Abstract
Human pyruvate dehydrogenase complex (PDC) is down-regulated by pyruvate dehydrogenase kinase (PDK) isoforms 1–4. PDK4 is overexpressed in skeletal muscle in type 2 diabetes, resulting in impaired glucose utilization. Here we show that human PDK4 has robust core-free basal activity, which is considerably higher than activity levels of other PDK isoforms stimulated by the PDC core. PDK4 binds the L3 lipoyl domain, but its activity is not significantly stimulated by any individual lipoyl domains or the core of PDC. The 2.0-Å crystal structures of the PDK4 dimer with bound ADP reveal an open conformation with a wider active-site cleft, compared with that in the closed conformation epitomized by the PDK2-ADP structure. The open conformation in PDK4 shows partially ordered C-terminal cross-tails, in which the conserved DW (Asp394–Trp395) motif from one subunit anchors to the N-terminal domain of the other subunit. The open conformation fosters a reduced binding affinity for ADP, facilitating the efficient removal of product inhibition by this nucleotide. Alteration or deletion of the DW-motif disrupts the C-terminal cross-tail anchor, resulting in the closed conformation and the nearly complete inactivation of PDK4. Fluorescence quenching and enzyme activity data suggest that compounds AZD7545 and dichloroacetate lock PDK4 in the open and the closed conformational states, respectively. We propose that PDK4 with bound ADP exists in equilibrium between the open and the closed conformations. The favored metastable open conformation is responsible for the robust basal activity of PDK4 in the absence of the PDC core.
The human pyruvate dehydrogenase complex (PDC)2 catalyzes the oxidative decarboxylation of pyruvate to produce acetyl-CoA and NADH, linking glycolysis to the Krebs cycle and lipogenic pathways. The human PDC is a 9.5 × 106-dalton macromolecular machine organized around a structural core formed by 60 combined transacetylase (E2p) and E3-binding protein (E3BP) subunits (1, 43). The 60-meric E2p/E3BP core of human PDC is a dodecahedron made up of 12 pentagonal faces; each of the 20 vertices of the cage-like hollow dodecahedron is occupied by a trimer comprising the E2p and E3BP inner core domains, as illustrated in recent cryo-electron microscopy reconstructions of the full-length and truncated human E2p cores (2). Each human E2p subunit consists of two lipoyl domains (L1 and L2), one E1p-binding domain (E1BD), and one inner core domain that contains the E2p-active site and is responsible for core assembly. Each E3BP subunit harbors a single lipoyl domain (L3), an E3-binding domain (E3BD), and a catalytically inactive inner core domain (3). In addition, one to two copies of pyruvate dehydrogenase kinase (PDK) and pyruvate dehydrogenase phosphatase are associated mainly with the inner lipoyl domain (L2) of the E2p subunit (4, 5).
The human PDC is primarily regulated by reversible phosphorylation through pyruvate dehydrogenase kinase (PDK) (6, 7). The phosphorylation of specific serine residues in E1p by PDK results in the inactivation of PDC, whereas the dephosphorylation by pyruvate dehydrogenase phosphatase restores PDC activity. To date, four PDK (1–4) isoforms in mammalian mitochondria have been identified (8). Phosphorylation of E1p occurs at three serine residues of the α subunit (Ser264, site 1; Ser271, site 2; and Ser203, site 3) (9–11). Although phosphorylation of each site alone inactivates PDC, site 1 is most rapidly modified, and phosphorylation of site 3 is the slowest among the three sites (11, 12). Interestingly, each PDK isoform exhibits different site specificity. Using E1p mutants with single functional phosphorylation sites, it was shown that sites 1 and 2 are phosphorylated by all four isoforms, but site 3 is only modified by PDK1 (13, 14).
The binding of PDK isoforms to the L2 domain requires a lipoyl group covalently attached to the Lys173 of L2 (15). Colocalization of PDK with the E1p substrate bound to the E1-binding domain (immediately downstream to L2) of E2p accounts, in part, for enhanced PDK activity. Individual isoforms exhibit different binding affinities for L2 with PDK3 > PDK1 ≈ PDK2 > PDK4 (16). PDK3 binds to L2 most tightly among the four PDK isoforms and is robustly activated by the E2p/E3BP core; this activation is largely achieved through binding to isolated L2 (17, 18). PDK2 activity is augmented by the E2p/E3BP core, but not by the L2 domain alone (17, 19). PDK4 has the lowest affinity for L2 and is only modestly stimulated by the E2p/E3BP core (19).
Mitochondrial protein kinases including the four PDK isoforms and the related BCKD kinase (BCK) constitute a novel family of protein kinases (3), in which motifs that normally occur in eukaryotic Ser/Thr/Tyr kinases (20) are absent. Structural studies of PDK and BCK have revealed that these kinases consist of two distinct domains, the N- and the C-terminal domains (21–23). The N-terminal domain of PDK (or B domain in BCK) consists of eight α-helices with a four-helix bundle-like structure forming the core. The C-terminal domain (or K domain in BCK) contains the phosphotransfer catalytic site whose architecture is conserved in the GHKL ATPase/kinase superfamily (24). Members of this superfamily share four conserved motifs (N-, G1-, G2- and G3-boxes), which form a unique ATP-binding fold (24–27). This fold includes a common structural element known as the “ATP lid,” whose conformational change is coupled to both ATP hydrolysis and protein-protein interactions (22, 28, 29). Crystal structures of the PDK3-L2-nucleotide complexes revealed that L2 binding to the lipoyl-binding pocket in the N-terminal domain of PDK3 caused an open conformation with a wider active-site cleft in PDK3 (23), compared with the closed conformation present in the L2-free rat PDK2-ADP structure (21, 23). The open activesite cleft destabilizes the ATP lid located inside the ATP-binding pocket of PDK3. This promotes ATP/ADP exchange and markedly stimulates PDK3 activity by facilitating the removal of product inhibition exerted by ADP (23, 30).
PDK isoforms exhibit tissue-specific expression; PDK1 is detected in heart, pancreatic islets, and skeletal muscles; PDK2 is expressed in all tissues; PDK3 is present in testes, kidney, and brain; and PDK4 is abundant in heart, skeletal muscle, kidney, and pancreatic islets (31). The expression of PDK2 and PDK4 is induced in starvation and diabetes, which is reversed by insulin treatment (6, 32, 33). PDK4 knock-out mice show reduced blood glucose level and improved glucose tolerance and insulin sensitivity in diet-induced obesity (34). Peroxisome proliferator-activated receptor-α, WY-14,643, a potent agonist of peroxisome proliferator-activated receptor-α, and fatty acids increase the expression of PDK4 (35, 36). Protein kinase B-α inhibits the induction of the PDK4 gene by dexamethasone through inactivation of FOXO transcription factors (18). Impaired insulin-induced down-regulation of PDK4 (because of the lack of insulin or insensitivity to insulin) leads to the overexpression of PDK4 and shuts off glucose oxidation in diabetic animals (7, 18). The overexpression of PDK4 led to the discovery of this PDK isoform by positional cloning in the chromosomal region 7q21.3 of the Pima Indians inflicted with congenital obesity and insulin-resistant type 2 diabetes (37). Therefore, PDK4 is a potential drug target for the treatment of obesity and type 2 diabetes by inhibiting PDK4 activity.
Dichloroacetate (DCA) is a long-standing PDK inhibitor (38) that has been shown to reduce glucose levels in type 2 diabetic rats (39, 40). Recently, the structures of human PDK1 (41) and PDK2 (42) in complex with DCA have been determined. The PDK1-DCA structure showed that DCA bound to the N-terminal domain promotes local conformational changes that are communicated to both nucleotide-binding and lipoyl-bearing domain of PDK1, leading to the inactivation of kinase activity (41). DCA is disadvantageous as a drug because of its low affinity for PDK isoforms (31, 43) and potential toxicity to humans (44). A recently developed glucose-lowering compound AZD7545 (2R-N-{4-[4-(dimethylcarbamoyl)phenylsulfonyl]-2-chlorophenyl}-3,3,3-trifluoro-2-hydroxy-2-methylpropanamide) (45) functions as a dihydrolipoamide mimetic (46) and binds to the lipoyl domain-binding pocket in the PDK1-AZD7545 structure (41). Paradoxically, AZD7545 serves as a “dual role” ligand for PDK3. This compound is an inhibitor for the E2p/E3BP-stimulated activity by preventing the binding of PDK3 to the E2p/E3BP core. On the other hand, AZD7545 robustly augments core-free basal PKD3 activity, similar to the isolated L2 domain (41). AZD7545 as a potent activator offers a useful nonproteinaceous ligand for investigating the allosteric mechanism underlying the stimulation of core-free PDK3 activity.
