Abstract
Rho GTPases (20 human members) comprise a major branch of the Ras superfamily of small GTPases, and aberrant Rho GTPase function has been implicated in oncogenesis and other human diseases. Although many of our current concepts of Rho GTPases are based on the three classical members (RhoA, Rac1, and Cdc42), recent studies have revealed the diversity of biological functions mediated by other family members. A key basis for the functional diversity of Rho GTPases is their association with distinct subcellular compartments, which is dictated in part by three posttranslational modifications signaled by their carboxyl-terminal CAAX (where C represents cysteine, A is an aliphatic amino acid, and X is a terminal amino acid) tetrapeptide motifs. CAAX motifs are substrates for the prenyltransferase-catalyzed addition of either farnesyl or geranylgeranyl isoprenoid lipids, Rce1-catalyzed endoproteolytic cleavage of the AAX amino acids, and Icmt-catalyzed carboxyl methylation of the isoprenylcysteine. We utilized pharmacologic, biochemical, and genetic approaches to determine the sequence requirements and roles of CAAX signal modifications in dictating the subcellular locations and functions of the Rho GTPase family. Although the classical Rho GTPases are modified by geranylgeranylation, we found that a majority of the other Rho GTPases are substrates for farnesyltransferase. We found that the membrane association and/or function of Rho GTPases are differentially dependent on Rce1- and Icmt-mediated modifications. Our results further delineate the sequence requirements for prenyltransferase specificity and functional roles for protein prenylation in Rho GTPase function. We conclude that a majority of Rho GTPases are targets for pharmacologic inhibitors of farnesyltransferase, Rce1, and Icmt.
Rho proteins are members of the Ras superfamily of small GTPases and function as GDP/GTP-regulated switches (1, 2). Much of our current understanding of the biochemistry and biology of the Rho family has come from the extensive evaluation of three classical members, RhoA, Rac1, and Cdc42 (3). Similar to Ras, Rho GDP/GTP cycling is regulated by guanine nucleotide exchange factors that promote the formation of the active GTP-bound form (4) and GTPase-activating proteins that catalyze the intrinsic GTPase activity and promote the formation of inactive GDP-bound Rho (5). Active, GTP-bound Rho GTPases bind preferentially to downstream effectors, stimulating diverse cytoplasmic signaling cascades that control actin reorganization and regulate cell shape, polarity, motility, adhesion, and membrane trafficking (6). As such, it is thought that activated Rho proteins contribute to cancer progression by influencing the ability of cells to migrate and thus to invade and metastasize. In addition to these alterations in cellular function, aberrant activation of Rho proteins has also been shown to contribute to other cancer phenotypes by promoting cell growth, proliferation, survival, and angiogenesis (7). Therefore, defining pharmacologic approaches for inhibition of Rho GTPase function represents an important direction for target-based anti-cancer drug discovery.
Similar to Ras, the majority of Rho family GTPases are known or anticipated to undergo a series of posttranslational modifications that promote proper subcellular localization to the plasma membrane and/or endomembranes, which is required for biological activity. This series of modifications is initiated by the recognition of a carboxyl-terminal CAAX tetrapeptide motif (where C represents cysteine, A is an aliphatic amino acid, and X is any amino acid), which is found on 16 of 20 Rho GTPases (Table 1; canonical CAAX motifs are not present in the Wrch-1, Chp/Wrch-2, RhoBTB1, or RhoBTB2). The first step, mediated by farnesyltransferase (FTase)2 and/or geranylgeranyltransferase type I (GGTase-I), results in the covalent addition of a farnesyl or geranylgeranyl isoprenoid lipid, respectively, to the cysteine residue of the CAAX sequence. Next, the -AAX peptide is cleaved from the carboxyl terminus by the Rce1 (Ras-converting enzyme 1) endoprotease. Finally, isoprenylcysteine-O-carboxyl methyltransferase (Icmt) catalyzes the addition of a methyl group to the prenylated cysteine residue (8). Together, these modifications increase protein hydrophobicity and facilitate membrane association. Where studied, mutation of the cysteine residue of the CAAX motif, which prevents all three modifications, renders Rho GTPases inactive due to mislocalization to the cytosol (9). Thus, pharmacological inhibitors of protein prenylation are anticipated to be effective inhibitors of Rho GTPase activity. Recent observations upon genetic ablation of GGTase-I activity support this possibility. Transient genetic depletion of GGTase-I caused mouse embryonic fibroblasts to undergo growth arrest, cell rounding, impaired cell migration, and reduced actin polymerization, and these phenotypic alterations were partially rescued by GGTase-I-independent, farnesylated variants of RhoA and Cdc42 (10). These phenotypic consequences are consistent with loss of Rho GTPase function but additionally suggest that multiple GGTase-I substrates are important for regulation of cell morphology and actin organization. Similarly, loss of GGTase-I activity was lethal in the budding yeast Saccharomyces cerevisiae, and the combined expression of GGTase-I-independent, farnesylated variants of RhoA and Cdc42 suppressed this lethality (11).
TABLE 1.
Carboxyl-terminal membrane-targeting sequence elements of human Ras and Rho GTPases
a Polybasic residues are in boldface type; putative or known palmitoylated (palm) cysteines are shaded gray; known or putative CAAX prenylation motifs are in boldface type and underlined; underlined sequences comprise a GDP/GTP binding motif.
b Compiled from references cited in Ref. 67; RhoG and RhoH (68); Wrch-1 (23); Chp (28). PM, plasma membrane; ER, endoplasmic reticulum; NE, nuclear envelope; MT, mitochondria.
c The GTPase domains are followed by carboxyl-terminal tandem BTB domains.
Although the CAAX-signaled posttranslational modifications are necessary for Ras and Rho GTPase function and membrane association, these three modifications alone are not sufficient to promote full membrane association or to target the proteins to the specific cellular subdomains required for proper GTPase function (12). Instead, at least two distinct sequence elements positioned immediately upstream of the CAAX motif serve as additional signals that are required to promote efficient membrane association and biological function. One element is composed of clusters of polybasic amino acid residues, as seen in K-Ras4B, that provide a positive charge that facilitates association with acidic membrane-associated lipids. The second sequence element present upstream of CAAX in some Rho GTPases is one or two cysteine residues that undergo post-translational modification by the fatty acid palmitate. Palmitoylated cysteines comprise the additional targeting signal for H-Ras and N-Ras proteins as well as for some Rho family GTPases (RhoB and TC10). Mutant Ras proteins that undergo the CAAX-signaled modifications but lack either the polybasic residues or palmitoylated cysteine(s) are mislocalized and are significantly compromised in their biological activities. Finally, additional sequences flanking these elements form a largely uncharacterized third signal that also contributes to dictating the precise subcellular localization of Ras and Rho GTPases (13–15). These locations can vary significantly; whereas some Rho GTPases are found predominantly at the plasma membrane (e.g. Rac1), some are associated mainly with endomembranes (e.g. RhoH), and still others are associated with endosomes (e.g. RhoD) (Table 1).
Because of the importance of CAAX-signaled modifications for small GTPase localization and function, farnesyltransferase inhibitors (FTIs) were developed initially as anti-Ras therapies for cancer treatment. Unfortunately, K-Ras and N-Ras (the two Ras isoforms most commonly mutated in human cancers) undergo alternative prenylation by GGTase-I when in the presence of FTIs and therefore escape FTI-mediated inhibition of membrane association (16, 17). Nevertheless, FTIs have exhibited anti-tumor activity in preclinical and clinical trial analyses, presumably due to the inhibition of function of other FTase substrates (8). In light of the role of aberrant Rho GTPase function in oncogenesis, Rho family GTPases (e.g. RhoB) are logical candidates for key targets of FTIs (18). Although GGTase-I modifies the classical Rho GTPases, the nature of the CAAX sequences of other members suggests that they may be FTase substrates.
The observation that K-Ras and N-Ras undergo alternative prenylation in response to FTI treatment has also stimulated interest in the development of inhibitors that block other enzymes that facilitate Ras membrane association. First, GGTase-I inhibitors (GGTIs) were developed to block the function of the alternatively prenylated Ras proteins (19). Furthermore, with increasing evidence for the involvement of normally geranylgeranylated proteins in cancer (e.g. Ral and Rho GTPases) (7, 20), there is now additional interest in the development of GGTIs to target these GGTase-I substrates for cancer treatment. Second, efforts to develop inhibitors of Rce1 and Icmt as novel anti-cancer agents have recently intensified (9). However, there is concern regarding their effectiveness, since Ras proteins that fail to undergo these two modifications do retain partial localization and function (21, 22). Additionally, since many FTase and GGTase-I substrates are also substrates for these two enzymes, there is also concern that such inhibitors will affect a broad array of cellular proteins and cause significant cell toxicity in normal cells. Support for this latter concern is provided by the observed embryonic lethality in mice deficient in either Rce1 or Icmt. Whether similar toxicity would be seen in adult animals is an important area of investigation.
In light of the essential function of Rho family GTPases in normal cell physiology and their aberrant activation in oncogenesis (7, 20), establishing the sensitivity of Rho GTPases to FTI and GGTI inhibitors and the contribution of Rce1- and Icmt-catalyzed modifications to their cellular functions will be critical to the successful development of inhibitors of CAAX-signaled modifications. Therefore, we have utilized pharmacologic and genetic approaches to establish the importance of CAAX-signaled modifications for the functions of the less studied Rho GTPases. We found that, in contrast to the common perception based on the study of the classical Rho GTPases, farnesylation is a lipid modification that is equally important as geranylgeranylation for Rho GTPase function. Furthermore, we conclude that the rules governing palmitoylation of cysteine-containing signal sequences and even CAAX tetrapeptide prenyltransferase specificity derived from structural studies are imprecise and that experimental analyses are still required to establish the lipid modification status of a particular CAAX-terminating protein. Finally, our observations that Rho GTPase subcellular localization and/or function depend on Rce1 and/or Icmt enzymatic activity support the value of developing inhibitors of these two enzymes as therapeutic strategies to block Rho GTPase function.
