Abstract
Glutaredoxins are small heat-stable proteins that act as glutathione-dependent disulfide oxidoreductases. Two genes, designated GRX1 and GRX2, which share 40–52% identity and 61–76% similarity with glutaredoxins from bacterial and mammalian species, were identified in the yeast Saccharomyces cerevisiae. Strains deleted for both GRX1 and GRX2 were viable but lacked heat-stable oxidoreductase activity using β-hydroxyethylene disulfide as a substrate. Surprisingly, despite the high degree of homology between Grx1 and Grx2 (64% identity), the grx1 mutant was unaffected in oxidoreductase activity, whereas the grx2 mutant displayed only 20% of the wild-type activity, indicating that Grx2 accounted for the majority of this activity in vivo. Expression analysis indicated that this difference in activity did not arise as a result of differential expression of GRX1 and GRX2. In addition, a grx1 mutant was sensitive to oxidative stress induced by the superoxide anion, whereas a strain that lacked GRX2 was sensitive to hydrogen peroxide. Sensitivity to oxidative stress was not attributable to altered glutathione metabolism or cellular redox state, which did not vary between these strains. The expression of both genes was similarly elevated under various stress conditions, including oxidative, osmotic, heat, and stationary phase growth. Thus, Grx1 and Grx2 function differently in the cell, and we suggest that glutaredoxins may act as one of the primary defenses against mixed disulfides formed following oxidative damage to proteins.
INTRODUCTION
Glutaredoxin from Escherichia coli was first discovered as a small, heat-stable protein required for the glutathione-dependent synthesis of deoxyribonucleotides catalyzed by ribonucleotide reductase (Holmgren, 1976). Glutaredoxin 1 is a 9-kDa protein that acts as a reduced glutathione (GSH)-dependent disulfide oxidoreductase by virtue of the two cysteine residues in its active site (Holmgren and Aslund, 1995). Later studies in mutants that lacked both glutaredoxin and thioredoxin revealed that E. coli actually contains three glutaredoxins (Grx1–3), with glutaredoxin 3 also able to function in ribonucleotide synthesis (Aslund et al., 1994). In contrast, glutaredoxin 2 was proposed to be the first member of a novel class of glutaredoxins that lack activity as hydrogen donors for ribonucleotide reductase (Vlamis-Gardikas et al., 1997). Glutaredoxins have subsequently been identified and isolated from various eukaryotes, including human, bovine, pig, yeast, and rice (Minakuchi et al., 1994; Holmgren and Aslund, 1995). The structure of these proteins has been highly conserved throughout evolution, particularly in the region of the active site (Wells et al., 1993; Holmgren and Aslund, 1995). However, despite extensive structural analysis, little is known regarding the biochemical function of these eukaryotic glutaredoxins in vivo.
There appears to be considerable functional overlap between the glutaredoxin and thioredoxin systems. Similar to glutaredoxin, thioredoxin is a small, heat-stable protein that contains two redox-active cysteines in its active site and can serve as an electron donor for ribonucleotide reductase. The oxidized disulfide form of thioredoxin is reduced by NADPH and thioredoxin reductase, an enzyme that is a member of the FAD-containing pyridine disulfide oxidoreductase class of proteins. In contrast, the glutaredoxin system consists of NADPH, GSH, and glutathione reductase with electrons being transferred from NADPH to glutaredoxin via GSH (Holmgren, 1990). Utilization of GSH results in its conversion to the disulfide form, and it is regenerated in an NADPH-dependent reaction catalyzed by glutathione reductase (Grant and Dawes, 1996). Glutaredoxin shows no activity with thioredoxin reductase, and similarly, thioredoxins are not reduced by GSH and glutathione reductase (Holmgren, 1979). In addition to their function in ribonucleotide reduction, glutaredoxins and thioredoxins have proposed roles in many cellular processes, including reduction of dehydroascorbate, protein folding and regulation, and sulfur metabolism (Holmgren, 1989; Wells et al., 1993).
The yeast Saccharomyces cerevisiae contains two genes encoding thioredoxins, designated TRX1 and TRX2, which are dispensable under normal growth conditions (Gan, 1991; Muller, 1991). However, deletion of TRX1 and TRX2 affects the cell cycle, resulting in a prolonged S phase and a shortened G1 phase, which does not occur as a result of alterations in the levels of deoxyribonucleotides (Muller, 1991, 1995). In addition, a trx1 trx2 double mutant cannot grow in the absence of methionine or cysteine, presumably because of a defect in sulfate assimilation, indicating that thioredoxin is the only hydrogen donor for 3′-phosphoadenosine 5′-phosphosulfate reductase in yeast (Muller, 1991). Thioredoxins are also required to maintain the redox balance of GSH, and loss of TRX1 and TRX2 results in elevated levels of oxidized glutathione (GSSG), indicating a link between thioredoxin and GSH with the redox status of the cell (Muller, 1996). In yeast, a single thioltransferase (glutaredoxin) has been identified and purified, which was later cloned, sequenced, and designated TTR1 (Gan et al., 1990; Gan, 1992). Here, we show that yeast actually contains two genes encoding glutaredoxins, and that their gene products are required for protection during conditions of oxidative stress. This is the first in vivo demonstration of a requirement for glutaredoxins in protection against reactive oxygen species.