To understand the structure and function of PDK4 and to provide the structural basis for inhibitor design for this isoform, our laboratory has determined the crystal structures of the human PDK4-ADP complex at 2.0-Å resolution. These structures reveal an open conformation in the active-site cleft, similar to that observed in the structures of PDK3-L2-ADP (23) and the PDK2-L2-(AMP-PNP) complex (47). The open conformation in the PDK4 structure is accompanied by partially ordered C-terminal crossed-tails anchoring at the conserved DW (Asp394/Trp395)-motif. Moreover, the core-free PDK4 shows robust basal activity, which is significantly higher than the E2p/E3BP-stimulated levels exhibited by other PDK isoforms. A structural model, based on the metastable open conformation observed in its crystal structures, is proposed to explain the robust basal activity of human PDK4.
EXPERIMENTAL PROCEDURES
Reagents and Recombinant Proteins—Compound AZD7545 (45) was a generous gift from Dr. Rachel Mayers, Astrazeneca, UK. Dichloroacetate was purchased from Sigma. Human PDK isoforms (1–3) were expressed either as N-terminal His6 tagged small ubiquitin modifier (SUMO) or MBP (maltose-binding protein) fusions with a tobacco etch virus (TEV) protease recognition sequence (Glu-Asn-Leu-Tyr-Phe-Gln↓Gly, the arrow showing the cleavage site) engineered into the linker region as described previously (23, 41). His6 tagged human L1 (residues 1–127), L2 (residues 128–266), and L3 (residues 1–124) were expressed also as described previously (23). The expression plasmids for human E1p protein (48) and the E2p/E3BP core (3) were kindly supplied by Dr. Krill Popov, University of Alabama at Birmingham. N-terminal His6 tagged human E3 protein was expressed and purified as described previously (49). Single and double variants of human PDK4 were produced using the QuickChange™ site-directed mutagenesis kit from Stratagene (La Jolla, CA), and the mutations were confirmed by DNA sequencing. All recombinant human proteins were expressed in Escherichia coli BL-21 cells, which, in the case of E1p and PDK isoforms, were co-transformed with pGroESL plasmid overexpressing chaperonins GroEL and GroES. Bacterial lysates were obtained by sonication, and recombinant proteins were extracted by Ni-NTA resin. The isolated proteins were further purified by fast protein liquid chromatography with Resource Q and Superdex 200 or Sephacryl S-400 columns (for E2p/E3BP) to apparent homogeneity.
Expression and Purification of Human PDK4—Human PDK4-coding sequence (residues 10–411) was inserted into the pSUMO vector after the TEV protease recognition site as described previously (41). The His6 tagged SUMO-PDK4 fusion protein was co-expressed with chaperonins GroEL/GroES in the BL21 cell line. Following the addition of 0.5 mm isopropyl thiogalactopyranoside, the cells were grown at 23 °C overnight. Harvested cells were lysed with lysozyme followed by sonication, and the cell homogenate was extracted with Ni-NTA resin. The PDK4-bound resin was washed with a wash buffer (50 mm potassium phosphate (pH 7.5), 200 mm KCl, 5% glycerol, 20 mm β-mercaptoethanol (BME), 20 mm imidazole, and 0.05% octyl β-d-glucopyranoside). SUMO-PDK4 was then eluted with the wash buffer containing 250 mm imidazole and 0.1% octyl β-d-glucopyranoside. The eluted SUMO-PDK4 was further purified on a Superdex 200 column equilibrated with 50 mm potassium phosphate (pH 7.5), 200 mm KCl, 5% glycerol, and 20 mm BME; octyl β-d-glucopyranoside was added to peak fractions to a final concentration of 0.05%.
To remove the SUMO tag for crystallization, the His6SUMO-PDK4 protein was mixed with the TEV protease (25:1 mass ratio) and incubated at 4 °C for 2–3 days. The completeness of digestion was determined by SDS-PAGE. The digestion mixture was cycled through a Ni-NTA column several times. The flow-through and washes that contain tag-free PDK4 were combined, concentrated, and further purified on Superdex 200 column as described above. Tag-free PDK4 was concentrated to 110 mg/ml in 0.05% octyl β-d-glucopyranoside and stored at –80 °C in small aliquots.
Microcentrifuge-based Assay for Kinase Activity—The reaction mixture (25 μl in total volume) in a 0.5-ml microcentrifuge tube contained the following: 20 mm Tris-HCl (pH 7.5), 0.2 μm SUMO-PDK1–3 or 0.1 μm SUMO-PDK4, variable concentrations of the E1p heterotetramer (30 μm for PDK1 to 3 or 20 μm for PDK4), with or without 1.2 μm E2p/E3BP (based on the 60-meric core), or 100 μm L2 domain (based on the monomer), 10 mm KCl, 2 mm dithiothreitol, 5 mm MgCl2, and 25 μg of acetylated bovine serum albumin. The E1p, the L2 domain, and the E2p/E3BP core proteins were dialyzed against the 20 mm Tris-HCl buffer (pH 7.5) prior to addition to the reaction mixture. The concentration of the 60-meric E2p/E3BP core at 1.2 μm contained the 58 μm E1p-binding domain, based on the hypothetical subunit ratio of E2p/E3BP = 48:12 (43). Thus, all E1p heterotetramers would be immobilized on the E2p/E3BP core, when the latter was present in the reaction mixture. Following preincubation at 23 °C for 15 min, the phosphorylation reaction was initiated by adding [γ-32P]ATP (specific activity 200–300 cpm/pmol) to the reaction mixture to a final concentration of 0.5 mm. The reaction carried out at 23 °C was linear for up to 90 s and 0.4 μm PDK concentration (supplemental Fig. S1, A and B). At 1 min, the reaction was terminated by the addition of 150 μl of 20% trichloroacetic acid and 50 mm sodium pyrophosphate. Upon centrifugation, the pellet was washed three times with 10% trichloroacetic acid. Microcentrifuge tubes without the caps were transferred to 5-ml scintillation vials containing 3 ml of scintillation mixture, and radioactivity incorporated into E1p was counted. The specific activities of PDK isoforms were expressed as nanomoles of 32P incorporated/min/mg kinase (after subtracting the molecular mass of the SUMO moiety).
Binding Studies by Isothermal Titration Calorimetry—The isolated lipoyl domain (L1, L2, or L3) or the E2p/E3BP core, along with MBP-PDK3 or His6 tagged PDK4, were dialyzed exhaustively against a buffer containing 50 mm potassium phosphate (K-Pi) (pH 7.5) and 50 mm KCl. For measuring binding of the lipoyl domain to PDK4, the solution of 500 μm L1, L2, or L3 in a syringe was injected in 8-μl increments into the reaction cell containing 1.8 ml of 80 μm PDK4 (based on the monomer) at 20 °C in a VP-ITC microcalorimeter (MicroCal, Northampton, MA). For measuring PDK4 binding to the E2p/E3BP core, the syringe contained 270 μm E2p/E3BP core and the reaction cell 35 μm PDK4. For quantifying nucleotide binding to PDK isoforms, 320 μm Mg-ADP or the ATP analogue Mg-(AMP-PNP) in the syringe was injected into the reaction cell containing 20 μm SUMO-PDK3 or SUMO-PDK4 (based on monomers) at 15 °C. Binding constants (Kd) were calculated with the ORIGIN version 7.0 software (MicroCal). The concentrations for MBP-PDK3, SUMO-PDK3, His6 tagged PDK4, and SUMO-PDK4 were determined by absorbance at 280 nm using the calculated extinction coefficients (mg–1 ml ·cm–1) of 1.24, 0.89, 0.73, and 0.67, respectively.
Crystallization and Structural Determination—Crystals were obtained using the hanging-drop vapor-diffusion technique by mixing 2 μl of protein solution (10 mg/ml in 50 mm potassium phosphate (pH 7.5), 200 mm KCl, 5% glycerol, 20 mm BME, 5 mm ADP, and 5 mm MgCl2) and 2 μl of reservoir solution containing 1.8–1.9 m (NH4)2SO4, 2–4% polyethylene glycol (PEG)-400, 100 mm potassium phosphate, (pH 7.5). Crystals were flash-frozen in 2 m (NH4)2SO4, 4% PEG-400, 100 mm potassium phosphate (pH 7.5), 5 mm ADP, 5 mm MgCl2, and 20% glycerol. To grow crystals in HEPES buffer, a concentrated PDK4 solution (81 mg/ml) was diluted in a HEPES buffer containing 50 mm HEPES (pH 7.5), 200 mm KCl, 5% glycerol, and 20 mm BME. The final concentrations of PDK4 and phosphate ions were 5 mg/ml and 3 mm, respectively. This protein solution was mixed with an equal volume of reservoir solution containing 1.7 m (NH4)2SO4, 4% PEG-400, and 100 mm HEPES (pH 7.5). Crystals grown in the HEPES buffer were soaked and then flash-frozen in 2 m (NH4)2SO4, 4% PEG-400, 100 mm HEPES (pH 7.5), 5 mm ADP, 5 mm MgCl2, and 20% glycerol. Diffraction data were collected at the 19ID beamline in Structural Biology Center at the Advanced Photon Source (Argonne, IL). The data were processed with HKL2000 (50).