EXPERIMENTAL PROCEDURES
Expression Constructs and Cell Culture Lipid Inhibitor Analyses—cDNAs for human Ras and Rho GTPases (H-Ras, K-Ras, RhoA, Rnd1, and Rnd2 and Rnd3, RhoD, RhoH, TC10, TCL, and Rif) and rat RhoB were cloned into pEGFP mammalian expression vectors (Clontech) as previously described (23, 24) or constructed for this study. All constructs were sequence-verified, and cloning details are available upon request.
HEK 293T cells were maintained in Dulbecco's modified minimum essential medium supplemented with 10% fetal calf serum (Sigma), 100 units/ml penicillin, and 100 μg/ml streptomycin. NIH 3T3 cells were maintained in Dulbecco's modified minimum essential medium supplemented with 10% calf serum (Sigma) and 100 units/ml penicillin and 100 μg/ml streptomycin (“complete growth medium”). Spontaneously immortalized mouse embryonic fibroblasts (MEFs) were originally prepared from Icmt–/– and Rce1–/– mouse embryos, along with control fibroblasts (Icmt+/+ and Rce1+/+) from littermate embryos (25) and were kindly provided by Stephen G. Young (UCLA, Los Angeles, CA). MEF cultures were maintained in Dulbecco's modified minimum essential medium supplemented with 15% calf serum (Colorado Serum, Denver, CO), nonessential amino acids, and l-glutamine.
The highly selective inhibitors of FTase (FTI-2153) and of GGTase-I (GGTI-2417) were provided by Saïd Sebti (Moffitt Cancer Center) and Andrew Hamilton (Yale) and were dissolved in DMSO (26, 27). The palmitate analog 2-bromopalmitate (2-BP), an inhibitor widely used to evaluate the role of protein palmitoylation in protein targeting (28–30), was purchased from Sigma and dissolved in ethanol. Control cultures were treated with the equivalent final concentration of ethanol or DMSO (designated vehicle). In the inhibitor assays, cells were transfected as described below, washed, and incubated for 20 h with growth medium supplemented with 10 μm FTI-2153, 10 μm GGTI-2417, or 100 μm 2-BP.
Transfection, Immunofluorescence, and Microscopy—For live cell microscopy, cells were plated, transfected, and imaged in a 35-mm culture dish that incorporated a number 1.5 glass coverslip-sealed 15-mm cut-out on the bottom (MatTek, Ashland, MA). Uncoated dishes were used for NIH 3T3 cells, and poly-d-lysine-coated dishes were used for all experiments using MEFs. DNA transfections were performed with Lipofectamine Plus reagent according to the manufacturer's instructions (Invitrogen). Three h after transfection, cells were washed, grown in phenol red-free Dulbecco's modified minimum essential medium/F-12 supplemented with 10% calf serum, and treated with inhibitors where indicated.
For immunofluorescence, cells transiently transfected with plasmid DNAs encoding GFP fusion constructs of small GTPases were fixed 24 h after transfection with 4% paraformaldehyde, permeabilized with Triton X-100, stained with Alexa 594-phalloidin (Molecular Probes, Inc., Eugene, OR), and mounted with FluorSave (Calbiochem).
For both live cell imaging and immunofluorescence studies, cells were examined with an inverted laser-scanning confocal microscope (Zeiss 510 LSM) using an oil immersion ×63 numerical aperture 1.4 objective. Images were captured by scanning with the 488 nm spectral line of an argon-ion laser using the LP 505 emission filter (for live cell imaging; GFP) or sequential scanning with the 488 nm argon laser and the 543 nm HeNe1 laser and the BP 505–530 (for GFP) or LP 585 (for Alexa 594) emission filters. 0.3-μm confocal z-sections that show both nuclear and membrane/cytosolic localization of GFP fusion proteins were obtained and analyzed. Brightness and contrast of JPEG images were adjusted using Adobe Photoshop CS2 software.
Transformation Assays—For soft agar colony formation analyses of anchorage-independent growth, NIH 3T3 cells stably expressing activated Rac or Rac C178S mutants were seeded at a density of 105 cells/60-mm dish in a solution of complete growth medium containing 0.4% bacto-agar over a layer of complete growth medium containing 0.6% bacto-agar. Colonies were allowed to form for 2 weeks, after which viable colonies were stained with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide tetrazolium salt. Plates were scanned, and the number of 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide-positive colonies was quantified using ImageJ software. Results for transformation assays are representative of at least three experiments from independently generated sets of stable cell lines.
1-Biotinamido-4-(4′-(maleimidomethyl Cyclohexanecarboxamido) Butane (Biotin-BMCC) Labeling—Analyses of protein palmitoylation were done as described in Refs. 31 and 32). Briefly, 293T cells were transfected with 7 μg of the indicated pEGFP construct using a calcium phosphate transfection technique. Forty-eight h after transfection, cells were lysed and incubated with 5 μg of anti-GFP monoclonal antibody (JL-8; Clontech) at 4 °C for 1 h, at which point 20 μg of protein G (Invitrogen) was added to the lysates and incubated at 4 °C for 1 h. Bound protein was washed and incubated with lysis buffer containing 50 mm N-ethylmaleimide (Sigma) for 48 h at 4 °C. Bound protein was then washed and treated with 1 m hydroxylamine, pH 7.4, to cleave thioester bonds for 1 h at 25 °C, washed again, and treated with biotin-BMCC (Pierce), which recognizes free sulfhydryl groups, for 2 h at 25 °C. Bound protein was washed again, resuspended in 50 μl of 2 × sample loading buffer, resolved by 12% SDS-PAGE, and transferred to polyvinylidene difluoride membrane. Labeled protein was detected by incubation with streptavidin-horseradish peroxidase (Pierce), and the membrane was washed and exposed to x-ray film. Twenty μg of lysate was resolved by SDS-PAGE, transferred, incubated with anti-GFP primary antibody and anti-mouse IgG-horseradish peroxidase secondary antibody, and exposed to x-ray film to verify the presence of each GFP-tagged protein.
In Vitro Prenylation Analyses—Full-length human Rnd, RhoB, and TC10 proteins were expressed as fusion proteins containing six histidine residues at the amino terminus. The cDNA coding regions were amplified from appropriate cell lines by PCR and subcloned into the pQE bacterial expression vector (Qiagen). The identity of all plasmids was confirmed by restriction mapping and DNA sequencing of the PCR-amplified fragments. Six-histidine-tagged Rnd1, Rnd2, Rnd3, and TC10 were expressed and purified from Escherichia coli by nickel affinity chromatography, as we have described previously for Ras proteins (33). Expression and purification of recombinant human FTase and GGTase-I from SF9 insect cells (∼50% pure) were performed as we have described elsewhere (33). FTase and GGTase-I activity were determined by measuring the transfer of [3H]farnesyl or [3H]geranylgeranyl to the Rho GTPase substrate in reaction mixtures containing (in 200 μl) 50 mm Tris·HCl (pH 7.5), 1 mm dithiothreitol, 20 mm KCl, 5 mm MgCl2. The concentration of recombinant FTase and GGTase-I was 0.5 μm, whereas the GTPase protein substrate concentrations were varied from 0 to 1.0 μm. Reactions were started by the addition of 20 ng of FTase or GGTase-I and proceeded for 4 min at 37 °C.
Statistical Analysis—Data were analyzed by the use of Student's t test. In all analyses, p < 0.05 was considered statistically significant, and data are presented as mean ± S.D.
RESULTS
Inhibition of Farnesyltransferase Blocks Rho Family GTPase Localization and Function—Substrate specificity of FTase and GGTase-I toward small GTPases is determined primarily by the sequence of the CAAX tetrapeptide motif (Table 1). Biochemical and structural studies of CAAX peptides in complex with FTase and GGTase-I have defined rules that govern substrate selectivity (34). Whereas the specific isoprenoid modification of the classical Rho GTPases and of their highly related isoforms (RhoA/B/C, Rac1/2/3, and Cdc42) has been confirmed in vivo, the precise isoprenoid modification of the majority of Rho family GTPases has not been tested. Where studied, the prenylation of Ras and Rho small GTPases has been found to be essential for proper subcellular localization. Therefore, to determine the importance of farnesylation or geranylgeranylation to Rho GTPase localization, we ectopically expressed GFP-fusion Rho GTPase proteins to visualize their subcellular localization in live cells and determined the ability of treatment with the potent and highly selective FTI (FTI-2153) and/or GGTI (GGTI-2417) to alter their subcellular location (35). Exogenous expression of GFP fusion proteins has been used extensively to evaluate the subcellular localization of small GTPases and has been validated as an accurate reflection of the subcellular location of the endogenous protein (24, 36, 37). For these analyses, we utilized wild-type (and therefore predominantly GDP-bound) human Rho GTPases to avoid potential complications in Rho GTPase localization due to effector association or to altered interaction with Rho GDP dissociation inhibitor proteins seen with the activated forms of some Rho GTPases (24, 36, 37).