MATERIALS AND METHODS
Yeast Strains and Media
The S. cerevisiae strains used in this study were CY4 (Grant et al., 1996b) and its isogenic derivatives Y70 (grx1), Y100 (grx2), and Y117 (grx1 grx2) described below.
Strains were grown in rich YEPD medium [2% (wt/vol) glucose, 2% (wt/vol) bactopeptone, and 1% (wt/vol) yeast extract] or minimal SD medium [0.17% (wt/vol) yeast nitrogen base without amino acids, 5% (wt/vol) ammonium sulfate, and 2% (wt/vol) glucose (Sherman et al., 1974)] supplemented with appropriate amino acids and bases: 2 mM leucine, 4 mM isoleucine, 1 mM valine, 0.3 mM histidine, 0.4 mM tryptophan, 0.15 mM adenine, and 0.2 mM uracil. For growth on nonfermentable carbon sources, YEPGE contained 3% (vol/vol) glycerol and 1% (vol/vol) ethanol. Media were solidified by the addition of 2% (wt/vol) agar.
Cloning and Disruption of GRX1 and GRX2
The GRX1 and GRX2 genes were isolated by PCR amplification of total yeast DNA with oligonucleotides specific for GRX sequences. For GRX1, a 1617-bp fragment was amplified using oligonucleotides that hybridized 735 bp upstream of the putative ATG start codon (5′-GCCTCGAGAGATGAACAGATCCAAG-3′) and 549 bp downstream of the TAG stop codon (5′-ACTACTCGTGTTCATCTTGGACA-3′) respectively. A 1246 bp fragment was cloned into the polylinker region of plasmid pRS426 (Christianson et al., 1992) using an XhoI restriction site introduced by the 5′ oligonucleotide (underlined) and a naturally occurring EcoRI site. The resulting construct was called pG501 and was verified by DNA sequence analysis. For GRX2, a 1436 bp fragment was amplified using oligonucleotides that hybridized 941 bp upstream of the putative ATG start codon (5′-GTTGCACAAAGATATCGATAACCCG-3′) and 163 bp downstream of the TAG stop codon (5′-CGGAATTCAGCGGGTCTCATTGGT-3′), respectively. The amplicon was cloned into the polylinker region of plasmid pRS424 (Christianson et al., 1992) using an EcoRI restriction site introduced by the 3′ oligonucleotide (underlined) and a naturally occurring ClaI site. The resulting construct was called pL4 and was verified by DNA sequence analysis.
The GRX1 disruption construct pG507 was made by insertion of a 1.6-kb BamHI fragment containing the yeast LEU2 gene, isolated from plasmid YDp-L (Berben et al., 1991), into the BglII restriction site in pG501, which lies 53 bp downstream from the GRX1 ATG start codon. The 2.8-kb PstI fragment from pG507 was used to direct homologous recombination at the GRX1 locus, creating strain Y100 (grx1). A null allele of GRX2 (strain Y100) was generated in strain CY4 by a one-step PCR amplification protocol that replaced the entire GRX2 open reading frame (ORF) with the yeast HIS3 gene (Baudin et al., 1993). The oligonucleotides used for the PCR disruption of GRX2 were GRX2-D1 and GRX2-D2, the sequences of which were 5′-TTTGCCACAAGAATTATTGCTAAAAGATTTTTATCTACTCCAAAAAGCGCTAGGAGTCACTGCCA-3′ and 5′-TATATATATGTAAATATTATGAAGGGGATATTAGCGTAATTTAAAGGAAAGCGCGCCTCGTTCAG-3′ respectively. The underlined regions correspond to HIS3 sequences. The grx1 grx2 double mutant strain (Y117) was generated by disrupting GRX1, using plasmid pG507, in strain Y100.
Sensitivity to Oxidants
Sensitivity to H2O2, tert-butyl hydroperoxide, cumene hydroperoxide, menadione, and diamide was determined by spotting strains onto YEPD plates containing various concentrations of oxidants (Grant et al., 1997). Cells were grown to stationary phase in YEPD, and 10-μl aliquots of each culture, diluted to an A600 of 3.0 and 0.3, were spotted onto appropriate plates. Sensitivity was determined by comparison of growth with the wild-type strain after 3 d. Dose-response curves were obtained by growing cells to exponential phase (1–2 × 107 cells/ml) in SD medium at 30°C and treating with 4 mM H2O2 or 18 mM menadione for 1 h. Aliquots of cells were diluted in fresh YEPD medium at 20-min intervals and plated in triplicate on YEPD plates to obtain viable counts after 3 d of growth.