The PDK4 structures were solved by molecular replacement using the program Phaser (51) in the CCP4 package (52). A search template model for PDK4 was constructed based on the published PDK3 structure (PDB code 1Y8N) (23) using a homology-modeling technique in the Swiss-Model program (53). After rigid-body refinements with REF-MAC5 (54), the electron density map was improved using program DM (55). The Fo – Fc map unambiguously showed the electron density of the bound ADP. After modeling the adenine nucleotide, phosphate ion, magnesium ion, and water molecules were gradually added in the subsequent refinement cycles. Data processing and refinement statistics are summarized in Table 1. Molecular graphics for structural representations was rendered by PyMOL (DeLano Scientific LLC, Palo Alto, CA).
TABLE 1.
Data collection and refinement statistics
| PDK4-ADP (K-Pi) | PDK4-ADP (HEPES) | |
|---|---|---|
| PDB code | 2ZKJ | 3D2R |
| Data collectiona | ||
| Space group | P21 | P21 |
| Unit cell (Å) | ||
| a | 71.62 | 70.60 |
| b | 68.74 | 69.37 |
| c | 81.55 | 82.33 |
| β | 100.14 | 99.26 |
| Wavelength (Å) | 0.98 | 1.54 |
| Resolution (Å) | 2.0 | 2.03 |
| Unique reflections | 52,354 | 50,510 |
| Completeness (%) | 99.6 (100) | 99.5 (99.4) |
| R-merge (%)b | 5.6 (66.5) | 3.8 (25.5) |
| 〈I〉/〈σ(I)〉 | 23.4 (2.4) | 61.4 (4.7) |
| Multiplicity | 4.1 (4.2) | 3.6 (3.3) |
| Refinementa | ||
| No. of reflections (work/test) | 49,635/2668 | 47,925/2564 |
| No. of atoms (mean B value (Å2)) | ||
| Protein | 5737 (40.2) | 5756 (38.3) |
| Solvents | 225 (44.2) | 287 (40.5) |
| Nucleotides | 54 (30.0) | 54 (27.8) |
| Metal and phosphate ions | 12 (34.7) | 14 (46.4) |
| Missing residue regions | Chain A: 46-48, 182-188, 317-324, 388-392, 398-411; Chain B: 46-48, 147-148, 320-324, 387-393, 396-411 | Chain A: 21, 48, 146-147, 183-188, 317-324, 388-392, 398-411; Chain B: 318-325, 387-393, 398-411 |
| R-work (%)c | 18.8 (20.4) | 18.3 (19.5) |
| R-free (%)c | 24.3 (28.6) | 23.0 (25.7) |
| Root mean square deviation | ||
| Bond length (Å) | 0.020 | 0.020 |
| Bond angle (°) | 1.801 | 1.817 |
| Ramachandran plot | ||
| Most favored (%) | 94.9 | 94.3 |
| Allowed (%) | 5.1 | 5.7 |
| Disallowed (%) | 0 | 0 |
Values in parentheses refer to data in the highest resolution shell unless otherwise indicated.
R-merge = Σhkl Σj|Ij — 〈I〉|/Σhkl Σj Ij, where 〈I〉 is the mean intensity of j observations from a reflection hkl and its symmetry equivalents.
R-work = Σhkl||Fobs| — k|Fcalc||/Σhkl|Fobs|. R-free = R-work for 5% of reflections that were omitted from refinement.
Fluorescence Quenching Measurements—Ligand-induced release of the inter-subunit crossed tail conformation is indicated by the fluorescence quenching of Trp395 in the conserved DW-motif of PDK4 (56). Fluorescence spectra for PDK4 with or without ligands in a 3-ml quartz cuvette were recorded at 23 °C in a PTI (Photon Technology International) fluorescence spectrometer operated with the Felix 32 software. Excitation wavelength was set at 290 nm. Emission spectra were averaged from three scans conducted between 320 and 420 nm at a constant 0.49 μm protein concentration in 50 mm potassium phosphate buffer (pH 7.5), 0.2 mm EDTA, and 2 mm MgCl2. The slit width for both excitation and emission was set at 3 nm. Initial scans with PDK4 or PDK4 in the presence of 10 μm AZD7545 showed that fluorescence spectra were stable in 5 min. However, in the presence of 250 μm DCA, stable spectra were obtained only after 30–35 min. At this point, ADP aliquot from a 5 mm stock solution was added, and spectra were recorded immediately afterward. The fluorescence change at 350 nm was used to calculate fluorescence quenching. The L0.5 value, the ligand (ADP) concentration causing half-maximum fluorescence quenching, was calculated as described previously (56). The %Qmax value, the maximal quenching at saturating ADP concentration, was initially estimated by extrapolation using the double-reciprocal plot. The detailed data processing and curve-fitting procedures for %Qmax and L0.5 have also been described previously (56).
RESULTS AND DISCUSSION
Interaction of PDK4 with L1, L2, and L3 Domains and E2p/E3BP Core—The ability of PDK4 to bind the isolated L1, L2, and L3 domains was deciphered by isothermal titration calorimetry (ITC). Individual domains were titrated into the reaction cell containing either PDK3 or PDK4. Fig. 1 shows that PDK3 binds L2 with high affinity (Kd = 1.3 μm) with a subunit stoichiometry of PDK3/L2 = 1:0.75. This result serves as a positive control for the ITC measurements. Under identical conditions, no detectable enthalpy changes were detected when the same concentration range of L1 or L2 was titrated into PDK4. These results suggest that individual L1 and L2 domains are unable to bind to PDK4 with affinities high enough to be detected by ITC. Nevertheless, titration of PDK4 with the L3 domain shows small but detectable enthalpy changes (Fig. 1), which corresponds to a Kd value of 5 μm. The ITC data provide direct evidence to support the thesis that PDK4 binds the L3 domain stronger than L2 and L1 (18). Titration of the E2p/E3BP core with PDK4 produced an average Kd value of 18.8 μm with appreciable enthalpy changes, indicating weak binding also occurs between PDK4 and the E2p/E3BP core (data not shown).
FIGURE 1.
Measurements for binding of the three distinct lipoyl domains to PDK3 and PDK4 by isothermal titration calorimetry. For measuring binding of the lipoyl domain to PDK4, the solution containing 500 μm of L1, L2, or L3 in a syringe was injected in 8-μl increments into the reaction cell containing 1.8 ml of 80 μm PDK3 or PDK4 (based on the monomer) at 20 °C in a VP-ITC microcalorimeter. The L3 domain binds to PDK4 weakly with a Kd of 5 μm.
Robust Core-free Basal Activity of PDK4—Activities of PDK isoforms vary with the buffers used with the highest levels of PDK2 or PDK3 obtained with the Tris-HCl, over the K+-phosphate, MOPS-K+, and Tris-HEPES buffers (17). The phosphate and potassium ions were shown to be inhibitory to PDK2 (47, 57, 58) and PDK4 activities3 (59). To assess optimal PDK isoform activities, we recently developed a microcentrifuge-based assay in Tris-HCl buffer (pH 7.5) in the absence of phosphate ions. The assay mixture in 0.5-ml microcentrifuge tubes contains 10 mm KCl, which is required for nucleotide and L2 binding (22, 47, 58), and shows only slight inhibition of basal PDK activities. The microcentrifuge-based method offers robust assay efficiency, which greatly facilitated enzyme kinetic studies. The levels of 32P incorporation into E1p obtained with the microcentrifuge-based assay are in good agreement with those determined by the SDS-PAGE-based assays described in the previous studies from this laboratory (23, 45).
Using the microcentrifuge-based assay, activities for the four His6-SUMO tagged PDK isoforms were measured in the absence (basal activity) and presence of the lipoyl domain or the E2p/E3BP core. The presence of the His6-SUMO tag enhances solubility of all PDK isoforms without affecting kinase activity. At saturating 0.5 mm ATP concentration (13, 17, 31), E1p levels were maintained at 30 μm for PDK1–3 and 20 μm for PDK4. These E1p concentrations are 5–10-fold higher than the Km values of these PDK isoforms for the E1p substrate (data not shown). When the E1p concentration becomes higher than 30 μm, PDK2 and PDK4 basal activities slightly decline,3 consistent with substrate inhibition (59). Under these assay conditions, PDK4 shows the highest basal activity (690 ± 60 nmol of 32P incorporation/min/mg kinase) among the four PDK isoforms, including the L2- and E2p/E3BP-stimulated activities of PDK1, PDK2, and PDK3 (Fig. 2). In the presence of the L2 domain at a saturating concentration of 100 μm, PDK4 basal activity is slightly stimulated (1.1-fold) to 720 ± 60 nmol/min/mg. No discernible change of PDK4 activity was observed (670 ± 50 nmol/min/mg), when assayed in the presence of 1.2 μm E2p/E3BP core. PDK3 shows a basal activity of 60 ± 0.5 nmol/min/mg, which is stimulated by the L2 domain and the E2p/E3BP core by 3.3- and 3.7-fold, respectively, to 210 ± 2 and 230 ± 20 nmol/min/mg. PDK2 with a basal activity of 270 ± 10 nmol/min/mg was stimulated by 1.4-fold by the L2 domain and 1.9-fold by the E2p/E3BP core, respectively, to 370 ± 60 and 500 ± 50 nmol/min/mg. Under similar assay conditions, PDK1 exhibits basal activity at 190 ± 30 nmol/min/mg, which is activated by the L2 domain by 1.2-fold (230 ± 10 nmol/min/mg). PDK1 activity is not significantly stimulated when assayed in the presence of the E2p/E3BP core.