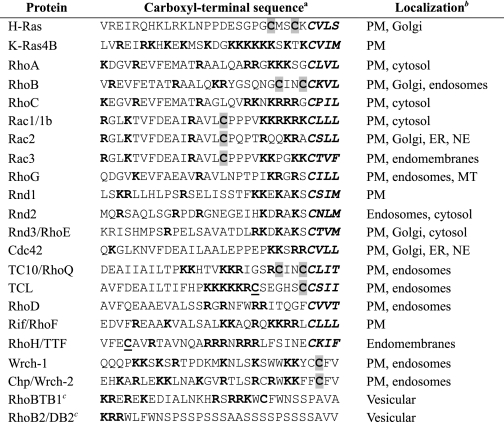
First, we verified the specificity and potency of the prenyltransferase inhibitors by determining their ability to disrupt the membrane association and subcellular location of small GTPases that are well known substrates of FTase and/or GGTase-I (Fig. 1). Whether due to structural mutations or to treatment with prenyltransferase inhibitors, nonprenylated GFP-tagged small GTPases localize to the cytoplasm and nucleus, similar to the distribution of GFP protein alone (Fig. 1) (24, 35–39). H-Ras is normally localized to the plasma membrane and has been shown previously to be solely farnesylated. As expected, we found that treatment with FTI but not GGTI caused mislocalization of GFP-H-Ras to the cytoplasm and nucleus. RhoA is a substrate for GGTase-I but not FTase (40), and, as seen previously, treatment with GGTI but not FTI caused accumulation of GFP-tagged RhoA in the nucleus. In contrast, RhoB has been found to exist in both farnesylated and geranylgeranylated forms (41), and FTI treatment results in further accumulation of the geranylgeranylated form (42). A previous study showed RhoB localization at the plasma membrane and endosomes (43), and our results confirm these observations. It has also been reported that FTI treatment of cells that express RhoB caused loss of the plasma membrane-associated pool, but not the endosome-associated pool, suggesting that farnesylated RhoB is plasma membrane-associated, whereas geranylgeranylated RhoB is endosome-associated (43). Surprisingly, treatment of cells with either FTI or GGTI alone did not cause a significant loss of either plasma membrane- or endosome-associated RhoB. Instead, a loss of RhoB from both membrane compartments, resulting in cytosolic and nuclear accumulation, was observed only upon treatment with inhibitors of both prenyltransferases. This result is consistent with the ability of RhoB to undergo alternative prenylation when either FTase or GGTase-I activity is inhibited. However, our results suggest that subcellular localization is not influenced by the specific isoprenoid modification. Finally, K-Ras4B is normally farnesylated but can be alternatively modified by GGTase-I in vivo when cells are challenged with FTI (16, 17). Consistent with these observations, we found that only concurrent treatment with FTI and GGTI caused relocalization of K-Ras4B from the plasma membrane to the cytoplasm and nucleus. These results verify that treatment with FTI and GGTI can be used to predict the species of isoprenyl group that is added to Rho GTPases in vivo.
FIGURE 1.
The specific isoprenoid modifications of Ras and Rho GTPases in vivo are accurately defined using specific pharmacologic inhibitors of FTase and GGTase-I. NIH 3T3 cells were transiently transfected with expression constructs for GFP alone or GFP-tagged fusion proteins of the indicated Ras or Rho GTPases and treated with FTI-2153, GGTI-2417, or both (10 μm each) or DMSO. Live cells were visualized using confocal microscopy. Images shown are representative of three independent experiments with >80 cells examined per assay. Scale bar, 10 μm.
Unlike the classical Rho GTPases, Rnd3/RhoE and the related Rnd1 and Rnd2 isoforms are predicted to be substrates for FTase, and Rnd3 has been demonstrated to be farnesylated in vivo (44). However, Rnd proteins contain carboxyl-terminal sequences similar to that seen in K-Ras4B, terminating with a CXXM motif and possessing upstream polybasic sequences like those that dictate the prenyltransferase interactions of K-Ras4B (45, 46). Consequently, it has been speculated and assumed that Rnd proteins may also undergo FTI-induced alternative prenylation (8).
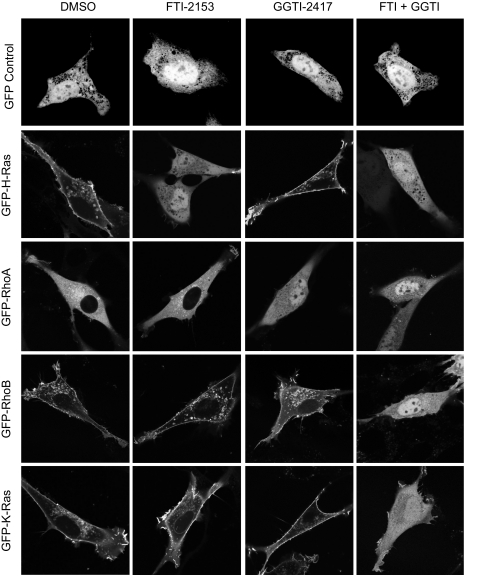
To determine the prenylation specificity of Rnd proteins, we first evaluated the ability of bacterially expressed proteins to serve as substrates for FTase and GGTase-I in vitro. These assays were done as we described previously (33), where each GTPase was expressed in E. coli as amino-terminally Histagged proteins and purified to 90–95% purity (data not shown). Insect cell-expressed purified recombinant FTase or GGTase-I was incubated with increasing concentrations of each Rnd protein and with saturation concentrations of [3H]FPP or [3H]GGPP (0.5 μm), respectively. For these analyses, we included RhoB as a control, since previous studies determined that RhoB is modified by both FTase-I and GGTase-I in vitro and in vivo (41, 42). In agreement with these previous observations, we found that FTase and GGTase-I catalyzed the farnesylation and geranylgeranylation of RhoB, respectively, in vitro (Fig. 2A).
FIGURE 2.
Rnd proteins are farnesylated in vitro and in vivo. A, in vitro RhoB is a strong substrate for both FTase and GGTase-I, whereas Rnd proteins are strong substrates for FTase and weak substrates for GGTase-I. Purified recombinant RhoB and Rnd proteins were added to our standard FTase or GGTase-I reaction mixture containing radiolabeled FPP or GGPP, respectively, and processed as described under “Experimental Procedures.” B, both membrane association and function of Rnd proteins are inhibited by FTI treatment. NIH 3T3 cells were transiently transfected with expression constructs for GFP-tagged fusion proteins with the indicated Rho GTPases and treated with FTI-2153, GGTI-2417, both (10 μm each), or DMSO. Live cells were visualized using confocal microscopy. Images shown are representative of three independent experiments with >80 cells examined per assay. Scale bar, 10 μm.
As expected from the CAAX sequence, all three Rnd proteins were efficient substrates for FTase. Consistent with the possibility that Rnd proteins may undergo alternative prenylation in the absence of FTase activity, we found that Rnd proteins were capable of serving as minor substrates for GGTase-I in vitro, although with much lower affinity than seen with FTase (Fig. 2A). This result is similar to our previous observations with K-Ras4B, where we found that K-Ras4B, but not H-Ras, was a substrate for GGTase-I in vitro, albeit with a 140-fold lower affinity than that for FTase (33). Thus, like K-Ras4B, we anticipated that Rnd proteins are normally farnesylated, but with FTI treatment, Rnd proteins may become modified by geranylgeranylation.
Consistent with published data, we found that Rnd1 and Rnd3 showed plasma membrane association, whereas Rnd2 lacked this association and was found in the cytosol (Table 1). In addition, we observed that Rnd1 and Rnd3, but not Rnd2, caused cell rounding (47). Interestingly, localization of all three Rnd proteins was exquisitely sensitive to treatment with FTI alone (Fig. 2B). FTI and not GGTI treatment resulted in the loss of plasma membrane localization, accompanied by increased cytoplasmic and nuclear accumulation of all three Rnd proteins. In vitro analysis of both K-Ras (33) and Rnd proteins (Fig. 2A) suggested that these proteins were strong substrates for FTase but could also serve as weak GGTase substrates. However, unlike K-Ras, which undergoes alternative prenylation and therefore displays unaltered subcellular localization when treated with FTI, Rnd proteins were not found to be alternatively prenylated in vivo to a detectable degree when cells were treated with FTI (Fig. 2B).
In addition to disrupting subcellular localization, FTI treatment reversed the cell rounding phenotype caused by ectopic expression of Rnd1 and Rnd3. Although treatment with GGTI had no effect on the subcellular localization of Rnd proteins, interestingly, it induced an exaggerated rounding phenotype in the majority of cells expressing ectopic Rnd1 (>90%), probably due to RhoA inactivation secondary to loss of RhoA geranylgeranylation. Thus, unlike K-Ras4B, the weak GGTase-I activity seen in vitro may not be physiologically significant, and Rnd proteins do not appear to undergo significant, detectable FTI treatment-induced alternative prenylation in vivo.