β-Galactosidase Assays
The GRX1::lacZ fusion construct was made by inserting the 744-bp PstI-BglII fragment of pG501 into the same sites of YIp358R (Berben et al., 1991). The resulting fusion construct, designated pL1, contains the ATG codon and 17 codons from the GRX1 coding region inserted in frame with the lacZ gene. The GRX2::lacZ fusion construct was made by insertion of a 1-kb PCR fragment amplified using oligonucleotides 5-GTTGCACAAAGAATTCGATAACCCG-3′ and 5′-CCTTGGATCCGGGAACGTTCAATTC-3′ into YIp357 (Berben et al., 1991). The amplicon was cloned into the polylinker region of YIp357 using EcoRI and BamHI restriction sites introduced by the oligonucleotides (underlined). The resulting fusion construct, designated pL2, contains the ATG codon and 44 codons from the GRX2 coding region inserted in frame with the lacZ gene.
For the determination of β-galactosidase activity, transformants were assayed essentially as described previously (Rose and Botstein, 1983). Cells were grown to early exponential phase (A600 = 1) at 30°C before treatment with various stress conditions. Cells were exposed to 0.3 mM H2O2, 18 mM menadione, or 1.5 mM diamide for 1 h. Heat shock conditions were 39°C for 30 min, and osmotic shock conditions were 0.4 M sodium chloride for 30 min. β-Galactosidase activity is expressed as nanomoles of o-nitrophenyl-β-d-galactopyranoside hydrolyzed per minute per microgram of total protein.
RNA Analysis
For Northern analysis, cell cultures were grown to an A600 of 1.0, and RNA was extracted by the method of Schmitt et al. (1990). Total RNA (10 μg) was separated by electrophoresis in a 1% formaldehyde gel. Nitrocellulose filters were probed for GRX1 using a 744-bp PstI-BglII DNA fragment from pG501 and for GRX2 using the 1-kb PCR fragment used in the construction of pL2. Loading controls were probed with a 1.7-kb HindIII fragment of the PDA1 gene (Wenzel et al., 1995). Quantitation of transcript levels was made using a Molecular Dynamics (Sunnyvale, CA) PhosphorImager.
Determination of Thiol Levels and Glutathione Reductase Activity
Glutathione reductase activity was determined by the method of Casalone et al (1988) and is expressed as nanomoles of NADPH oxidized per minute per milligram of protein. Total glutathione, GSH, and GSSG were determined by a microtiter plate assay method (Vandeputte et al., 1994). Free thiol groups were measured using 5,5′-dithiobis(2-nitrobenzoic acid) (DTNB; Rice-Evans et al., 1991) and are expressed as micromoles per milligram of total protein.
GSH-dependent Disulfide Oxidoreductase Activity
Glutaredoxin activity was measured by the reduction of the mixed disulfide formed between β-hydroxyethylene disulfide (HED) and GSH (Holmgren and Aslund, 1995). Cell-free extracts were prepared by breaking cells with glass beads using a Minibead beater (Biospec Scientific, Bartlesville, OK) for 30 s at 4°C. Where indicated, extracts were heat treated at 85°C for 5 min to inactivate enzymes such as glutathione reductase, thioredoxin reductase, and other interfering, non-heat–stable activities (Holmgren, 1976). This was confirmed by the fact that the heat-treated extracts were entirely dependent on exogenous glutathione reductase for activity. The components of the glutaredoxin system, NADPH (0.4 mM), GSH (1 mM), and glutathione reductase (6 μg/ml), as well as HED (0.7 mM) were added to a reaction volume of 1 ml in 0.1 M Tris-Cl (pH 7.4). A mixed disulfide between HED and GSH is formed within 2 min, and the reaction was started by the addition of 10–100 μl of cell extracts. The reaction was followed by the decrease in A340 attributable to the oxidation of NADPH.
RESULTS
Identification of GRX1 Encoding Glutaredoxin 1
Analysis of the sequence of yeast chromosome III revealed an ORF of 110 codons (YCL35c, GenBank accession number x59720), the putative protein product of which exhibited significant similarity to known glutaredoxins. The predicted protein (named Grx1) shares 40–52% identity and 61–76% similarity over the entire sequence with glutaredoxins from E. coli, rice, and humans (Figure 1). In addition, GRX1 is homologous to the previously identified yeast gene TTR1, encoding thioltransferase 1 (Gan et al., 1990; Gan, 1992). TTR1 shares 64% identity and 85% similarity with GRX1 (Figure 1), and we propose renaming TTR1 as GRX2, in accordance with the standard nomenclature suggested by Holmgren and Aslund (1995) for the thioltransferase class of proteins.
Figure 1.
Comparison of the predicted amino acid sequence of GRX1 and GRX2 with glutaredoxins from rice, human, and E. coli. Amino acid sequences were aligned for maximal homology, with dashes used to denote gaps introduced for maximal alignment. Identical amino acid residues are boxed, and conserved residues are shaded. Alignments were performed using the program ClustalW, version 1.4 (Thompson et al., 1994) and displayed using the graphic program seqVu 1.01 (J. Gardner, The Garvan Institute, Sydney, New South Wales, Australia). The active site sequence (CPYC) is underlined.