FIGURE 2.
Robust core-free PDK4 activity in comparison with activities of other PDK isoforms. The reaction mixture in 0.5-ml microcentrifuge tubes contained 20 mm Tris-HCl (pH 7.5), 10 mm KCl, 2 mm dithiothreitol, 2 mm MgCl2, and 30 μm E1p (based on the tetramer) for PDK1, PDK2, and PDK3 or 20 μm for PDK4, 0.2 μm SUMO-PDK isoform (1–3) (based on the dimer) or 0.1 μm SUMO-PDK4, with and without 1.2 μm E2p/E3BP (based on the 60-meric core) or 100 μm L2 domain (based on the monomer). The phosphorylation reaction was initiated by the addition of 0.5 mm [γ-32P]ATP (specific activity 200–300 cpm/pmol). The reaction was allowed to proceed for 1 min at 23 °C and stopped by the addition of 20% trichloroacetic acid and sodium pyrophosphate. Following centrifugation and washing with 10% trichloroacetic acid, 32P incorporation into the E1p α-subunit was determined by scintillation counting. Specific activities of PDK isoforms were expressed as nanomoles of 32P incorporated/min/mg kinase (after subtracting the mass of the SUMO moiety). The bars represent averages ± S.D. (n = 3–5).
The above results show that PDK4 possesses robust basal activity in the absence of the E2p/E3BP core. The higher core-free PDK4 activity compared with the E2p/E3BP-stimulated activities of other PDK isoforms was also observed, when the MOPS-K+ buffer was used (data not shown). The slight activation of PDK4 basal activity by the L2 domain is of interest in light of the weak interactions. PDK4 is unstable at dilute concentrations3 (59), and the presence of the L2 domain may play a stabilizing role during the assay, resulting in the modest elevation of kinase activity. However, it is perplexing that the L3 domain with higher affinity for PDK4 than the L1 and L2 domains shows slight inhibition of PDK4 activity,3 similar to that reported previously (59). The 3.3- and 3.7-fold stimulation of PDK3 basal activity by L2 and E2p/E3BP, respectively, is significantly lower than that we previously reported (23, 41, 45). In our earlier studies, up to 50 mm KCl and/or 20 mm potassium phosphate were present in the assay mixtures. The presence of high concentrations of the potassium and/or phosphate ions markedly inhibit basal PDK activity (57, 58). In this study, the higher PDK3 basal activity than previously obtained (41) may account for the reduced fold activation of PDK3 by L2 and the E2p/E3BP core. The non-concordance in fold activation of PDK2 and PDK3 by L2 and E2p/E3BP between the present and previous studies from other laboratories (16, 17) may also be explained by enzyme instability and the different assay conditions used. In this study, significantly higher E1p and lower potassium ion concentrations were employed to measure activities of various PDK isoforms.
A picture emerges from the above activity data, i.e. PDK1 and PDK4 are similar in that their basal activities are not significantly stimulated by either the isolated L2 domain or the E2p/E3BP core. In contrast, PDK2 and PDK3 basal activities are markedly enhanced by the E2p/E3BP core. It is of interest that PDK3 activity, but not PDK2, is significantly elevated in the presence of the L2 domain. These results may indicate that the augmentation of PDK3 activity by the E2p/E3BP core results largely from allosteric activation upon binding to the L2 domain. On the other hand, the stimulation of PDK2 activity by the E2p/E3BP core is mainly achieved through co-localization of the kinase and E1p substrate to the PDC core. The small enhancement of the PDK2 activity by the L2 domain, which was not observed in a previous study (17), may suggest that the allosteric activation of PDK2 by the L2 domain is a minor mechanism.
Crystal Structures of Human PDK4—Structural determination was undertaken to offer insight into the robust core-free basal activity of PDK4. Crystals of human PDK4 were grown with Mg-ADP either in the K-Pi or the HEPES buffer. PDK4 structures containing bound ADP were solved at 2.0-Å resolution by molecular replacement using the human PDK3 structure (PDB code 1Y8N) (23) as a template (Fig. 2A and Table 1). The two PDK4 structures determined with crystals produced in the K-Pi and HEPES buffer are virtually identical. Thus, unless indicated otherwise, only the PDK4 structure derived from crystals grown in the K-Pi buffer is described. The PDK4 monomer structure consists of the N-terminal and C-terminal domains, with the chain-fold of each domain similar to the corresponding domains in the known structure of other PDK isoforms. The root mean square deviation of the overall structure of PDK4 from other isoforms, i.e. human PDK1 (PDB code 2Q8F) (41), human PDK2 (PDB code 2BTZ) (42), and human PDK3, is less than 1.2 Å for 326–336 Cα atoms in PDK4. The asymmetric unit of the PDK4 crystal contains one dimer. The C-terminal tails from both subunits are partially ordered in the regions harboring invariant Asp394 and Trp395, which we designate the DW-motif. The DW-motif from one subunit interacts with the N-terminal domain of the other subunit, resulting in an apparent cross-tail configuration in the PDK4 dimer (Fig. 3A), reminiscent of the fully ordered cross-tails present in the PDK3-L2-ADP structure (23). The partially ordered C-terminal cross-tails are absent in a comparable structure of human PDK4 harboring a bound nonhydrolyzable ATP analogue AMP-PNP recently deposited in the PDB (accession code 2E0A). The basis for the differences between these two PDK4 structures is not known. The partially ordered C-terminal cross-tail configuration, similar to that in the present PDK4-ADP structure, is also observed in apo-PDK1 (41) and apo-PDK2 (42) as well as in apo-PDK3.4 The cross-tail conformations in the latter structures are derived from crystals grown in the absence of ammonium sulfate (41, 42). Thus, the partially ordered cross-tail conformation in the PDK4-ADP structure is not an artifact of ammonium sulfate included in the crystallization well solution, which may have enhanced hydrophobic interactions involving tryptophan residues in the DW-motif.
FIGURE 3.
The 2.0-Å crystal structure of human PDK4. A, overall structure of PDK4 dimer derived from crystals grown in the K-Pi buffer. Subunits A and B are colored in green and brown, respectively. The DW-motifs and bound ADP + PO2–4 ion are shown as ball-and-stick models. The ATP lid is shown in yellow. Gray triangles indicate the locations of the active-site clefts between the N-terminal and C-terminal domains. B, refined electron (2Fo – Fc) density of the DW-motif of subunit A (brown) bound to the N-terminal domain of subunit B (green). The residues in the subunit A and subunit B are shown as ball-and-stick and stick models, respectively. The density is contoured at 1-σ level. Dashed lines indicate disordered regions flanking the DW-motif. C, superimposition of the subunits in PDK4 dimer. Subunit A (brown) is superimposed onto subunit B (green) based on the residues in the C-terminal domain. The bound ADP and PO2–4 ion are shown as stick models. Gray triangle indicates the entrance of the wedge-shaped active-site cleft. D, refined electron (2Fo – Fc) density of the bound ADP and PO2–4 ion. The density is contoured at a 1-σ level. The triphosphate part of ATP bound to PDK3 in complex with L2 is superimposed and shown as a transparent stick model.
The structures of both subunits in the PDK4 dimer are virtually identical. However, small but significant differences were observed. First, the extent of the ordered region harboring the DW-motif was different between the two subunits. The region between Asp393 and Leu397 is visible in subunit A (Fig. 3B), whereas a shorter segment from residues Asp394′ to Trp395′ is observed in subunit B (supplemental Fig. S1A). Second, the conformation of the linker region between helices α7 and α8 in the N-terminal domain is different between the two subunits (Fig. 3C). The linker in subunit B is longer than that of subunit A because of an unwinding of the first turn of helix α8 and protrudes into the nucleotide-binding site in the C-terminal domain. As a result, the linker interacts with the ATP lid through van der Waals contacts. These interactions appear to result in a more ordered ATP lid in subunit B (Fig. 3C). The significance of this interaction is unknown. However, the PDK inhibitor Pfz3 developed by Pfizer binds to the pocket formed between helices α4/α5 and helices α7/α8, the latter being adjacent to the above linker region (42). The latter result suggests that the vicinity of the linker region may be critical for kinase activity.