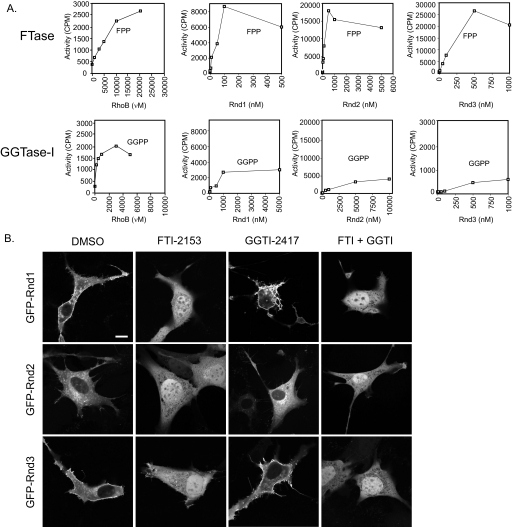
TC10 (CAAX = CLIT) and the closely related Rho GTPase, TCL (CAAX = CSII) are predicted to be modified by farnesyl and geranylgeranyl isoprenoids, respectively (34). In agreement with this prediction, we found that recombinant TC10 was an excellent substrate for FTase in vitro (Fig. 3A). However, TC10 can also serve as a weak substrate for GGTase-I, suggesting that it may undergo FTI-induced alternative prenylation. To address this possibility, we ectopically expressed wild-type TC10 and evaluated sensitivity to FTI and GGTI treatment. In agreement with previous findings (48), we found that ectopically expressed TC10 exhibited perinuclear endosomal localization and induced formation of filopodia (Fig. 3B). We found that treatment with FTI alone was sufficient to disrupt the vesicular localization of TC10, resulting in a diffuse cytosolic and nuclear accumulation, and additionally prevented filopodia formation. GGTI treatment alone did not cause mislocalization of TC10 or block filopodia formation, and the combined treatment with both inhibitors did not alter localization beyond what was seen with FTI treatment alone. Ectopically expressed TCL also exhibited perinuclear endosomal localization and induced filopodia formation. Unexpectedly, although the CAAX sequence suggested that this GTPase would be a GGTase-I substrate (34), we found that FTI treatment alone caused mislocalization and nuclear accumulation, although significant vesicular staining and filopodia formation were retained. The additional treatment with GGTI did not disrupt this vesicular localization or filopodia formation, suggesting that these activities may be prenylation-independent.
FIGURE 3.
TC10 and TCL are farnesylated in vitro and in vivo. A, TC10 is a substrate for FTase and GGTase-I in vitro. Assays were done as described in the legend to Fig. 2A and under “Experimental Procedures.” B, TC10 subcellular localization is partially dependent on farnesylation. NIH 3T3 cells transfected with expression constructs for GFP-tagged fusion proteins of human TC10 or TCL were treated with FTI-2153, GGTI-2417, both (10 μm each), or DMSO. Live cells were visualized using confocal microscopy. Images shown are representative of three independent experiments with >80 cells examined per assay. Scale bar, 10 μm.
Like TC10, RhoD terminates in X = T, and therefore is predicted to be farnesylated (34). We found that RhoD endosome association was reduced but not abolished by treatment with FTI alone (Fig. 4). Although we consistently observed an increase in nuclear accumulation of RhoD upon treatment with FTI alone, additional treatment with both inhibitors did not disrupt the endosomal localization or impair filopodia formation (Fig. 4). Thus, this localization may be prenylation-independent. In summary, in contrast to the classical Rho GTPases, we determined that the localization of six other Rho GTPases (Rnd1, Rnd2, Rnd3, TC10, TCL, and RhoD) can be disrupted with FTI treatment, whereas GGTI treatment had little to no effect on the localization of these GTPases.
FIGURE 4.
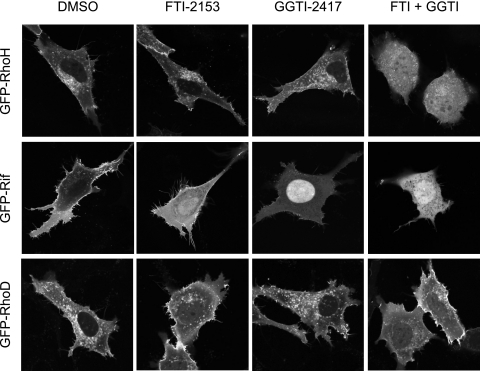
Atypical Rho GTPases are not simple substrates for FTase or GGTase-I. RhoH and Rif are farnesylated and/or geranylgeranylated in vivo. NIH 3T3 cells were transfected with expression constructs for the indicated GFP-tagged Rho GTPase and treated with FTI-2153, GGTI-2417, or both (10 μm each) or DMSO. Live cells were visualized using confocal microscopy. Images shown are representative of three independent experiments with >80 cells examined per assay. Scale bar, 10 μm.
Some Rho GTPases Are Substrates for Both FTase and GGTase-I—RhoH/TTF terminates in a CKIF CAAX motif and, similar to TC21/R-Ras2 (CVIF) (49), is therefore predicted to be modified by both farnesyl and geranylgeranyl isoprenoid groups (34). In agreement with this prediction, we found that treatment with either FTI or GGTI alone had no effect on the perinuclear vesicular subcellular localization of RhoH (Fig. 4). Instead, dual treatment with FTI and GGTI resulted in complete relocalization of RhoH to the cytoplasm and nucleus, indicating that RhoH, like RhoB, may normally be modified by both isoprenoids or that it is alternatively prenylated when challenged by FTI treatment. In addition to altered subcellular localization of RhoH, we found that dual treatment also resulted in a cell rounding phenotype in cells expressing this Rho GTPase.
In agreement with previous observations (50), we found that ectopically expressed wild-type Rif exhibited a plasma membrane localization and induced filopodia formation (Fig. 4). Rif/RhoF terminates in a CLLL motif and is predicted to be modified exclusively by GGTase-I. Consistent with this possibility, we found that treatment with GGTI caused a complete and striking nuclear accumulation of Rif. Unexpectedly, FTI treatment alone caused a partial relocalization of Rif to the cytoplasm and nucleus (Fig. 4). Interestingly, the Rif-induced filopodia seen in untreated cells were not blocked by FTI treatment. Although single treatment with either FTI or GGTI impaired Rif localization, dual treatment had a synergistic effect on Rif mislocalization, as judged by increased cytoplasmic and nuclear fluorescence and lack of filopodia. These results indicate that Rif is naturally prenylated by both FTase and GGTase-I and exists in independent F- and GG-modified pools. Collectively, our results also indicate that, of the 16 Rho GTPases that possess carboxyl-terminal CAAX motifs, nine can serve as substrates for FTase.
The Membrane Association and/or Function of Rho GTPases Is Dependent on Rce1- and Icmt-mediated Processing—Since the majority of Rho GTPases (16 of 20) terminate in CAAX motifs, they are also likely substrates for Rce1-catalyzed removal of the -AAX residues and Icmt-mediated carboxyl methylation of the prenylated cysteine. A recent study found that the membrane association and biological function of farnesylated Ras proteins, but not of geranylgeranylated Rho GTPases, were impaired in the absence of Rce1 or Icmt expression (22). However, other studies using pharmacologic inhibitors suggested that geranylgeranylated protein function also depends on these modifications (51, 52). Therefore, to elucidate whether the specific isoprenoid modification of a Rho GTPase dictates a dependence on Rce1-catalyzed -AAX cleavage and Icmt-mediated carboxyl methylation, we evaluated the subcellular localization of Rho GTPases in MEF cell lines deficient in Rce1 (Rce1–/–) or Icmt (Icmt–/–).
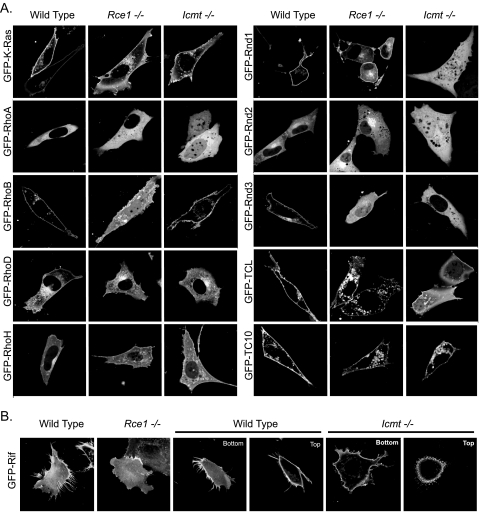
For these analyses, we transiently transfected wild-type, Rce1–/–, and Icmt–/– MEFs with expression constructs for K-Ras4B or Rho GTPases as GFP fusion proteins. Plasma membrane localization of K-Ras4B has been demonstrated to be partially dependent on Rce1- and Icmt-mediated carboxyl-terminal modifications (22, 53). In agreement with earlier studies, we found that GFP-K-Ras4B was partially localized to the cytoplasm in both Rce1–/– and Icmt–/– cells (Fig. 5A). However, this shift was subtle, and significant association with the plasma membrane was retained in each deficient cell line. Although RhoA localization was unchanged in Rce1–/– MEFs, we unexpectedly found that RhoA localization, shown previously to be independent of Rce1- and Icmt-mediated modifications (22), displayed increased nuclear and cytosolic accumulation in Icmt–/– cells (Fig. 5A). The differential sensitivity to loss of Rce1 compared with Icmt is surprising, since proteolytic cleavage of the AAX residues to expose the prenylated cysteine to Icmt is thought to be required for methylation to occur.
FIGURE 5.
Differential requirements for Rce1- and Icmt-mediated postprenyl processing in the subcellular localization and function of Rho family proteins. Wild-type, Rce1–/–, and Icmt–/– MEFs were transiently transfected with expression constructs for GFP-tagged fusion proteins of the indicated Rho GTPases. A, live cells were visualized using confocal microscopy. Images shown are representative of three independent experiments with >80 cells examined per assay. B, Icmt-mediated processing of Rif is not required for membrane association but contributes to subcellular membrane distribution. GFP-Rif localization was examined by Z-sectioning. The bottom and top stacks of each cell were examined for the presence of filopodia, and images shown are representative of three independent experiments with >50 cells examined per assay. Scale bar, 10 μm.
Although RhoA and RhoB share significant sequence (83%) and biochemical identity, RhoB can be modified by both farnesylation and geranylgeranylation, probably in distinct pools. In contrast to RhoA, we found that the absence of Rce1 expression resulted in decreased RhoB plasma membrane localization and increased cytoplasmic and nuclear accumulation in Rce1–/– MEFs, whereas normal localization was maintained in Icmt–/– MEFs (Fig. 5A). Our observations contrast with previous analyses that found that RhoB localization was not dependent on either Rce1 or Icmt expression (22).