The active site of glutaredoxins contains two redox-active cysteine residues that are conserved in GRX1 and GRX2 (positions 27 and 30 in yeast numbering). Unlike microbial and plant glutaredoxins, animal glutaredoxins contain an additional half-cysteine pair in the pentapeptide Cys79-Ile-Gly-Gly-Cys83 (human numbering) (Hopper et al., 1989) that are not conserved in the yeast proteins. Interestingly, the intervening tripeptide (Ile-Gly-Gly) is conserved in plant and microbial glutaredoxins, as well as in GRX1 and GRX2, and the C-terminal cysteine has been replaced by a conserved histidine residue (Figure 1; Hopper et al., 1989). Conserved regions are also found at positions 74–76 and 84–87 (yeast numbering), which as well as the region around the active site have been proposed to play a role in interactions with other proteins, including ribonucleotide reductase (Xia et al., 1992).
Yeast Strains Deleted for GRX1 and GRX2 Are Viable
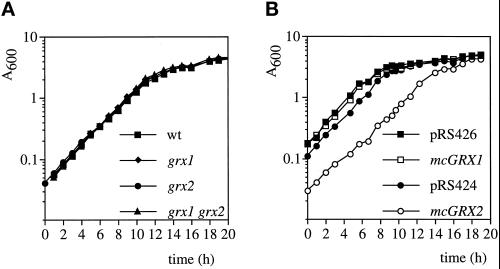
A strain carrying a disruption of GRX1 was generated by insertion of the yeast LEU2 gene within the coding region of GRX1 in the haploid wild-type strain CY4 (see Materials and Methods). A strain containing a total gene deletion of GRX2 was generated by replacement of the entire GRX2 ORF with the yeast HIS3 gene. Finally, a double mutant strain lacking both GRX1 and GRX2 was generated (grx1 grx2). The resulting strains were all viable, indicating that GRX1 and GRX2 are not essential for normal aerobic growth. In addition, the glutaredoxin mutants showed wild-type growth rates on rich glucose-based medium (YEPD) and on nonfermentable carbon sources such as glycerol. The glutaredoxin mutants were also able to grow at the same growth rate on minimal medium (SD), indicating that sulfate can be assimilated as the sole source of sulfur (Figure 2A). The effect of overexpressing GRX1 or GRX2 on the growth of wild-type cells (CY4) was examined by growing cells containing multicopy GRX1 (mcGRX1) or GRX2 (mcGRX2) on minimal SD medium (Figure 2B). mcGRX1 did not affect the growth of CY4 (compare pRS426 with mcGRX1), whereas mcGRX2 resulted in a marked lag phase and reduced growth rate (compare pRS424 with mcGRX2). In addition, when the growth of these strains was compared by streaking for single colonies on SD plates, cells containing mcGRX2 grew extremely slowly, forming small pinpoint colonies with occasional faster-growing revertants.
Figure 2.
Growth of glutaredoxin strains. (A) Strains CY4 (wt), Y70 (grx1), Y100 (grx2), and Y117 (grx1 grx2) and (B) CY4 transformed with pRS426, mcGRX1, pRS424, and mcGRX2 were grown in minimal medium (SD), and growth was monitored by optical density at 600 nm (A600). Strains were inoculated to the same cell density to start each experiment (our unpublished results).
Grx1 and Grx2 Display GSH-Disulfide Oxidoreductase Activity
The oxidoreductase activity of glutaredoxins can be measured by their ability to reduce the mixed disulfide formed between GSH and HED (Holmgren and Aslund, 1995). Briefly, the assay contains HED, GSH, and glutathione reductase, and the reaction rate is followed by the oxidation of NADPH. Both the wild-type and grx1 mutant strains displayed similar GSH-dependent oxidoreductase activities during stationary phase growth (Table 1). In contrast, the grx2 and grx1 grx2 double mutants displayed only 20 and 15% of the wild-type activity. To ensure specificity of the assay for glutaredoxins, extracts were heat treated at 85°C for 5 min to inactivate enzymes such as glutathione reductase, thioredoxin reductase, and other interfering, non-heat–stable activities (Holmgren, 1976). This treatment should not affect glutaredoxins or thioredoxins, which are heat stable, and thioredoxin shows no activity in the HED assay because of its dependence on thioredoxin reductase. Exponential phase wild-type cells displayed GSH-dependent disulfide oxidoreductase activity (47 nmol · min−1 · mg−1), which was elevated twofold in stationary phase cells (Table 1). The heat-stable oxidoreductase activity was unaffected in the grx1 mutant during both exponential and stationary phase growth. In contrast, the activity fell to 17 and 18% of the wild-type levels in the grx2 mutant during exponential and stationary phase growth, respectively. In the grx1 grx2 double disruption strain, oxidoreductase activity was undetectable during exponential phase growth, and a low 18% activity was detectable during stationary phase growth. These results indicate that Grx2 accounts for the majority of GSH-dependent oxidoreductase activity in yeast.