Both PDK4 subunits show good densities for bound ADP in the nucleotide-binding sites. Interestingly, an inorganic phosphate ion (PO2–4) is trapped near the β-phosphate of the bound ADP in both subunits (Fig. 3D). When PDK4 was crystallized in the HEPES buffer with a low phosphate concentration, the corresponding density for the PO2–4 ion is absent from the structure (supplemental Fig. S1B). Instead, water molecules cluster in the same vicinity and form a hydrogen-bond network with a magnesium ion, the β-phosphate of the bound ADP and the ATP lid. This result indicates that the additional density in the PDK4 structure deduced from crystals grown in the K-Pi buffer is a phosphate ion and not a sulfate used as a precipitant under the crystallization conditions. When the PDK4 structure is superimposed onto that of PDK3-L2-ATP (23), the position of the PO2–4 ion is very close to that of the γ-phosphate of the bound ATP in the PDK3-L2-ATP structure (23). Therefore, the hydrogen-bonding interactions of the exogenous PO2–4 ion with the ATP lid and the vicinity are similar to those of the γ-phosphate in the PDK3-L2-ATP structure. Moreover, the ATP lid in the present PDK4-ADP structure is ordered to an extent similar to that in the PDK4-(AMP-PNP) structure (2E0A) deposited in the PDB.
Open Conformation with Widened Active-site Cleft in the PDK4-ADP Structure—The structure of human PDK4-ADP is compared with those of human PDK3-L2-ADP (23), rat PDK2-L2-(AMP-PNP) (47), and rat PDK2-ADP (21) (Fig. 4). The C-terminal domains of these structures are superimposed, with the N-terminal domains omitted from the calculation. In the structures of PDK3-L2-ADP and PDK2-L2-(AMP-PNP), PDK3 and PDK2 exist in the open conformation with a wider active-site cleft than that present in the PDK2-ADP structure, which assumes the closed conformation. We have shown previously that L2 binding imparts the C-terminal cross-tail conformation in the PDK3 dimer, resulting in a widening of the active-site cleft (23). On the other hand, the C-terminal tail in the PDK2-ADP structure is completely disordered, which is associated with the closed conformation. Significantly, PDK4-ADP assumes the open conformation in the active-site cleft, when comparing the positions of helices α6 and α7 in the N-terminal domain with those in the structures of the PDK3-L2-ADP and PDK2-L2-(AMP-PNP) complexes (Fig. 4). Moreover, the open conformation in the current PDK4-ADP structure coincides with the presence of the partially ordered C-terminal cross-tails, similar to those observed in the nucleotide-free structures of both apo-PDK1 (41) and apo-PDK2 (42).
FIGURE 4.
Superimpositions of the open and the closed conformations present in different PDK isoforms. The structures of rat PDK2-ADP (pink) (PDB code 1JM6), rat PDK2-L2-(AMP-PNP) (blue) (3CRL), and human PDK3-L2-ADP (yellow) (1Y8O) are superimposed on the present human PDK4-ADP structure (green) based on the residues in the C-terminal domains. The bound ADP + PO2–4 ion in the PDK4-ADP structure is shown as stick model. The gray triangle indicates the entry of the active-site cleft between the N-terminal and the C-terminal domains. The disordered region of the ATP lid in PDK4 is shown as dotted line.
We reported previously that the ATP lid is completely disordered in the open conformation of the PDK3-L2-ADP structure, whereas the same lid is largely ordered in the PDK3-L2-ATP structure (23). The nucleotide-induced conformational changes in the ATP lid account for the weaker binding affinity of PDK3 for ADP than ATP, as measured by ITC. The differential nucleotide-binding affinities likely facilitate an ADP/ATP exchange, removing the product inhibition imparted by ADP (30, 60). In variance with the PDK3-L2-ADP structure, partially ordered ATP lids are present in the PDK4-ADP structures obtained with crystals grown in both the K-Pi and HEPES buffer, despite the fact that both PDK4 structures are in the open conformation (Fig. 4). These results suggest that the ATP lid in PDK4 is intrinsically more stable than the equivalent in PDK3.
Reduced Affinities of PDK4 for Adenine Nucleotide Binding—We showed previously that L2 domain-induced transition from the closed to the open conformation of PDK3 is accompanied by the significant reduction of affinity of PDK3 for adenine nucleotides ATP and ADP (23). As described above, the dihydrolipoamide mimetic AZD7545 (61) binds to the lipoyl domain-binding pocket of PDK1, fostering an open conformation in the PDK1-AZD7545 structure (41). AZD7545 robustly augments basal PDK3 activity to a level similar to that obtained with the PDK3-L2 complex (41). The result supports the notion that the PDK3-AZD7545 complex also assumes the open conformation, although the structure of the PDK3-AZD7545 complex remains to be deciphered. Despite its very low affinity for the L2 domain (Fig. 1), PDK4 binds AZD7545 with robust affinity (Kd = 48 nm).5 Thus, to probe the active-site cleft conformation in PDK4, affinities of SUMO-PDK3 and SUMO-PDK4 for adenine nucleotides ADP and the ATP analogue AMP-PNP were determined by ITC in the presence and absence of AZD7545. In the absence of AZD7545, apo-PDK3 is in the closed conformation and shows Kd values of 0.7 and 1.4 μm for ADP and AMP-PNP, respectively (Table 2). When PDK3 is in the AZD7545-induced open conformation, affinities of PDK3-AZD7545 for ADP and AMP-PNP are significantly decreased, analogous to those exhibited by the PDK3-L2 complex (23), to Kd values of 5.6 and 3.9 μm for ADP and AMP-PNP, respectively. In parallel, PDK4 exhibits a Kd value of 3.3 μm in the absence of AZD7545, which is comparable with those obtained with PDK3-AZD7545 or the PDK3-L2 complex in the open conformations (Table 2). The addition of AZD7545 results in a slight but noticeable increase in Kd value for ADP to 5.9 μm. Increases in Kd values of PDK4 for AMP-PNP are also observed when AZD7545 is added. These results suggest that apo-PDK4, unlike apo-PDK3, is in a metastable open conformation with reduced affinities for nucleotides. The presence of bound AZD7545 further enhances the DW-motif anchoring, effectively locking PDK4 in the stable open conformation. The reduced affinity of PDK4 for ADP facilitates the removal of product inhibition by this nucleotide, resulting in augmented kinase activity, similar to that observed in the PDK3-L2 complex.
TABLE 2.
Binding affinities of PDK3 and PDK4 for ATP/ADP in the closed and open conformations
For ITC measurements, 0.32 mm Mg-ADP or Mg-(AMP-PNP) in the syringe was injected into the reaction cell containing 20 μm SUMO-PDK3 or SUMO-PDK4 (based on monomers) at 15 °C in the absence and presence of 50 μm AZD7545. One-site binding model was used to fit the binding isotherms. Values are expressed as average ± S.D. (n = 3).
| Kinase | Nucleotide | AZD7545 | Kd value | Conformation |
|---|---|---|---|---|
| mm | ||||
| PDK3 | ADP | - | 0.7 ± 0.2 | Closed |
| AMP-PNP | - | 1.4 ± 0.4 | Closed | |
| ADP | + | 5.6 ± 0.2 | Open | |
| AMP-PNP | + | 3.9 ± 0.4 | Open | |
| PDK4 | ADP | - | 3.3 ± 0.7 | Open |
| AMP-PNP | - | 7.2 ± 0.9 | Open | |
| ADP | + | 5.9 ± 0.9 | Open | |
| AMP-PNP | + | 10 ± 0.4 | Open |
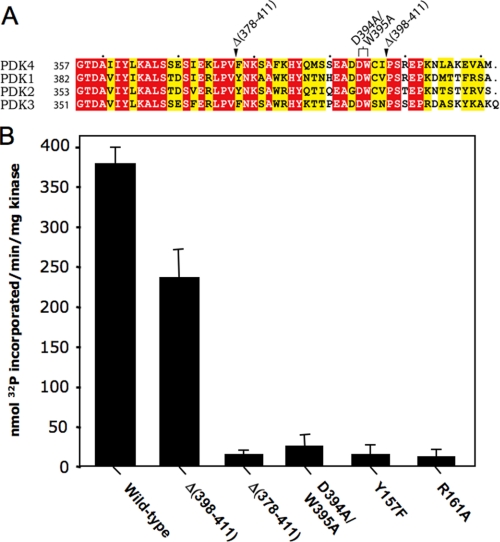
The DW-motif Anchor Is Indispensable for PDK4 Activity—The DW-motif in the C-terminal tail region is conserved in all four PDK isoforms (Fig. 5A). As described above, side chains of the DW-motif (Asp394 and Trp395) from subunit A interact with those of Tyr157′ and Arg161′ in the N-terminal domain of subunit B (Fig. 3B). To dissect the significance of the DW-motif for PDK4 activity, C-terminal truncations and site-directed mutagenesis of the DW-motif were carried out; kinase activities of these PDK4 mutants were assayed. As shown in Fig. 5B, the deletion of the C-terminal tail downstream of the DW-motif, Δ(398–411), has no major effect on PDK4 activity. The Δ(378–411) variant with a deletion, including the DW-motif, shows very low residual kinase activity. More importantly, only a small amount (6% of wild type) of kinase activity is detected in the D394A/W395A double mutant of the DW-motif itself. Similarly, low residual activities are present in the reciprocal DW-motif mutants of Y157F and R161A. These data provide, for the first time, direct evidence that the DW-motif anchor is indispensable for PDK4 activity.