Based on previous observations of farnesylated and geranylgeranylated Ras proteins, the subcellular localization of farnesylated Rho GTPases was expected to be dependent on Rce1 and Icmt activity (22). Surprisingly, the farnesylated RhoD and RhoH GTPases displayed subtle changes in their subcellular distribution in the null MEFs. Expression of RhoD in Rce1–/– and Icmt–/– MEFs resulted in reduced association with the plasma membrane, whereas vesicular localization was unchanged (Fig. 5A). Although loss of Rce1 function did not impair RhoH plasma membrane association, it did result in reduced RhoH association with endomembranes (Fig. 5A). In Icmt–/– cells, RhoH displayed a loss of endomembrane association and was found on vesicles throughout the cytoplasm.
We found that even closely related isoforms differed substantially in their degree and manner of dependence on Rce1 and Icmt. When expressed in Rce1–/– cells, Rnd1 consistently demonstrated a weak increase in nuclear staining yet retained significant plasma membrane association. Rnd2, which is normally cytoplasmic, showed increased endomembrane localization as well as increased negative imaging of internal organelles in Rce1–/– cells (Fig. 5A). Finally, Rnd3 expression in Rce1–/– cells showed a significant decrease in plasma membrane association, accompanied by substantial cytosolic distribution and some nuclear accumulation (Fig. 5A). Therefore, we concluded that all three Rnd proteins require Rce1-mediated AAX proteolysis for proper localization but to very different degrees, with Rnd3 being the most sensitive. Interestingly, Rnd1 and Rnd2 displayed far greater sensitivity to the absence of Icmt-mediated methylation than to the absence of Rce1-mediated proteolysis, with both proteins showing significant diffuse cytosolic staining and extensive nuclear accumulation. Rnd3 also exhibited a complete loss of plasma membrane and endomembrane localization that was accompanied by increased cytosolic but not nuclear distribution (Fig. 5A). Finally, consistent with the lack of Rnd1 plasma membrane association seen upon loss of Icmt but not upon loss of Rce1 activity, Rnd1-induced rounding was abrogated completely in Icmt-deficient but not at all in Rce1-deficient cells.
Proper subcellular localization of the highly related TC10 and TCL proteins exhibited differential requirements for Rce1-mediated proteolysis versus Icmt-mediated carboxyl methylation (Fig. 5A). Both of these farnesylated GTPases displayed reduced plasma membrane association while maintaining their vesicular localization in Rce1–/– cells. In contrast, in the absence of Icmt, TCL was strongly mislocalized to the cytosol and nucleus, whereas TC10 protein localization demonstrated only a mild loss of membrane association with no nuclear accumulation. Thus, the loss of carboxymethylation is more critical for membrane association of TCL than of TC10.
Last, despite the loss of Rif-induced filopodia in Rce1-deficient cells, we found that Rif maintained its plasma membrane localization (Fig. 5B). Expression of Rif in Icmt–/– cells did not cause mislocalization of Rif as determined by increased cytoplasmic or nuclear localization. However, the absence of Icmt resulted in an altered appearance and spatial redistribution of Rif-induced filopodia from the cell periphery to the dorsal side of the cell (Fig. 5B).
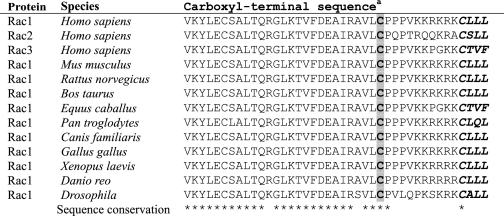
Complexity of Predicting Palmitoylation of Carboxyl-terminal Cysteines—In addition to CAAX processing, a second carboxyl-terminal sequence motif is required for proper subcellular localization and membrane association. Ras (H-Ras and N-Ras) and Rho (RhoB and TC10) GTPases with cysteine residues upstream of the CAAX motif undergo posttranslational modification by palmitate that is critical for their proper subcellular localization (24, 54). The three Rac isoforms (Rac1, -2, and -3) contain an invariant cysteine residue at position 178 in the hypervariable domain, several residues amino-terminal to the cysteine of the CAAX motif (Table 1). Furthermore, this cysteine residue and its flanking sequences are conserved in evolution and are found in Rac1 orthologs in Drosophila, Danio, and Xenopus (Table 2). Since there are no known consensus palmitoylation motifs that can predict whether a given cysteine residue is a substrate for palmitoylation (55), experimental analyses are required to establish definitively whether these sites within Rac1, Rac2, Rac3, and TCL are palmitoylated. Therefore, we determined whether these proteins were palmitoylated in vivo.
TABLE 2.
Evolutionary conservation of the Rac1 carboxyl-terminal cysteine
a Conserved cysteine residue is shaded; CAAX motif is in boldface type and in italics.
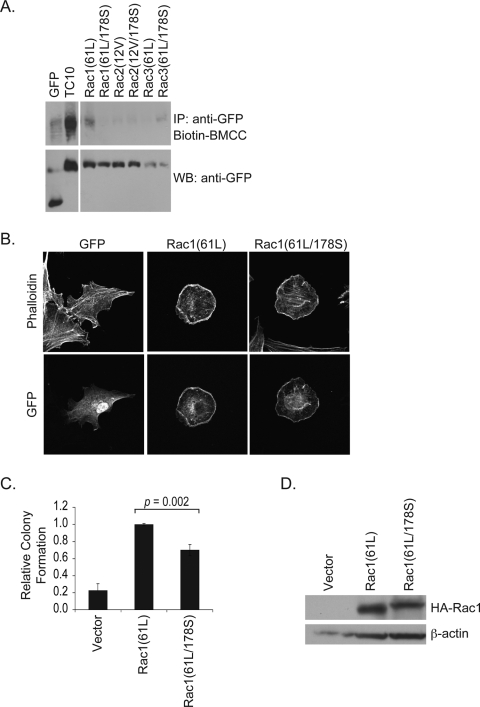
To evaluate the palmitoylation status of the Rac proteins, we used biotin-BMCC labeling to analyze protein palmitoylation. This assay involves hydrolyzing the thioester bond that links fatty acid groups to cysteines and treating with a biotinylated compound that recognizes and binds to the free sulfhydryl group generated upon cleavage of the thioester bond (31). We recently applied this assay to determine that the Chp and Wrch-1 Rho GTPases are palmitoylated (23, 28). Using this technique, we found efficient labeling of TC10 (Fig. 6A), which is consistent with the previous suggestion that TC10 undergoes palmitoylation (24). Surprisingly, none of the Rac proteins was labeled, indicating that Rac isoforms are not palmitoylated (Fig. 6A). Nevertheless, although the Rac C178 does not appear to undergo palmitoylation, the strong conservation of this cysteine in Rac orthologs and paralogs through evolution supported an important functional role for this residue. To address this possibility, we generated missense mutants containing a Cys178 to Ser substitution for all three Rac isoforms. Surprisingly, this substitution did not affect subcellular localization or impair lamellipodia formation of activated Rac1, Rac2, or Rac3 when these proteins were expressed in NIH 3T3 cells (Fig. 6B). However, this substitution did lead to a modest but reproducible reduction in Rac1(61L)-induced soft agar colony formation (Fig. 6C), suggesting that this conserved cysteine is of functional importance despite not being a site of palmitoylation. We noted a similar limited reduction in the transforming activity of activated Rac2 and Rac3 (data not shown).
FIGURE 6.
Distinct roles of conserved cysteines in palmitoylation and protein function of Rac. A, 293T cells were transiently transfected with expression constructs for GFP-tagged activated Rac1(61L), Rac2(12V), and Rac3(61L) with or without an additional C178S mutation; TC10; or empty vector and subjected to the biotin-BMCC labeling assay. Levels of biotin-labeled GTPases were measured by Western blotting with streptavidin-horseradish peroxidase (top). Expression of GFP-tagged constructs was confirmed by immunoblotting with anti-GFP antibodies (bottom). B, NIH 3T3 cells were transiently transfected with expression vectors encoding GFP-tagged activated Rac1(61L) or mutant Rac1(61L/178S) proteins and stained with Alexa 594-conjugated phalloidin to visualize the actin cytoskeleton. Images shown are representative of three independent experiments with >80 cells examined per assay. C, mutation of C178 causes a limited reduction in Rac transforming activity. Single cell suspensions of NIH 3T3 cells stably expressing activated Rac1 proteins were suspended in soft agar, and colony formation was monitored after 2 weeks. The number of colonies was quantified as described under “Experimental Procedures.” Colony numbers were normalized to those seen with Rac1(61L). D, expression of hemagglutinin (HA)-tagged Rac constructs was confirmed by immunoblotting with anti-hemagglutinin antibodies. IP, immunoprecipitation; WB, Western blot.
TCL also possesses two carboxyl-terminal putative palmitoylation sites similar to TC10 that may serve as additional signals to control proper subcellular localization. More specifically, TCL possesses a cysteine residue immediately adjacent to the CAAX motif, a topology found in the palmitoylated TC10 and RhoB, suggesting that it may undergo palmitoylation. However, similar to Rac, we found that TCL did not exhibit significant biotin-BMCC labeling (Fig. 7A). These results highlight the complex and highly unpredictable nature of sequences that dictate carboxyl-terminal palmitoylation of prenylated proteins.
FIGURE 7.
