generalizable strategy for studying many ion transport proteins. Site-directed placement of cysteine residues will continue to be relied upon as an attachment point for tethering reagents; however, recent scientific advances have provided alternative chemical handles for tethering. Unnatural amino incorporation into ion channels, once relegated to the Xenopus oocyte expression system (46), has transitioned to mammalian cells (47) and may ease the cysteine dependence of chemical tethering. Alternatively, the increase in effective molarity of a tethering reagent makes the exploration of nonthiol-based electrophiles feasible. Recently, a less specific electrophile, acrylamide, has been tethered to a pore blocker to target native K+ channels in cultured neurons (22). Ironically, the electrophiles in most tethering reagents preferentially react with water-exposed residues, leaving much of the interesting K+ channel biology, that happening in the membrane itself, hidden from currently available chemical probes. New chemistries that rapidly and quantitatively label membrane-embedded residues in living cells would open up a new frontier for chemical tethering to K+ channels.
Supplementary Material
Acknowledgments
We thank J. Rocheleau and N. Rhind for critical comments on the manuscript.
This work was supported, in whole or in part, by National Institutes of Health Grants GM-070650 and DC-007669. This minireview will be reprinted in the 2008 Minireview Compendium, which will be available in January, 2009.
Footnotes
The abbreviations used are: E1–E5, KCNE1–KCNE5; MTS, methanethiosulfonate(s); QA, quaternary ammonium(s); SPARK, synthetic photoisomerizable azobenzene-regulated K+; Q1, KCNQ1; CTX, charybdotoxin; CTX-Mal, maleimidocharybdotoxin; CTX-Clv, cleavable maleimidocharybdotoxin; MTSET, 2-(trimethylammonium)ethyl-MTS; MTSES, sodium (2-sulfonatoethyl)-MTS.
References
- 1.Bezanilla, F. (2006) Biol. Res. 39 425–435 [DOI] [PubMed] [Google Scholar]
- 2.McCrossan, Z. A., and Abbott, G. W. (2004) Neuropharmacology 47 787–821 [DOI] [PubMed] [Google Scholar]
- 3.Nakajo, K., and Kubo, Y. (2007) J. Gen. Physiol. 130 269–281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rocheleau, J. M., and Kobertz, W. R. (2008) J. Gen. Physiol. 131 59–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roepke, T. K., Anantharam, A., Kirchhoff, P., Busque, S. M., Young, J. B., Geibel, J. P., Lerner, D. J., and Abbott, G. W. (2006) J. Biol. Chem. 281 23740–23747 [DOI] [PubMed] [Google Scholar]
- 6.Schroeder, B. C., Waldegger, S., Fehr, S., Bleich, M., Warth, R., Greger, R., and Jentsch, T. J. (2000) Nature 403 196–199 [DOI] [PubMed] [Google Scholar]
- 7.Gulbis, J. M., Zhou, M., Mann, S., and MacKinnon, R. (2000) Science 289 123–127 [DOI] [PubMed] [Google Scholar]
- 8.Shamgar, L., Ma, L., Schmitt, N., Haitin, Y., Peretz, A., Wiener, R., Hirsch, J., Pongs, O., and Attali, B. (2006) Circ. Res. 98 1055–1063 [DOI] [PubMed] [Google Scholar]
- 9.Marx, S. O., Kurokawa, J., Reiken, S., Motoike, H., D'Armiento, J., Marks, A. R., and Kass, R. S. (2002) Science 295 496–499 [DOI] [PubMed] [Google Scholar]
- 10.Pioletti, M., Findeisen, F., Hura, G. L., and Minor, D. L., Jr. (2006) Nat. Struct. Mol. Biol. 13 987–995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Karlin, A., and Winnik, M. (1968) Proc. Natl. Acad. Sci. U. S. A. 60 668–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karlin, A., and Akabas, M. H. (1998) Methods Enzymol. 293 123–145 [DOI] [PubMed] [Google Scholar]
- 13.Larsson, H. P., Baker, O. S., Dhillon, D. S., and Isacoff, E. Y. (1996) Neuron 16 387–397 [DOI] [PubMed] [Google Scholar]
- 14.Liu, Y., Holmgren, M., Jurman, M. E., and Yellen, G. (1997) Neuron 19 175–184 [DOI] [PubMed] [Google Scholar]
- 15.Liu, Y., Jurman, M. E., and Yellen, G. (1996) Neuron 16 859–867 [DOI] [PubMed] [Google Scholar]
- 16.Neale, E. J., Elliott, D. J., Hunter, M., and Sivaprasadarao, A. (2003) J. Biol. Chem. 278 29079–29085 [DOI] [PubMed] [Google Scholar]
- 17.Kobertz, W. R., Williams, C., and Miller, C. (2000) Biochemistry 39 10347–10352 [DOI] [PubMed] [Google Scholar]
- 18.Mannuzzu, L. M., Moronne, M. M., and Isacoff, E. Y. (1996) Science 271 213–216 [DOI] [PubMed] [Google Scholar]