FIGURE 5.
The conserved DW-motif is indispensable for PDK4 activity. A, conservation of the DW-motif in the C-terminal region of PDK isoforms. Sequence alignments of human PDK isoforms were carried out with ClustalW (65) and drawn by ESPript (66). Conserved residues are colored in red and similar residues in yellow. B, kinase activities in wild type, C-terminally truncated variants, the DW-motif-modified (D394A/W395A), and the reciprocal DW-motif (Y157F and R161A) mutants of PDK4. Kinase activity was assayed as described under “Experimental Procedures.” Sequences truncated in the deletion mutants Δ378–411 and Δ398–411 are indicated in A.
The present results with the deletion mutants confirm our earlier studies with PDK3, where truncations of the C-terminal sequence encompassing the DW-motif nullify PDK3 activity (45). In contrast, deletions of the C-terminal sequence harboring the DW-motif in rat PDK2 still retain significant residual kinase activity (62). The reason for the disagreements between results obtained with PDK2 and those with PDK3 and PDK4 is not clear.
Presence of Metastable C-terminal Partial Cross-tails in PDK4—Tryptophan fluorescence quenching has been used as an indicator for the ligand-induced release of the inter-subunit cross-tail in PDK2 (56). Trp383 (mature sequence numbering) in the DW-motif also plays a critical and essential anchoring role for PDK2, and the release of the DW-motif by ADP and/or the PDK inhibitor DCA results in the quenching of the tryptophan fluorescence largely from Trp383 (56). PDK4 contains two tryptophan residues, Trp91 and Trp395, with the latter located in the conserved DW-motif. To ascertain whether Trp395 is directly responsible for ligand-induced fluorescence quenching, emission spectra of wild type and D394A/W395A mutant PDK4 were determined. In the presence of 0.5 mm DCA and 0.05 mm ADP, large tryptophan fluorescence quenching occurs in wild-type PDK4 with a blue shift of the peak fluorescence intensity from 347 to 340 nm (Fig. 6A). Intermediate fluorescence quenching and blue shift was observed when 0.5 mm DCA alone is present. In contrast, with the D394A/W395A mutant devoid of Trp395, only slight fluorescence quenching is obtained without a blue shift when DCA or DCA/ADP is present. In the PDK1-DCA structure, the bound DCA dislocates the side chain of a histidine residue by more than 6 Å toward the outside of the helical bundle (41). In PDK4, this side-chain movement would bring the corresponding His127 in contact with Trp91 situated on the roof of the DCA-binding pocket. The interaction likely accounts for the rather small but detectable quenching of fluorescence from Trp91, as is the case suggested for PDK2 (56). Taken together, these results indicate that Trp395 in the DW-motif is largely responsible for DCA- and ADP-induced fluorescence quenching and reports the release of the DW-motif anchor to the bulk solvent.
FIGURE 6.
Tryptophan fluorescence quenching of the DW-motif in PDK4. Tryptophan fluorescence from the DW-motif was recorded in a Photon Technology International (Birmingham, NJ) spectrofluorometer, with excitation wavelength set at 290 nm and emission spectra recorded between 320 and 420 nm. Initial protein concentrations (based on dimers) were 0.49 μm for His6-tagged PDK4 in 50 mm phosphate buffer (pH 7.5) containing 2 mm MgCl2 and 0.2 mm EDTA, in the absence and presence of 0.25 mm DCA or 10 μm compound AZD7545. The % quenching is equal to 100 × (Fmax – Fq)/Fmax, where Fmax is the maximum fluorescence intensity recorded before titration with increasing concentrations of ADP, and Fq is the fluorescence intensity recorded after each addition of ADP. A, scans showing effects of ADP and/or DCA on the tryptophan fluorescence of wild type and D394A/W395A PDK4 between 320 and 420 nm wavelengths. Curves 1, PDK4; 2, PDK4 + 0.5 mm DCA; 3, PDK4 + 0.5 mm DCA + 50 μm ADP; 4, D394A/W395A PDK4; 5, D394A/W395A PDK4 + 0.5 mm DCA; 6, D394A/W395A PDK4 + 0.5 mm DCA + 50 μm ADP. B, % fluorescence quenching of PDK4 as a function of increasing ADP concentrations in the absence and presence of DCA or AZD7545. •, no addition; ▪, + 0.25 mm DCA; ▴, + 10 μm AZD7545.
Fig. 6B shows that Trp395 fluorescence in the partially ordered C-terminal cross-tail region of PDK4 is significantly quenched when titrated with increasing concentrations of ADP, in the absence and presence of 0.25 mm DCA or 10 μm AZD7545. Fluorescence quenching measurements were carried out in potassium phosphate buffer to enhance ADP binding. Curve fitting as described under “Experimental Procedures” produce an L0.5 value of 20 μm for ADP alone (Table 3). The ADP-induced fluorescence quenching reflects the displacement of the DW-motif (56), which exists in equilibrium between the closed (21) and open conformations (21, 23). The reduction of the kcat value from 35.1 min–1 in the absence of DCA to 2.6 min–1 in the presence of 0.25 mm DCA supports the presence of the mostly inactive closed conformation induced by this PDK inhibitor. The L0.5 value of 2.3 μm for ADP in the presence of DCA is an order of magnitude lower than that in the absence of the inhibitor (20 μm) (Table 3). The results strongly suggest that interactions involving the DW-motif anchor are weaker in the presence of DCA, leading to the weakening or absence of the cross-tail conformation, as observed in the PDK2-DCA-ADP structure (42). Notably, when 10 μm AZD7545 is present, the L0.5 value of PDK4 for ADP is elevated to 44 μm (Table 3). The PDK4-AZD7545 complex shows a higher kcat value (98.3 min–1) than PDK4 alone (35.1 min–1), in concordance with the presence of an active open conformation. The fluorescence quenching results support the thesis that the AZD7545 locks PDK4 in the stable open conformation by enhancing interactions involving the DW-motif anchor.
TABLE 3.
Fluorescence quenching measurements and enzyme kinetic parameters of PDK4 in the absence and presence of DCA and AZD7545
For tryptophan fluorescence, quenching by ADP of untagged PDK4 was carried out at 23 °C in the absence and presence of 0.25 mm DCA or 10 μm AZD7545. The L0.5 value, the ligand (ADP) concentration causing half-maximum fluorescence quenching, was estimated from the equation: L0.5 = (Lf)·(%Qmax – %Q)/(%Q), where %Q = 100%·(F0 – Fq)/F0, where F0 and Fq are the fluorescence at 350 nm in the absence of ADP and after each addition of the ADP aliquot, respectively; Lf is the free ligand concentration; %Qmax is the maximal quenching at saturating ADP concentration initially estimated by extrapolation using the double-reciprocal plot (56). The L0.5 and Qmax values are expressed as averages ± S.D. (n = 3). The turnover number, kcat, was obtained at 0.5 mm ATP with increasing E1P concentrations (0.25–40 μm). The kcat values are averages of triplicates.
| Addition | L0.5 | Qmax | kcat |
|---|---|---|---|
| μm | % | min-1 | |
| None | 20 ± 3 | 48 ± 4 | 35.1 |
| DCA | 2.3 ± 0.2 | 31 ± 3 | 2.6 |
| AZD7545 | 44 ± 6 | 24 ± 2 | 98.3 |
Physiological Significance of the Metastable Open Conformation in PDK4—We previously proposed that PDK3 exist in the classic T ↔ R equilibrium (63), with the T (tight) state and the R (relaxed) state corresponding to the basal closed and the active open conformations, respectively (Fig. 7) (23). The closed conformation with clamped active-site clefts and disordered C-terminal cross-tails was originally described in the rat PDK2-ADP structure (21). Subsequently, nucleotide-free apo-PDK2 (42) and apo-PDK1 (41) were shown to possess open active-site clefts with partially ordered C-terminal cross-tails containing the conserved DW-motif. The intermediate-open conformation is termed the R′ state and exists in the T ↔ R′ equilibrium. It is conceivable that apo-PDK isoforms exist in T ↔ R′ equilibrium, and the presence of bound ADP/ATP and/or DCA favors the T state. The human apo-PDK2 crystals when soaked with ADP and DCA shows disordered C-terminal cross-tails consistent with the closed conformation or the T state (42). However, the active-site cleft in the PDK2-ADP-DCA structure (PDB code 2BU8) remains in the open conformation, presumably because of crystal packing restrictions. In the T ↔ R′ equilibrium, the product ADP accumulated in a given phosphorylation cycle shifts the equilibrium toward the closed conformation or the T state, resulting in low basal activity. AZD7545 is able to robustly stimulate PDK3 (41) and PDK4 (this study) activities by locking these kinase isoforms in the R′ state. Similarly, by reference to results obtained with PDK2 (56), the isolated L2 domain binds to the R′ state, but not the T state, of PDK2 or PDK3 and locks the kinase in the open conformation or the R state. In the case of PDK3, interactions between the L2 protein moiety and the N-terminal domain further order the segment downstream of the DW-motif, resulting in the complete C-terminal tail conformation (41). In this study, we showed that PDK4 is present in a metastable intermediate open conformation or the active R′ state. The metastable interactions involving the DW-motif imparts the active R′ state despite the presence of bound ADP, as depicted in the PDK4-ADP structure. The favored R′ state in the T ↔ R′ equilibrium allows PDK4 to remove ADP efficiently without significantly reverting to the T state. The net result is robust kinase activity in the absence of the E2p/E3BP core.