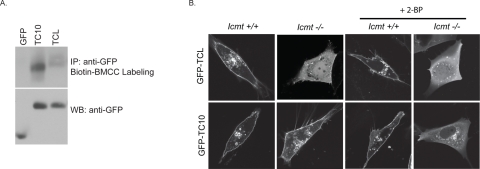
Methylation and palmitoylation act as redundant membrane targeting signals for TC10 localization. A, 293T cells were transiently transfected with expression constructs for GFP-tagged TC10, TCL, or empty vector and subjected to the biotin-BMCC labeling assay. Levels of biotin-labeled GTPases were measured by Western blotting with streptavidin-horseradish peroxidase (top). Expression of GFP-tagged constructs was confirmed by immunoblotting with anti-GFP antibodies (bottom). B, wild-type or Icmt–/– MEFs were transiently transfected with expression constructs for the indicated GFP-tagged Rho GTPase. Cells expressing GFP-TCL and GFP-TC10 were treated with 100 μm 2-BP and imaged alive. Images shown are representative of three independent experiments with >80 cells examined per assay. Scale bar, 10 μm. IP, immunoprecipitation; WB, Western blot.
Our results suggest that TC10, but not TCL, can be palmitoylated. Therefore, we speculated that the increased membrane affinity of TC10 due to palmitoylation may account for its insensitivity to the loss of Icmt-mediated processing that we described above (Fig. 5). To address this possibility, we treated cells expressing TC10 and TCL with 2-BP, a well characterized inhibitor of palmitoylation in vivo (24, 28–30), and determined subcellular localization of TC10 and TCL. As anticipated from the lack of biotin-BMCC labeling, treatment with 2-BP had no effect on the subcellular localization of TCL in wild-type or Icmt–/– MEFs (Fig. 7B). Unexpectedly, and in contrast to a previous study using COS-1 cells (24), we found that plasma membrane association of TC10 was insensitive to 2-BP treatment in wild-type MEFs, suggesting that loss of palmitoylation is not sufficient to mislocalize TC10 (Fig. 7B). However, we did find that plasma membrane association of GFP-TC10 was lost in Icmt–/– MEFs treated with 2-BP, leading to an increase in cytosolic accumulation (Fig. 7B). These results suggest that carboxyl methylation and palmitoylation modifications are partially redundant and cooperate to target TC10 to the plasma membrane. Furthermore, only prior farnesylation and not full CAAX-mediated processing, including carboxyl methylation, is a necessary prerequisite for palmitoylation.
DISCUSSION
Due to the critical role of Rho family small GTPases in oncogenesis and their dependence on CAAX processing for proper localization and function, pharmacological inhibitors of the CAAX-processing enzymes FTase, GGTase-I, Rce1, and Icmt are considered promising approaches to block Rho small GTPase function for cancer treatment (9). In the current study, we utilized pharmacologic and genetic approaches to determine the sequence requirements and roles of CAAX-signaled modifications in dictating the subcellular location and function of atypical members of the Rho GTPase family. Our results demonstrate the complexity of sequence requirements that direct prenyltransferase specificity and uncover functional roles for protein prenylation and postprenyl processing in Rho GTPase biology. Additionally, we have shown that the localization (and presumably the activity) of a majority of human Rho family GTPases may be blocked effectively by pharmacologic inhibitors of FTase, Rce1, and/or Icmt (Table 3). Finally, we found that the carboxyl-terminal sequences that signal palmitoylation of Rho GTPases are complex and not defined simply by the presence of a cysteine residue.
TABLE 3.
Contribution of prenyltransferase, Rce1, and Icmt activities to subcellular membrane association and localization
|
Name
|
Sensitivitya
|
Dependencea
|
|||
|---|---|---|---|---|---|
| FTI | GGTI | FTI + GGTI | Rce1 | Icmt | |
| K-Ras4B | - | - | + | ± | ± |
| RhoA | - | + | + | - | + |
| RhoB | - | - | + | + | - |
| RhoC | - | + | NDb | ND | ND |
| Rac1 | - | + | + | - | - |
| Rac2 | - | + | + | ND | ND |
| Rac3 | - | + | + | ND | ND |
| RhoG | ND | ND | ND | ND | ND |
| Rnd1 | + | - | + | ± | + |
| Rnd2 | + | - | + | ± | + |
| Rnd3 | + | - | + | + | + |
| Cdc42 | - | + | + | ND | ND |
| TC10 | + | - | + | ± | ± |
| TCL | + | - | + | ± | + |
| RhoD | ± | - | ± | ± | ± |
| Rif | ± | + | + | + | + |
| RhoH | - | - | + | ± | ± |
| Wrch-1c | - | - | - | ND | ND |
| Chpc | - | - | - | ND | ND |
Our analyses of the specificity of CAAX-directed prenylation uncovered unexpected results, in light of the previously published rules governing substrate selectivity derived from in vitro prenylation assays (56, 57) and from recent crystallographic analyses of FTase and GGTase-I in complex with validated CAAX-containing prenyltransferase substrate peptides (34). These prior studies determined that the X residue of the CA1A2X motif is the primary determinant of prenyltransferase specificity, such that FTase favors motifs where X is a hydrophobic (Met), polar (Gln), or small (Cys, Ser, Thr, or Ala) residue. In contrast, GGTase-I favors motifs where X is Leu but can also accommodate Met, Phe, Ile, and Val. The nature of the A1 and A2 residues alone does not dictate enzyme selectivity. In contrast to the classical Rho family members that are known to be prenylated by GGTase-I, these rules predict that some Rho family members are farnesylated in vivo. In general agreement with these predictions, we found that a majority of the noncanonical Rho GTPases are substrates for FTase in vivo: Rnd1–3, TC10, TCL, RhoD, and Rif (Table 3). However, our results also suggest that these rules are imperfect. Thus, although previous analyses with peptide substrates indicated that CAAX motifs where X is Ile can be recognized efficiently by GGTase-I in vitro (56, 58), we found that at least some proteins terminating in such a motif (e.g. TCL) are still more effective substrates for FTase in vivo. Finally, two Rho GTPases exhibited complex responses to prenyltransferase inhibition. Rif terminates in the same CAAX motif as Rac1 (CLLL), and, as expected, GGTI treatment caused complete mislocalization and nuclear accumulation of this GTPase and a block of Rif-induced filopodia formation. However, FTI treatment caused only partial disruption of Rif subcellular localization, increasing nuclear accumulation, although filopodia induction was not blocked. Perhaps, like RhoB (CKVL), a subset of Rif is farnesylated, or alternatively, Rif subcellular localization is dependent on another farnesylated protein. Finally, whereas RhoD subcellular localization was disrupted partially with FTI treatment, surprisingly, the combined treatment with FTI and GGTI did not cause further disruption. One logical interpretation of these data is that RhoD subcellular localization is not dependent exclusively on prenylation and may instead be regulated by palmitoylation, as found for other endosomal GTPases, Wrch-1 and Chp (23, 28).
In addition to defining the preferred prenyl modification of the Rho family members, we were also interested in determining whether these proteins would be alternatively prenylated when challenged with prenyltransferase inhibitors. Of the nine Rho family members evaluated, only RhoH showed the potential to be alternatively prenylated. This result is in agreement with the prediction that CAAX motifs where X is Phe are substrates for both FTase and GGTase-I (34). Surprisingly, the Rnd proteins, which have been speculated to undergo FTI-induced alternative prenylation (8), were exquisitely sensitive to FTI treatment alone and did not undergo detectable FTI-induced alternative prenylation by GGTase-I. This result was unexpected, because the carboxyl-terminal sequences of Rnd proteins are similar to that of K-Ras4B, which does undergo FTI-induced alternative prenylation. Together, these data suggest that alternative prenylation of Rho proteins is a rare event and that most Rho targets of FTase inhibitors will probably be sensitive to inhibition of FTase alone. Finally, we recently determined that Rnd3 expression is up-regulated in human melanomas (59), and a recent study suggested that Rnd3 promotes melanoma invasion (60). Perhaps Rnd3 may represent an important target for the anti-tumor activity seen with FTIs in melanomas and other cancers that overexpress Rnd3.
Efforts to develop inhibitors for the postprenyl processing steps regulated by Rce1 and Icmt have intensified since K-Ras4B was determined to be alternatively prenylated when treated with FTI (9). The possibility that pharmacologic inhibitors of these two enzymes may exhibit anti-Ras activity is supported by recent genetic studies using Rce1- or Icmt-deficient MEFs (53). These studies showed that K-Ras and H-Ras-mediated transformation is indeed impaired by a deficiency in either enzyme. However, in light of the significant number of CAAX-terminating proteins encoded in the human genome, including many proteins with established roles in normal cell proliferation and survival, there is significant concern that such inhibitors will be intolerable to patients due to normal cell toxicity. The observation that transformation caused by the B-Raf oncoprotein, which is not a substrate for Icmt, was also dependent on Icmt activity supports this concern (61). This result suggested that the perceived Icmt deficiency-induced inhibition of K-Ras activity is not due simply to impaired K-Ras function; indeed, the authors provided evidence that the phenotype may instead be due to impaired RhoA function.