- 19.Perozo, E., Cortes, D. M., and Cuello, L. G. (1999) Science 285 73–78 [DOI] [PubMed] [Google Scholar]
- 20.Doyle, D. A., Morais Cabral, J., Pfuetzner, R. A., Kuo, A., Gulbis, J. M., Cohen, S. L., Chait, B. T., and MacKinnon, R. (1998) Science 280 69–77 [DOI] [PubMed] [Google Scholar]
- 21.Blaustein, R. O., Cole, P. A., Williams, C., and Miller, C. (2000) Nat. Struct. Biol. 7 309–311 [DOI] [PubMed] [Google Scholar]
- 22.Fortin, D. L., Banghart, M. R., Dunn, T. W., Borges, K., Wagenaar, D. A., Gaudry, Q., Karakossian, M. H., Otis, T. S., Kristan, W. B., Trauner, D., and Kramer, R. H. (2008) Nat. Methods 5 331–338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blaustein, R. O. (2002) J. Gen. Physiol. 120 203–216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krishnamurthy, V. M., Semetey, V., Bracher, P. J., Shen, N., and Whitesides, G. M. (2007) J. Am Chem. Soc. 129 1312–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Long, S. B., Tao, X., Campbell, E. B., and MacKinnon, R. (2007) Nature 450 376–382 [DOI] [PubMed] [Google Scholar]
- 26.Schoppa, N. E., McCormack, K., Tanouye, M. A., and Sigworth, F. J. (1992) Science 255 1712–1715 [DOI] [PubMed] [Google Scholar]
- 27.Ahern, C. A., and Horn, R. (2004) Trends Neurosci. 27 303–307 [DOI] [PubMed] [Google Scholar]
- 28.Ruta, V., Chen, J., and MacKinnon, R. (2005) Cell 123 463–475 [DOI] [PubMed] [Google Scholar]
- 29.Jiang, Y., Lee, A., Chen, J., Ruta, V., Cadene, M., Chait, B. T., and MacKinnon, R. (2003) Nature 423 33–41 [DOI] [PubMed] [Google Scholar]
- 30.Posson, D. J., Ge, P., Miller, C., Bezanilla, F., and Selvin, P. R. (2005) Nature 436 848–851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chanda, B., Asamoah, O. K., Blunck, R., Roux, B., and Bezanilla, F. (2005) Nature 436 852–856 [DOI] [PubMed] [Google Scholar]
- 32.Aziz, Q. H., Partridge, C. J., Munsey, T. S., and Sivaprasadarao, A. (2002) J. Biol. Chem. 277 42719–42725 [DOI] [PubMed] [Google Scholar]
- 33.Darman, R. B., Ivy, A. A., Ketty, V., and Blaustein, R. O. (2006) J. Gen. Physiol. 128 687–699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lester, H. A., Krouse, M. E., Nass, M. M., Wassermann, N. H., and Erlanger, B. F. (1980) J. Gen. Physiol. 75 207–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Banghart, M., Borges, K., Isacoff, E., Trauner, D., and Kramer, R. H. (2004) Nat. Neurosci. 7 1381–1386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volgraf, M., Gorostiza, P., Numano, R., Kramer, R. H., Isacoff, E. Y., and Trauner, D. (2006) Nat. Chem. Biol. 2 47–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen, H., Kim, L. A., Rajan, S., Xu, S., and Goldstein, S. A. (2003) Neuron 40 15–23 [DOI] [PubMed] [Google Scholar]
- 38.Wang, K. W., and Goldstein, S. A. (1995) Neuron 14 1303–1309 [DOI] [PubMed] [Google Scholar]
- 39.Wang, W., Xia, J., and Kass, R. S. (1998) J. Biol. Chem. 273 34069–34074 [DOI] [PubMed] [Google Scholar]
- 40.Bendahhou, S., Marionneau, C., Haurogne, K., Larroque, M. M., Derand, R., Szuts, V., Escande, D., Demolombe, S., and Barhanin, J. (2005) Cardiovasc. Res. 67 529–538 [DOI] [PubMed] [Google Scholar]
- 41.Lundquist, A. L., Manderfield, L. J., Vanoye, C. G., Rogers, C. S., Donahue, B. S., Chang, P. A., Drinkwater, D. C., Murray, K. T., and George, A. L., Jr. (2005) J. Mol. Cell. Cardiol. 38 277–287 [DOI] [PubMed] [Google Scholar]
- 42.Morin, T. M., and Kobertz, W. R. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 1478–1482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chandrasekhar, K. D., Bas, T., and Kobertz, W. R. (2006) J. Biol. Chem. 281 40015–40023 [DOI] [PubMed] [Google Scholar]
- 44.Gutman, G. A., Chandy, K. G., Grissmer, S., Lazdunski, M., McKinnon, D., Pardo, L. A., Robertson, G. A., Rudy, B., Sanguinetti, M. C., Stuhmer, W., and Wang, X. (2005) Pharmacol. Rev. 57 473–508 [DOI] [PubMed] [Google Scholar]
- 45.Morin, T. M., and Kobertz, W. R. (2007) ACS Chem. Biol. 2 469–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beene, D. L., Dougherty, D. A., and Lester, H. A. (2003) Curr. Opin. Neurobiol. 13 264–270 [DOI] [PubMed] [Google Scholar]
- 47.Wang, W., Takimoto, J. K., Louie, G. V., Baiga, T. J., Noel, J. P., Lee, K. F., Slesinger, P. A., and Wang, L. (2007) Nat. Neurosci. 10 1063–1072 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.