FIGURE 7.
The metastable open conformation (the R ′ state) of PDK4 in equilibrium between the T state and the R state. The closed conformation (the T state) with (i) disordered C-terminal tails and (ii) the closed active-site clefts are observed in the rat PDK2-ADP structure (PDB code 1JM6). The active intermediate open conformation (the R′ state) with partially ordered cross-tails and the open active-site clefts is present in human apo-PDK2 (2BTZ), apo-PDK1 (2Q8F), and the present PDK4-ADP structures. The active open conformation (the R state) with fully ordered cross-tails and the open active-site clefts is present in the human apo-PDK3-L2 (1Y8N), PDK3-L2-ADP (1Y8O), PDK3-L2-ATP (1Y8P), PDK2-L2 (3CRK), and PDK2-L2-(AMP-PNP) (3CRL) structures. Only representative structures are shown in the figure. The presence of adenine nucleotides ADP/ATP and/or DCA/pyruvate shifts the T ↔ R′ equilibrium toward the T state (42). AZD7545 binds to PDK3 tightly (Kd = 5.5 nm) (see Footnote 5) and secures PDK3 in the active R′ state (41). The L2 domain binds to apo-PDK3 in the R′ state and maintains PDK3 in the fully active R state (23). The data suggest that a metastable open conformation is favored in PDK4 in equilibrium between the T and R′ states, resulting in robust basal activity independent of the lipoyl domain or the E2p/E3BP core. N, the N-terminal domain; C, the C-terminal domain; Circled N, the band nucleotide; Red dot, the conserved DW motif.
The hyperactivity of a core-free PDK4 may have severe pathological consequences in type 2 diabetes. It allows the overexpressed core-free PDK4 to efficiently phosphorylate both the unbound and E2p/E3BP-bound E1p substrate without the physical constraints imposed by the PDC core. This mechanism promotes the robust short term inactivation of PDC, resulting in impaired glucose utilization in the patients (7, 18). In contrast, PDK2 and PDK3 require the co-localization of these kinases with the E1p substrate on the same E2p/E3BP core for optimal kinase activity (5). On the core, the PDK dimer and the E1p heterotetramer bind to the L2 domain and the adjacent downstream E1BD domain, respectively, on the E2p subunit. Therefore, only one of the two E1p active sites facing the kinase may be efficiently phosphorylated, resulting in half-site phosphorylation, analogous to the related E1b component of the branched-chain α-ketoacid dehydrogenase complex (64).
The presence of the metastable open conformation in PDK4 corroborated in this study offers a potential target for structure-based inhibitor designs for a new generation of PDK inhibitors. It may be feasible to develop novel high affinity peptidomimetic inhibitors that specifically bind to the N-terminal domain region interacting with the DW-motif, which prevents the formation of the C-terminal cross-tails in this PDK isoform. A PDK4-peptidomimetic complex locked in the closed conformation would in turn reduce PDK4 activity and enhance the PDC flux. This strategy, if successful, would reinvigorate glucose oxidation in disease states such as obesity and type 2 diabetes.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant DK62306. This work was also supported by Grant I-1286 from the Welch Foundation. The costs of publication of this article were defrayed in part by the payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. Section 1734 solely to indicate this fact.
The atomic coordinates and structure factors (codes 2ZKJ and 3D2R) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1 and S2.
Footnotes
The abbreviations used are: PDC, pyruvate dehydrogenase complex; PDK, pyruvate dehydrogenase kinase; E1p, pyruvate dehydrogenase; E3, dihydrolipoamide dehydrogenase; E2p/E3BP, dihydrolipoyl transacetylase/E3-binding protein; L2, inner lipoyl domain of E2p; L3, lipoyl domain of E3BP; SUMO, small ubiquitin modifier, MBP, maltose-binding protein; BME, β-mercaptoethanol; ITC, isothermal titration calorimetry; TEV, tobacco-etch virus; MOPS, 3-(N-morpholino)propanesulfonic acid; DCA, dichloroacetic acid; AZD7545, 2R-N-{4-[4-(dimethylcarbamoyl)phenylsulfonyl]-2-chlorophenyl}-3,3,3-trifluoro-2-hydroxy-2-methylpropanamide; AMP-PNP, 5′-adenylylimidodiphosphate; PDB, Protein Data Bank; Ni-NTA, nickel-nitrilotriacetic acid.
J. L. Chuang, unpublished results.
M. Kato, unpublished results.
S.-C. Tso and R. M. Wynn, unpublished results.
References
- 1.Reed, L. J. (2001) J. Biol. Chem. 27638329–38336 [DOI] [PubMed] [Google Scholar]
- 2.Yu, X., Hiromasa, Y., Tsen, H., Stoops, J. K., Roche, T. E., and Zhou, Z. H. (2008) Structure (Lond.) 16104–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Harris, R. A., Bowker-Kinley, M. M., Wu, P., Jeng, J., and Popov, K. M. (1997) J. Biol. Chem. 27219746–19751 [DOI] [PubMed] [Google Scholar]
- 4.Patel, M. S., and Roche, T. E. (1990) FASEB J. 43224–3233 [DOI] [PubMed] [Google Scholar]
- 5.Liu, S., Baker, J. C., and Roche, T. E. (1995) J. Biol. Chem. 270793–800 [DOI] [PubMed] [Google Scholar]
- 6.Harris, R. A., Huang, B., and Wu, P. (2001) Adv. Enzyme Regul. 41269–288 [DOI] [PubMed] [Google Scholar]
- 7.Holness, M. J., and Sugden, M. C. (2003) Biochem. Soc. Trans. 311143–1151 [DOI] [PubMed] [Google Scholar]
- 8.Popov, K. M., Hawes, J. W., and Harris, R. A. (1997) Adv. Second Messenger Phosphoprotein Res. 31105–111 [PubMed] [Google Scholar]
- 9.Yeaman, S. J., Hutcheson, E. T., Roche, T. E., Pettit, F. H., Brown, J. R., Reed, L. J., Watson, D. C., and Dixon, G. H. (1978) Biochemistry 172364–2370 [DOI] [PubMed] [Google Scholar]
- 10.Teague, W. M., Pettit, F. H., Yeaman, S. J., and Reed, L. J. (1979) Biochem. Biophys. Res. Commun. 87244–252 [DOI] [PubMed] [Google Scholar]
- 11.Sale, G. J., and Randle, P. J. (1981) Biochem. J. 193935–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Korotchkina, L. G., and Patel, M. S. (1995) J. Biol. Chem. 27014297–14304 [DOI] [PubMed] [Google Scholar]
- 13.Korotchkina, L. G., and Patel, M. S. (2001) J. Biol. Chem. 2765731–5738 [DOI] [PubMed] [Google Scholar]
- 14.Kolobova, E., Tuganova, A., Boulatnikov, I., and Popov, K. M. (2001) Biochem. J. 35869–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Radke, G. A., Ono, K., Ravindran, S., and Roche, T. E. (1993) Biochem. Biophys. Res. Commun. 190982–991 [DOI] [PubMed] [Google Scholar]
- 16.Tuganova, A., Boulatnikov, I., and Popov, K. M. (2002) Biochem. J. 366129–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker, J. C., Yan, X., Peng, T., Kasten, S., and Roche, T. E. (2000) J. Biol. Chem. 27515773–15781 [DOI] [PubMed] [Google Scholar]
- 18.Roche, T. E., Hiromasa, Y., Turkan, A., Gong, X., Peng, T., Yan, X., Kasten, S. A., Bao, H., and Dong, J. (2003) Eur. J. Biochem. 2701050–1056 [DOI] [PubMed] [Google Scholar]