Table 1.
GSH-dependent disulfide oxidoreductase activity in glutaredoxin mutants
| Strain | Stationary phase | Heat-treateda stationary | Heat-treateda exponential |
|---|---|---|---|
| wt | 442 ± 40 | 118 ± 1 | 47 ± 10 |
| grx1 | 435 ± 29 | 134 ± 4 | 46 ± 4 |
| grx2 | 88 ± 22 | 21 ± 2 | 8 ± 2 |
| grx1 grx2 | 66 ± 13 | 22 ± 5 | ND |
Glutaredoxin activity is given as nanomoles of NADPH oxidized per minute per milligram of protein. Data are the means of triplicate experiments. ND, not detectable.
Extracts were heat treated at 85°C for 5 min (for details see text).
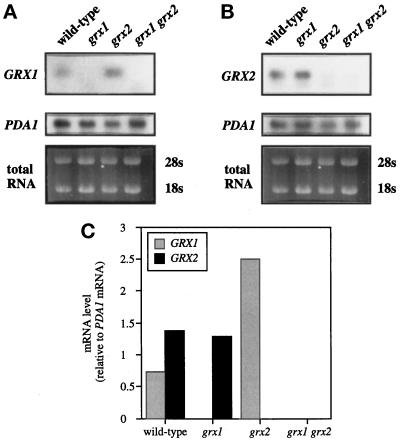
To determine whether the differences in GSH-disulfide oxidoreductase activity could be accounted for by differential expression of GRX1 and GRX2, transcript levels were measured by Northern blot analysis (Figure 3). The level of the GRX1 transcript was approximately half that of the GRX2 transcript in a wild-type strain, which is not sufficient to account for the differences in oxidoreductase activity observed. Interestingly, when GRX2 was deleted, the level of the GRX1 transcript increased by approximately threefold, which again did not correlate with the measured oxidoreductase activity. In agreement with the transcript analysis, the relative expression levels of GRX1::lacZ and GRX2::lacZ fusion constructs (see Figure 6) indicated that differences in expression of GRX1 and GRX2 could not account for the differences in oxidoreductase activity observed.
Figure 3.
Transcript analysis of GRX1 and GRX2. Total RNA was isolated from wild type, grx1, grx2, and the grx1 grx2 double mutant grown to exponential phase and subjected to Northern blot analysis. The resulting blots were probed with GRX1 (A) or GRX2 (B) and PDA1 as a loading control. mRNA hybridization signals were quantified for the representative experiments shown in A and B using a PhosphorImager, and the ratio of GRX1 or GRX2 mRNA to PDA1 mRNA is shown.
Figure 6.
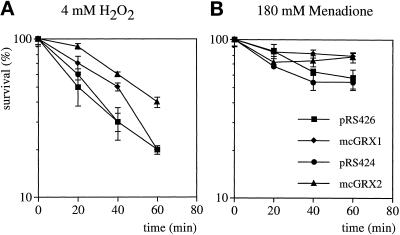
Overexpression of GRX1 and GRX2 increases resistance to oxidants. Wild-type strains containing pRS426, mcGRX1, pRS424, and mcGRX2 were grown to exponential phase in SD medium and treated with 4 mM H2O2 (A) or 180 mM menadione (B). Cells were diluted and plated in triplicate onto YEPD medium to monitor cell viability at 20-min intervals. Percent survival is expressed relative to the untreated control cultures.
To address the question of whether Grx1 lacked GSH-disulfide oxidoreductase activity, assays were performed on strains containing multicopy GRX1 (mcGRX1) or GRX2 (mcGRX2). In wild-type cells grown to stationary phase, mcGRX1 and mcGRX2 resulted in an approximate twofold to threefold increase in heat-stable oxidoreductase activity (Table 2, compare pRS426 with mcGRX1 and pRS424 with mcGRX2). Similarly, mcGRX1 and mcGRX2 resulted in a fourfold to sixfold increases in oxidoreductase activity during exponential phase growth. Because the increase in activity in the strain containing mcGRX1 may have arisen because of an indirect effect on Grx2, activity was next determined in the grx1 grx2 double mutant, which is devoid of glutaredoxin activity (Table 2). Again, both mcGRX1 and mcGRX2 resulted in significant increases in glutaredoxin activity; however, mcGRX2 resulted in a fivefold greater increase compared with mcGRX1. These results indicate that both Grx1 and Grx2 possess GSH-dependent disulfide oxidoreductase activity. To examine the differences between Grx1 and Grx2 further, we next examined the response of grx1 and grx2 mutants to various stress conditions during which mixed disulfides are likely to be formed.
Table 2.