A recent study of RhoA, as well as of Rac1 and Cdc42, suggested that these final two postprenyl modifications are not required for geranylgeranylated proteins (22). However, in general, we found that the function of most Rho family members, including geranygeranyl-modified GTPases, were dependent on both Rce1 and Icmt function for proper subcellular localization and/or function. In contrast to this previous report, we observed that the subcellular localization of RhoA was sensitive to loss of Icmt expression. In the previous study, an activated RhoA(63L) construct was used to evaluate the dependence of Icmt function on RhoA localization, whereas in the current study, we used wild-type RhoA. Therefore, to determine whether wild-type and activated RhoA are differentially dependent on Icmt, we also evaluated the localization of activated RhoA in Icmt–/– MEFs and found that, unlike wild-type RhoA, it was unaffected by Icmt deficiency (data not shown). This differential dependence may reflect a role for carboxyl methylation in the interaction of Rho proteins with Rho GDP dissociation inhibitors, an interaction that is sensitive to the GTP-bound state of the protein. Regardless of the mechanism behind the differential requirements for Icmt activity for wild-type versus mutant RhoA, we conclude that RhoA represents a functionally important anti-cancer target of Icmt inhibitors, since there are no known RhoA-activating mutations that naturally occur in cancer. In support of this conclusion, loss of Icmt has been shown to decrease the stability of RhoA protein within the cell (61, 62). In addition, inhibition of carboxyl methylation has been shown to increase permeability of the endothelial cell monolayer as well as to increase endothelial cell apoptosis through a RhoA-dependent mechanism (51). Thus, we conclude that the specific isoprenoid modification of a CAAX motif-containing protein will not be a reliable determinant of the requirement for Rce1 and Icmt activity. Furthermore, our results suggest that Icmt and Rce1 remain attractive targets for the development of anti-cancer therapeutics.
In addition to RhoA, we also found that RhoB was highly sensitive to loss of Rce1 and somewhat sensitive to loss of Icmt function. This result also differed from those of Philips and coworkers (22), who concluded that RhoB function was not dependent on the activity of either enzyme. Since both of these studies used enzyme-deficient MEFs, the basis for the difference in results is not clear. One possibility is subtle differences in cell culture conditions that alter the activity of other enzymes or interacting chaperone proteins, but this remains to be determined.
Rce1-mediated proteolysis is a prerequisite processing step essential for Icmt-mediated carboxyl methylation. As such, it might be expected that loss of Rce1 function would have more drastic consequences than loss of Icmt. However, whereas Rce1-deficient mice died during late gestation or soon after birth, Icmt-deficient mice showed a more severe phenotype, with virtually all of the knock-out embryos (Icmt–/–) dying by midgestation (25). This greater severity had been explained by the fact that there are additional Icmt substrates that are not Rce1 substrates and that it is the mislocalization of these additional proteins that results in the more severe phenotypic changes. In addition, conditional deletion of Icmt in mouse fibroblasts has been shown to cause a more dramatic reduction in Ras-induced transformation when compared with conditional deletion of Rce1. Interestingly, we found that many of the evaluated Rho family members were differentially sensitive to loss of Rce1 and Icmt. As mentioned previously, RhoB appeared to be more sensitive to loss of Rce1, whereas RhoA was more dependent on Icmt. In addition, we found that the Rnd proteins were sensitive to loss of both processing steps, although the loss of Icmt was much more dramatic. Finally, loss of Rce1 blocked Rif-induced filopodia but did not impair localization, whereas conversely loss of Icmt altered the spatial orientation of Rif without blocking filopodia formation. One explanation for the differential consequences of the loss of Rce1 and Icmt is that Rho GTPases may be less affected by the presence of the -AAX extension than they are by the presence of the carboxylate anion (63). In accordance with this hypothesis, protein stability as measured by cellular half-life is significantly altered by both pharmacologic and genetic disruption of Icmt (61, 62). The mechanism of these changes in protein stability is unknown, although it could be the result of altered recognition by degradative enzymes.
Our studies found unexpectedly that the Rac isoforms, as well as TCL, do not undergo palmitoylation. Although an important role for palmitoylated cysteines in facilitating the proper subcellular localization and membrane association of Ras and Rho GTPases is well established, the substrate specificity and enzymology of protein palmitoylation remain poorly understood (64). Aside from a conserved cysteine residue, a defined consensus sequence that directs protein palmitoylation has not been elucidated. Mutagenesis studies suggest that there will not be a strict requirement for these flanking sequences. Thus, although the conserved cysteine residue found in Rac small GTPases seemed ideally suited to be a substrate for palmitate addition, our results showed clearly that these cysteines were not palmitoylated and again highlight the difficulty of predicting palmitoylation simply by examining amino acid sequence. Future studies with specific protein acyltransferases (24 human members) will be required to determine if rules can be established for predicting substrates for palmitoylation.
In summary, our analyses of the Rho GTPase family reveal that the function of a majority of Rho GTPases will be sensitive to FTI treatment as well as to inhibitors of Rce1 and Icmt. Our observations also reveal the complex and varied roles for CAAX-mediated posttranslational modifications for the function of Rho family GTPases. FTIs are currently in phase II/III clinical trials, and our studies identify additional targets that may contribute to their anti-tumor activities. Recent preclinical studies in mouse models suggest that GGTIs will also exhibit anti-tumor activities (10, 65, 66), and our studies identify those Rho GTPases that may also be important targets for these inhibitors (10). Finally, although the development of pharmacologic inhibitors of Rce1 or Icmt has been limited to date (9), our findings provide further validation for these two enzymes as potentially important drug targets and support a therapeutic value for such inhibitors.
Acknowledgments
We thank Wendy Salmon and Michael Chua (University of North Carolina Michael Hooker Microscopy Facility), Ashutosh Tripathy (University of North Carolina Macromolecular Interaction Facility), Pontus Aspenström, Mark Philips, and John Sondek for cDNA expression vectors for Rho GTPases, Stephen G. Young for MEF cell lines deficient in Rce1 and Icmt, and Misha Rand for assistance with figure preparation.
This work was supported, in whole or in part, by National Institutes of Health Grants CA063071, CA67771, and CA92240 (to C. J. D.) and CA063071, CA67771, and CA109550 (to A. D. C.). The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
Footnotes
The abbreviations used are: FTase, farnesyltransferase; Rce1, Ras-converting enzyme 1; Icmt, isoprenylcysteine-O-carboxyl methyltransferase; GGTase-I, geranylgeranyltransferase type I; FTI, farnesyltransferase inhibitor; GGTI, geranylgeranyltransferase I inhibitor; MEF, mouse embryonic fibroblast; GFP, green fluorescent protein; 2-BP, 2-bromopalmitate; Biotin-BMCC, 1-biotinamido-4-(4′-(maleimidomethyl cyclohexanecarboxamido) butane; GTPase, guanine triphosphatase; GFP, green fluorescent protein.
References
- 1.Colicelli, J. (2004) Science's STKE 2004, RE13. [DOI] [PMC free article] [PubMed]
- 2.Wennerberg, K., Rossman, K. L., and Der, C. J. (2005) J. Cell Sci. 118 843–846 [DOI] [PubMed] [Google Scholar]
- 3.Wennerberg, K., and Der, C. J. (2004) J. Cell Sci. 117 1301–1312 [DOI] [PubMed] [Google Scholar]
- 4.Schmidt, A., and Hall, A. (2002) Genes Dev. 16 1587–1609 [DOI] [PubMed] [Google Scholar]
- 5.Bernards, A., and Settleman, J. (2004) Trends Cell Biol. 14 377–385 [DOI] [PubMed] [Google Scholar]
- 6.Etienne-Manneville, S., and Hall, A. (2002) Nature 420 629–635 [DOI] [PubMed] [Google Scholar]
- 7.Ridley, A. J. (2004) Breast Cancer Res. Treat. 84 13–19 [DOI] [PubMed] [Google Scholar]
- 8.Sebti, S. M., and Der, C. J. (2003) Nat. Rev. Cancer 3 945–951 [DOI] [PubMed] [Google Scholar]