- 19.Roche, T. E., and Hiromasa, Y. (2007) Cell. Mol. Life Sci. 64830–849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hanks, S. K., Quinn, A. M., and Hunter, T. (1988) Science 24142–52 [DOI] [PubMed] [Google Scholar]
- 21.Steussy, C. N., Popov, K. M., Bowker-Kinley, M. M., Sloan, R. B., Jr., Harris, R. A., and Hamilton, J. A. (2001) J. Biol. Chem. 27637443–37450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Machius, M., Chuang, J. L., Wynn, R. M., Tomchick, D. R., and Chuang, D. T. (2001) Proc. Natl. Acad. Sci. U. S. A. 9811218–11223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato, M., Chuang, J. L., Tso, S. C., Wynn, R. M., and Chuang, D. T. (2005) EMBO J. 241763–1774 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dutta, R., and Inouye, M. (2000) Trends. Biochem. Sci. 2524–28 [DOI] [PubMed] [Google Scholar]
- 25.Alex, L. A., and Simon, M. I. (1994) Trends Genet. 10133–138 [DOI] [PubMed] [Google Scholar]
- 26.Bergerat, A., de Massy, B., Gadelle, D., Varoutas, P. C., Nicolas, A., and Forterre, P. (1997) Nature 386414–417 [DOI] [PubMed] [Google Scholar]
- 27.Smirnova, I. N., Kasho, V. N., and Faller, L. D. (1998) FEBS Lett. 431309–314 [DOI] [PubMed] [Google Scholar]
- 28.Wigley, D. B., Davies, G. J., Dodson, E. J., Maxwell, A., and Dodson, G. (1991) Nature 351624–629 [DOI] [PubMed] [Google Scholar]
- 29.Ban, C., Junop, M., and Yang, W. (1999) Cell 9785–97 [DOI] [PubMed] [Google Scholar]
- 30.Bao, H., Kasten, S. A., Yan, X., Hiromasa, Y., and Roche, T. E. (2004) Biochemistry 4313442–13451 [DOI] [PubMed] [Google Scholar]
- 31.Bowker-Kinley, M. M., Davis, W. I., Wu, P., Harris, R. A., and Popov, K. M. (1998) Biochem. J. 329191–196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majer, M., Popov, K. M., Harris, R. A., Bogardus, C., and Prochazka, M. (1998) Mol. Genet. Metab. 65181–186 [DOI] [PubMed] [Google Scholar]
- 33.Wu, P., Sato, J., Zhao, Y., Jaskiewicz, J., Popov, K. M., and Harris, R. A. (1998) Biochem. J. 329197–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jeoung, N. H., and Harris, R. A. (2008) Am. J. Physiol. 295E46–E54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang, B., Wu, P., Bowker-Kinley, M. M., and Harris, R. A. (2002) Diabetes 51276–283 [DOI] [PubMed] [Google Scholar]
- 36.Muoio, D. M., MacLean, P. S., Lang, D. B., Li, S., Houmard, J. A., Way, J. M., Winegar, D. A., Corton, J. C., Dohm, G. L., and Kraus, W. E. (2002) J. Biol. Chem. 27726089–26097 [DOI] [PubMed] [Google Scholar]
- 37.Rowles, J., Scherer, S. W., Xi, T., Majer, M., Nickle, D. C., Rommens, J. M., Popov, K. M., Harris, R. A., Riebow, N. L., Xia, J., Tsui, L. C., Bogardus, C., and Prochazka, M. (1996) J. Biol. Chem. 27122376–22382 [DOI] [PubMed] [Google Scholar]
- 38.Whitehouse, S., and Randle, P. J. (1973) Biochem. J. 134651–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Aicher, T. D., Anderson, R. C., Bebernitz, G. R., Coppola, G. M., Jewell, C. F., Knorr, D. C., Liu, C., Sperbeck, D. M., Brand, L. J., Strohschein, R. J., Gao, J., Vinluan, C. C., Shetty, S. S., Dragland, C., Kaplan, E. L., DelGrande, D., Islam, A., Liu, X., Lozito, R. J., Maniara, W. M., Walter, R. E., and Mann, W. R. (1999) J. Med. Chem. 422741–2746 [DOI] [PubMed] [Google Scholar]
- 40.Liu, X., Perusse, F., and Bukowiecki, L. J. (1998) Am. J. Physiol. 274R1212–R1219 [DOI] [PubMed] [Google Scholar]
- 41.Kato, M., Li, J., Chuang, J. L., and Chuang, D. T. (2007) Structure (Lond.) 15992–1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Knoechel, T. R., Tucker, A. D., Robinson, C. M., Phillips, C., Taylor, W., Bungay, P. J., Kasten, S. A., Roche, T. E., and Brown, D. G. (2006) Biochemistry 45402–415 [DOI] [PubMed] [Google Scholar]
- 43.Hiromasa, Y., Fujisawa, T., Aso, Y., and Roche, T. E. (2004) J. Biol. Chem. 2796921–6933 [DOI] [PubMed] [Google Scholar]
- 44.Shroads, A. L., Guo, X., Dixit, V., Liu, H. P., James, M. O., and Stacpoole, P. W. (2008) J. Pharmacol. Exp. Ther. 3241163–1171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tso, S.-C., Kato, M., Chuang, J. L., and Chuang, D. T. (2006) J. Biol. Chem. 28127197–27204 [DOI] [PubMed] [Google Scholar]
- 46.Tuganova, A., Klyuyeva, A., and Popov, K. M. (2007) Biochemistry 468592–8602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Green, T., Grigorian, A., Klyuyeva, A., Tuganova, A., Luo, M., and Popov, K. M. (2008) J. Biol. Chem. 28315789–15798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hawes, J. W., Zhao, Y., Popov, K. M., Shimomura, Y., and Harris, R. A. (2000) Methods Enzymol. 324200–207 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brautigam, C. A., Chuang, J. L., Tomchick, D. R., Machius, M., and Chuang, D. T. (2005) J. Mol. Biol. 350543–552 [DOI] [PubMed] [Google Scholar]
- 50.Otwinowski, Z., and Minor, W. (1997) Methods Enzymol. 276307–326 [DOI] [PubMed] [Google Scholar]
- 51.Read, R. J. (2001) Acta Crystallogr. Sect. D Biol. Crystallogr. 571373–1382 [DOI] [PubMed] [Google Scholar]
- 52.Collaborative Computational Project, No. 4 (1994) Acta Crystallogr. Sect. D Biol. Crystallogr. 50760–76315299374 [Google Scholar]
- 53.Schwede, T., Kopp, J., Guex, N., and Peitsch, M. C. (2003) Nucleic Acids Res. 313381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murshudov, G. N., Vagin, A. A., and Dodson, E. J. (1997) Acta Crystallogr. Sect. D Biol. Crystallogr. 53240–255 [DOI] [PubMed] [Google Scholar]
- 55.Cowtan, K. D., and Main, P. (1996) Acta Crystallogr. Sect. D Biol. Crystallogr. 5243–48 [DOI] [PubMed] [Google Scholar]
- 56.Hiromasa, Y., Hu, L., and Roche, T. E. (2006) J. Biol. Chem. 28112568–12579 [DOI] [PubMed] [Google Scholar]
- 57.Hiromasa, Y., and Roche, T. E. (2008) Biochemistry 472298–2311 [DOI] [PubMed] [Google Scholar]
- 58.Hiromasa, Y., Yan, X., and Roche, T. E. (2008) Biochemistry 472312–2324 [DOI] [PubMed] [Google Scholar]
- 59.Dong, J. (2001) Expression, Purification, Characterization of Human Pyruvate Dehydrogenase Kinase Isoform 4 (PDK4) Ph.D. Dissertation, Kansas State University, Manhattan
- 60.Bao, H., Kasten, S. A., Yan, X., and Roche, T. E. (2004) Biochemistry 4313432–13441 [DOI] [PubMed] [Google Scholar]
- 61.Mayers, R. M., Leighton, B., and Kilgour, E. (2005) Biochem. Soc. Trans. 33367–370 [DOI] [PubMed] [Google Scholar]
- 62.Klyuyeva, A., Tuganova, A., and Popov, K. M. (2005) Biochemistry 4413573–13582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monod, J., Wyman, J., and Changeux, J. P. (1965) J. Mol. Biol. 1288–118 [DOI] [PubMed] [Google Scholar]
- 64.Li, J., Machius, M., Chuang, J. L., Wynn, R. M., and Chuang, D. T. (2007) J. Biol. Chem. 28211904–11913 [DOI] [PubMed] [Google Scholar]
- 65.Thompson, J. D., Higgins, D. G., and Gibson, T. J. (1994) Nucleic Acids Res. 224673–4680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gouet, P., Courcelle, E., Stuart, D. I., and Metoz, F. (1999) Bioinformatics (Oxf.) 15305–308 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.