Glutaredoxin activity in strains overexpressing GRX1 and GRX2
| Strain | Wild-type exponential | Wild-type stationary | grx1 grx2 stationary |
|---|---|---|---|
| pRS426 | 27 ± 4 | 152 ± 12 | 6 ± 0.4 |
| mcGRX1 | 107 ± 4 | 486 ± 18 | 115 ± 19 |
| pRS424 | 37 ± 7 | 184 ± 50 | 13 ± 5 |
| mcGRX2 | 215 ± 11 | 411 ± 55 | 573 ± 114 |
Glutaredoxin activity is given as nanomoles of NADPH oxidized per minute per milligram of protein. Data are the means of triplicate experiments.
Strains Lacking GRX1 or GRX2 Are Sensitive to Conditions of Oxidative Stress
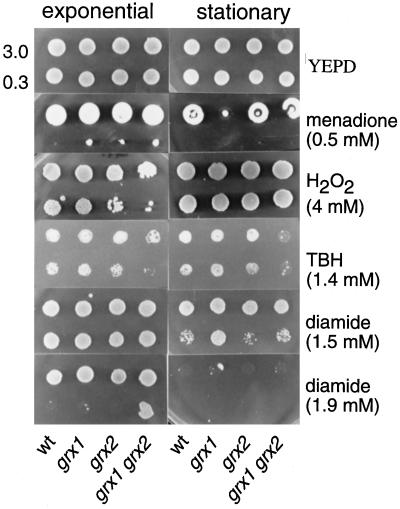
The glutaredoxin mutants were unaffected in resistance to a variety of stress conditions, including heat, heavy metal, and osmotic stress. However, loss of glutaredoxins resulted in altered sensitivity to oxidative stress induced by various reactive oxygen species (ROS). Specifically, exponential and stationary phase cells were tested for growth on plates containing various concentrations of menadione, hydrogen peroxide, tert-butyl hydroperoxide, and diamide (Figure 4). When added extracellularly, menadione (1,4-naphthoquinone) generates superoxide radicals through a redox-cycling mechanism (Hassan and Fridovich, 1979). No difference in sensitivity to menadione was seen for exponential phase cells exposed to various concentrations of menadione (Figure 4); however, the grx1 mutant was sensitive to 0.5 mM menadione during stationary phase. In contrast, exponential phase grx2 and grx1 grx2 double mutants were sensitive to 4 mM hydrogen peroxide, whereas the grx1 mutant and all stationary phase cells were unaffected by this oxidant. In addition, the grx1 grx2 double mutant was sensitive to tert-butyl hydroperoxide during both exponential and stationary phase growth (Figure 4). Diamide is a thiol-specific oxidant that can readily oxidize GSH (Kosower and Kosower, 1995), and stationary phase cells that lack TRX2, GSH1 or GSH2 show increased resistance to this oxidant (Muller, 1996; Grant et al., 1997). Similarly, the grx1 mutant was more resistant to 1.5 mM diamide than the wild-type strain during stationary phase. Interestingly, stationary phase cells were more sensitive to diamide than exponential phase cells and were unable to grow at higher concentrations. The grx1 grx2 double mutant was found to be resistant to 1.9 mM diamide during exponential phase growth (Figure 4). Hence, GRX1 and GRX2 appear to have different functions in the defense against diamide, depending on the growth phase. In exponential phase cells, the lack of both glutaredoxins resulted in resistance to diamide, whereas in stationary phase cells, the deletion of GRX1 alone led to resistance.
Figure 4.
Strains deleted for GRX1 or GRX2 are sensitive to reactive oxygen species. Sensitivity to menadione, hydrogen peroxide, tert-butyl hydroperoxide, and diamide was determined by spotting strains on YEPD plates containing various concentrations of oxidants. Cultures of wild-type, grx1, grx2, and grx1 grx2 double mutant strains were grown to exponential phase (A600 = 1) and into stationary phase in YEPD medium, adjusted to an A600 of 3.0 and 0.3 before spotting 10-μl aliquots onto appropriate plates. Plates were incubated at 30°C for 3 d before scoring growth.
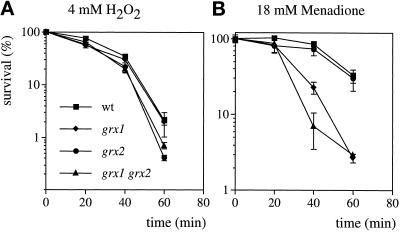
To compare the oxidant sensitivity of the glutaredoxin mutants quantitatively, dose-response curves to hydrogen peroxide and menadione were generated. In these experiments, wild-type cells and the grx1, grx2, and grx1 grx2 double mutant strains were grown to exponential phase in minimal media and then treated with 4 mM H2O2 or 18 mM menadione for 1 h during which cell viability was monitored (Figure 5). For the H2O2 treatment, both the grx2 and the grx1 grx2 double mutants were more sensitive compared with the wild-type strain and the grx1 mutant (Figure 5A). In contrast, grx1 and the grx1 grx2 double mutant were sensitive to menadione (Figure 5B). These results indicate that loss of GRX1 confers sensitivity to the superoxide anion, whereas loss of GRX2 confers sensitivity to H2O2. Plate test experiments indicated that the grx1 mutant was only sensitive to menadione during stationary phase, whereas the dose-response curves indicated that both grx1 and the grx1 grx2 double mutant were sensitive during exponential phase growth. This presumably reflects differences in the two assays, because in the case of the plate test, cells were exposed to the oxidant throughout their growth, whereas for the dose-response experiment, cells were exposed to the oxidant for just 1 h.