- 9.Winter-Vann, A. M., and Casey, P. J. (2005) Nat. Rev. Cancer 5 405–412 [DOI] [PubMed] [Google Scholar]
- 10.Sjogren, A. K., Andersson, K. M., Liu, M., Cutts, B. A., Karlsson, C., Wahlstrom, A. M., Dalin, M., Weinbaum, C., Casey, P. J., Tarkowski, A., Swolin, B., Young, S. G., and Bergo, M. O. (2007) J. Clin. Invest. 117 1294–1304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohya, Y., Qadota, H., Anraku, Y., Pringle, J. R., and Botstein, D. (1993) Mol. Biol. Cell 4 1017–1025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cox, A. D., and Der, C. J. (1992) Curr. Opin. Cell Biol. 4 1008–1016 [DOI] [PubMed] [Google Scholar]
- 13.Willumsen, B. M., Cox, A. D., Solski, P. A., Der, C. J., and Buss, J. E. (1996) Oncogene 13 1901–1909 [PubMed] [Google Scholar]
- 14.Parton, R. G., and Hancock, J. F. (2004) Trends Cell Biol. 14 141–147 [DOI] [PubMed] [Google Scholar]
- 15.Quatela, S. E., and Philips, M. R. (2006) Curr. Opin. Cell Biol. 18 162–167 [DOI] [PubMed] [Google Scholar]
- 16.Rowell, C. A., Kowalczyk, J. J., Lewis, M. D., and Garcia, A. M. (1997) J. Biol. Chem. 272 14093–14097 [DOI] [PubMed] [Google Scholar]
- 17.Whyte, D. B., Kirschmeier, P., Hockenberry, T. N., Nunez-Oliva, I., James, L., Catino, J. J., Bishop, W. R., and Pai, J. K. (1997) J. Biol. Chem. 272 14459–14464 [DOI] [PubMed] [Google Scholar]
- 18.Prendergast, G. C. (2000) Curr. Opin. Cell Biol. 12 166–173 [DOI] [PubMed] [Google Scholar]
- 19.Sebti, S. M., and Hamilton, A. D. (2000) Oncogene 19 6584–6593 [DOI] [PubMed] [Google Scholar]
- 20.Sahai, E., and Marshall, C. J. (2002) Nat. Rev. Cancer 2 133–142 [DOI] [PubMed] [Google Scholar]
- 21.Kato, K., Cox, A. D., Hisaka, M. M., Graham, S. M., Buss, J. E., and Der, C. J. (1992) Proc. Natl. Acad. Sci. U. S. A. 89 6403–6407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Michaelson, D., Ali, W., Chiu, V. K., Bergo, M., Silletti, J., Wright, L., Young, S. G., and Philips, M. (2005) Mol. Biol. Cell 16 1606–1616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berzat, A. C., Buss, J. E., Chenette, E. J., Weinbaum, C. A., Shutes, A., Der, C. J., Minden, A., and Cox, A. D. (2005) J. Biol. Chem. 280 33055–33065 [DOI] [PubMed] [Google Scholar]
- 24.Michaelson, D., Silletti, J., Murphy, G., D'Eustachio, P., Rush, M., and Philips, M. R. (2001) J. Cell Biol. 152 111–126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bergo, M. O., Leung, G. K., Ambroziak, P., Otto, J. C., Casey, P. J., Gomes, A. Q., Seabra, M. C., and Young, S. G. (2001) J. Biol. Chem. 276 5841–5845 [DOI] [PubMed] [Google Scholar]
- 26.Falsetti, S. C., Wang, D. A., Peng, H., Carrico, D., Cox, A. D., Der, C. J., Hamilton, A. D., and Sebti, S. M. (2007) Mol. Cell Biol. 27 8003–8014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun, J., Blaskovich, M. A., Knowles, D., Qian, Y., Ohkanda, J., Bailey, R. D., Hamilton, A. D., and Sebti, S. M. (1999) Cancer Res. 59 4919–4926 [PubMed] [Google Scholar]
- 28.Chenette, E. J., Abo, A., and Der, C. J. (2005) J. Biol. Chem. 280 13784–13792 [DOI] [PubMed] [Google Scholar]
- 29.Veit, M., Laage, R., Dietrich, L., Wang, L., and Ungermann, C. (2001) EMBO J. 20 3145–3155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Webb, Y., Hermida-Matsumoto, L., and Resh, M. D. (2000) J. Biol. Chem. 275 261–270 [DOI] [PubMed] [Google Scholar]
- 31.Drisdel, R. C., and Green, W. N. (2004) BioTechniques 36 276–285 [DOI] [PubMed] [Google Scholar]
- 32.Shutes, A., Berzat, A. C., Chenette, E. J., Cox, A. D., and Der, C. J. (2006) Methods Enzymol. 406 11–26 [DOI] [PubMed] [Google Scholar]
- 33.Zhang, F. L., Kirschmeier, P., Carr, D., James, L., Bond, R. W., Wang, L., Patton, R., Windsor, W. T., Syto, R., Zhang, R., and Bishop, W. R. (1997) J. Biol. Chem. 272 10232–10239 [DOI] [PubMed] [Google Scholar]
- 34.Reid, T. S., Terry, K. L., Casey, P. J., and Beese, L. S. (2004) J. Mol. Biol. 343 417–433 [DOI] [PubMed] [Google Scholar]
- 35.Berzat, A. C., Brady, D. C., Fiordalisi, J. J., and Cox, A. D. (2005) Methods Enzymol. 407 575–597 [DOI] [PubMed] [Google Scholar]
- 36.Choy, E., Chiu, V. K., Silletti, J., Feoktistov, M., Morimoto, T., Michaelson, D., Ivanov, I. E., and Philips, M. R. (1999) Cell 98 69–80 [DOI] [PubMed] [Google Scholar]
- 37.Heo, W. D., and Meyer, T. (2003) Cell 113 315–328 [DOI] [PubMed] [Google Scholar]
- 38.Joyce, P. L., and Cox, A. D. (2003) Cancer Res. 63 7959–7967 [PubMed] [Google Scholar]
- 39.Keller, P. J., Fiordalisi, J. J., Berzat, A. C., and Cox, A. D. (2005) Methods 37 131–137 [DOI] [PubMed] [Google Scholar]
- 40.Katayama, M., Kawata, M., Yoshida, Y., Horiuchi, H., Yamamoto, T., Matsuura, Y., and Takai, Y. (1991) J. Biol. Chem. 266 12639–12645 [PubMed] [Google Scholar]
- 41.Adamson, P., Marshall, C. J., Hall, A., and Tilbrook, P. A. (1992) J. Biol. Chem. 267 20033–20038 [PubMed] [Google Scholar]
- 42.Lebowitz, P. F., Casey, P. J., Prendergast, G. C., and Thissen, J. A. (1997) J. Biol. Chem. 272 15591–15594 [DOI] [PubMed] [Google Scholar]
- 43.Wherlock, M., Gampel, A., Futter, C., and Mellor, H. (2004) J. Cell Sci. 117 3221–3231 [DOI] [PubMed] [Google Scholar]
- 44.Foster, R., Hu, K. Q., Lu, Y., Nolan, K. M., Thissen, J., and Settleman, J. (1996) Mol. Cell Biol. 16 2689–2699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fiordalisi, J. J., Johnson, R. L., 2nd, Weinbaum, C. A., Sakabe, K., Chen, Z., Casey, P. J., and Cox, A. D. (2003) J. Biol. Chem. 278 41718–41727 [DOI] [PubMed] [Google Scholar]
- 46.James, G. L., Goldstein, J. L., and Brown, M. S. (1995) J. Biol. Chem. 270 6221–6226 [DOI] [PubMed] [Google Scholar]
- 47.Chardin, P. (2006) Nat. Rev. Mol. Cell Biol. 7 54–62 [DOI] [PubMed] [Google Scholar]
- 48.Vega, F. M., and Ridley, A. J. (2007) Cell 129 1430. [DOI] [PubMed] [Google Scholar]
- 49.Carboni, J. M., Yan, N., Cox, A. D., Bustelo, X., Graham, S. M., Lynch, M. J., Weinmann, R., Seizinger, B. R., Der, C. J., Barbacid, M., and Manne, V. (1995) Oncogene 10 1905–1913 [PubMed] [Google Scholar]
- 50.Ellis, S., and Mellor, H. (2000) Curr. Biol. 10 1387–1390 [DOI] [PubMed] [Google Scholar]
- 51.Lu, Q., Harrington, E. O., Hai, C. M., Newton, J., Garber, M., Hirase, T., and Rounds, S. (2004) Circ. Res. 94 306–315 [DOI] [PubMed] [Google Scholar]
- 52.Papaharalambus, C., Sajjad, W., Syed, A., Zhang, C., Bergo, M. O., Alexander, R. W., and Ahmad, M. (2005) J. Biol. Chem. 280 18790–18796 [DOI] [PubMed] [Google Scholar]
- 53.Bergo, M. O., Ambroziak, P., Gregory, C., George, A., Otto, J. C., Kim, E., Nagase, H., Casey, P. J., Balmain, A., and Young, S. G. (2002) Mol. Cell Biol. 22 171–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hancock, J. F., Paterson, H., and Marshall, C. J. (1990) Cell 63 133–139 [DOI] [PubMed] [Google Scholar]
- 55.Linder, M. E., and Deschenes, R. J. (2003) Biochemistry (Mosc.) 42 4311–4320 [DOI] [PubMed] [Google Scholar]
- 56.Moores, S. L., Schaber, M. D., Mosser, S. D., Rands, E., O'Hara, M. B., Garsky, V. M., Marshall, M. S., Pompliano, D. L., and Gibbs, J. B. (1991) J. Biol. Chem. 266 14603–14610 [PubMed] [Google Scholar]
- 57.Kinsella, B. T., Erdman, R. A., and Maltese, W. A. (1991) Proc. Natl. Acad. Sci. U. S. A. 88 8934–8938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Roskoski, R., Jr., and Ritchie, P. (1998) Arch. Biochem. Biophys. 356 167–176 [DOI] [PubMed] [Google Scholar]
- 59.Shields, J. M., Thomas, N. E., Cregger, M., Berger, A. J., Leslie, M., Torrice, C., Hao, H., Penland, S., Arbiser, J., Scott, G., Zhou, T., Bar-Eli, M., Bear, J. E., Der, C. J., Kaufmann, W. K., Rimm, D. L., and Sharpless, N. E. (2007) Cancer Res. 67 1502–1512 [DOI] [PubMed] [Google Scholar]
- 60.Klein, R. M., Spofford, L. S., Abel, E. V., Ortiz, A., and Aplin, A. E. (2008) Mol. Biol. Cell 19 498–508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bergo, M. O., Gavino, B. J., Hong, C., Beigneux, A. P., McMahon, M., Casey, P. J., and Young, S. G. (2004) J. Clin. Invest. 113 539–550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Backlund, P. S., Jr. (1997) J. Biol. Chem. 272 33175–33180 [DOI] [PubMed] [Google Scholar]
- 63.Silvius, J. R., and l'Heureux, F. (1994) Biochemistry (Mosc.) 33 3014–3022 [DOI] [PubMed] [Google Scholar]
- 64.Nadolski, M. J., and Linder, M. E. (2007) FEBS J. 274 5202–5210 [DOI] [PubMed] [Google Scholar]
- 65.Sun, J., Ohkanda, J., Coppola, D., Yin, H., Kothare, M., Busciglio, B., Hamilton, A. D., and Sebti, S. M. (2003) Cancer Res. 63 8922–8929 [PubMed] [Google Scholar]
- 66.El Oualid, F., Cohen, L. H., van der Marel, G. A., and Overhand, M. (2006) Curr. Med. Chem. 13 2385–2427 [DOI] [PubMed] [Google Scholar]
- 67.Ridley, A. J. (2006) Trends Cell Biol. 16 522–529 [DOI] [PubMed] [Google Scholar]
- 68.Aspenstrom, P., Fransson, A., and Saras, J. (2004) Biochem. J. 377 327–337 [DOI] [PMC free article] [PubMed] [Google Scholar]