Figure 5.
Sensitivity to hydrogen peroxide and menadione. Wild-type, grx1, grx2, and grx1 grx2 double mutant strains were grown to exponential phase in SD medium and treated with 4 mM H2O2 (A) or 18 mM menadione (B). Cells were diluted and plated in triplicate onto YEPD medium to monitor cell viability at 20-min intervals. Percent survival is expressed relative to the untreated control cultures.
We next examined the effect of overexpressing GRX1 and GRX2 on resistance to hydrogen peroxide and menadione. These results were complicated by the fact that multicopy vectors alone appeared to increase resistance to ROS, presumably because of induction of cellular stress responses. Overexpression of GRX2 resulted in increased resistance to 4 mM hydrogen peroxide throughout the 1 h time course (Figure 6A, compare mcGRX2 with pRS424). In contrast, mcGRX1 resulted in a somewhat elevated resistance to H2O2 during the first 40 min but fell to normal wild-type levels after 60 min (Figure 6A, compare mcGRX1 with pRS426). Yeast cells containing multicopy vectors (pRS426 and pRS424) were extremely resistant to menadione, and treatment with 180 mM resulted in ∼45% loss of viability (Figure 6B). mcGRX1 and mcGRX2 resulted in increased levels of resistance to menadione, with 78% survival after the 1 h treatment. Thus, overexpression of both GRX1 and GRX2 increased resistance to oxidative stress induced by hydrogen peroxide and the superoxide anion. Because the activity of glutaredoxin is dependent on glutathione, and alterations in glutathione levels and redox state have been shown to affect resistance to oxidative stress (Grant et al., 1996a–c), we examined whether the differences in oxidant sensitivity of the glutaredoxin mutants were attributable to differences in GSH metabolism.
Loss of GRX1 or GRX2 Does Not Affect Glutathione Redox State or Glutathione Reductase Activity
Glutathione reductase (Glr) is the primary enzyme that controls the redox state of GSH and is required for protection against oxidative stress (Grant et al., 1996a). Glr activity is regulated in response to ROS (Grant et al., 1996a) but was unaffected in the glutaredoxin mutants, indicating that there was no increased demand for the enzyme in these strains (Table 3). Cellular redox state, as measured by the reaction of DTNB with free thiol groups, was also unaffected in the glutaredoxin mutants (Table 3). Likewise, total GSH levels, as well as the ratio of GSH to GSSG was similar in the wild-type and glutaredoxin mutants (Table 3). These results indicate that the loss of glutaredoxin activity did not affect GSH metabolism, and hence their oxidant sensitivity was not as a result of alterations in GSH redox balance in the cell. Given the requirement for GRX1 and GRX2 in the cellular response to oxidative stress, we examined whether the expression of the two glutaredoxin genes was regulated in response to stress conditions.
Table 3.
Glutathione reductase activity and cellular redox state in glutaredoxin mutants
| Strain | Glr activity (nmol·min−1·mg−1) | Thiol contenta (μmol/mg) | Total GSHb (nmol/107 cells) | GSSGc (pmol/107 cells) | GSSG (%) |
|---|---|---|---|---|---|
| wt | 152 ± 8 | 243 ± 63 | 3.2 ± 0.1 | 47.6 ± 3.4 | 1.5 |
| grx1 | 158 ± 18 | 212 ± 11 | 3.0 ± 0.1 | 43.4 ± 1.4 | 1.5 |
| grx2 | 135 ± 20 | 210 ± 17 | 2.5 ± 0.6 | 50.4 ± 10.5 | 2 |
| grx1 grx2 | 149 ± 0 | 227 ± 18 | 3.0 ± 0.1 | 51.8 ± 9.1 | 1.7 |
Data are the means of duplicate experiments.
Total thiol content includes both protein and GSH.
Total GSH is the equivalent of GSH + 2GSSG.
Percent GSSG is the portion of GSSG present in the cell relative to the amount of total GSH.
GRX1 and GRX2 Expression Is Increased during Conditions of Stress
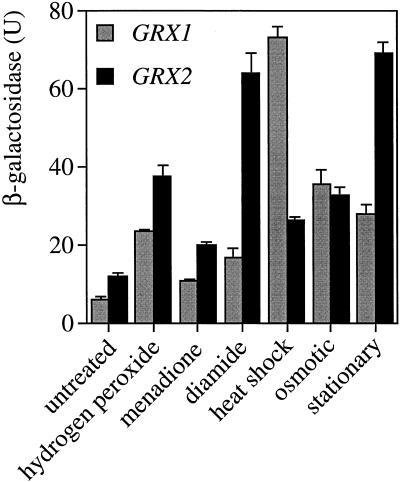
To test whether glutaredoxin expression responded to stress conditions, chimeric genes were constructed containing the lacZ gene fused to the GRX1 or GRX2 promoter. These fusion constructs were transformed into yeast, and expression was measured under a variety of stress conditions, including H2O2, superoxide, diamide, heat, and osmotic and stationary phase growth (Figure 7). Both GRX1 and GRX2 reporter constructs were expressed at relatively low levels during early exponential phase, with a slightly higher expression of GRX2 compared with GRX1 in agreement with the transcript analysis (Figure 3). Expression of both genes was induced in a growth phase–dependent manner, with a fivefold to sixfold increase as the cells progressed into stationary phase. In addition, expression of both GRX1::lacZ and GRX2::lacZ was induced by all of the stress conditions examined. There were, however, differences in the levels of induction, with GRX1 expression induced 12- and 6-fold under heat shock and osmotic shock conditions, respectively, compared with the 2-fold induction under both conditions for GRX2.
Figure 7.
Expression of both GRX1 and GRX2 is increased under a variety of stress conditions. GRX1::lacZ and GRX2::lacZ fusion constructs were assayed for β-galactosidase activity during exponential phase growth (untreated) and after treatment with 0.3 mM hydrogen peroxide, 9 mM menadione, 1.5 mM diamide, and 39°C heat shock and after entry into stationary phase. Values shown are the means of at least three independent determinations.
DISCUSSION
Glutaredoxin was originally identified as an electron donor for ribonucleotide reductase (Holmgren, 1979), and this remains its best characterized function. Nevertheless, not all glutaredoxins serve as hydrogen donors for ribonucleotide reductase, as shown for glutaredoxins from rabbit bone marrow and pig liver, and the recently characterized Grx2 from E. coli (Hopper et al., 1989; Vlamis-Gardikas et al., 1997). Similarly, it was shown that mouse fibroblasts depleted of glutaredoxin were not affected in growth rate, DNA synthesis, or the size of the deoxyribonucleotide pool (Spyrou and Holmgren, 1996). It remains to be established whether yeast Grx1, Grx2, or both can function in the reduction of ribonucleotides, but it seems likely given that the level of deoxynucleotide triphosphates in a mutant lacking both thioredoxin genes are unaltered, indicating the existence of an alternate hydrogen donor for ribonucleotide reductase in addition to thioredoxin in yeast (Muller, 1994). Confirmation of a role for the yeast glutaredoxins in deoxyribonucleotide synthesis will have to await purification of the yeast ribonucleotide reductase, which is an extremely unstable protein (Lammers and Follmann, 1984; Harder and Follmann, 1990). The activity with ribonucleotide reductase depends on a redox-active disulfide in the active site of glutaredoxin, whereas the activity as an oxidoreductase is only dependent on the N-terminal active site cysteine (Holmgren and Aslund, 1995). Results presented here indicate that both Grx1 and Grx2 display GSH-dependent disulfide oxidoreductase activity. However, despite the high degree of homology between these two proteins, Grx2 accounted for the majority of this oxidoreductase activity in the cell. The difference in activity did not arise as a result of differential expression of the two genes and may indicate, therefore, that Grx1 and Grx2 have different functions in yeast. In this view, subtle differences in primary or secondary structure would account for the differences in activity, either through effects on catalytic activity or on interactions with other components of the glutaredoxin system.
In this present study, eukaryotic glutaredoxins were shown to be required in vivo for protection against ROS. Grx1 was found to function in protection against the superoxide anion, whereas Grx2 was required for resistance to H2O2. It is unclear why the two glutaredoxins were required for protection against different forms of ROS, although it is consistent with the observation that the physiological mechanisms underlying adaptive responses to hydrogen peroxide and the superoxide anion are different (Jamieson, 1992; Flattery-O’Brien et al., 1993). The superoxide anion is a ubiquitous by-product of aerobic metabolism, which is itself relatively unreactive, but can serve as a precursor of highly reactive and deleterious ROS, including the hydroxyl free radical HO⋅ (Halliwell, 1991). In contrast, hydrogen peroxide is both freely diffusible and fairly reactive and may generate a different spectrum of cellular damage to the superoxide anion. In addition, it can generate the hydroxyl radical via the Fenton reaction. Detoxification of H2O2 is mediated by both catalase, which catalyses its breakdown to H2O2 and O2, and by glutathione peroxidases, which use GSH as a reductants. In yeast, GSH-mediated reactions appear to be the primary means of detoxifying this ROS (Grant and Dawes, 1996); however, catalases are required for the detoxification of H2O2 during stationary phase (Izawa et al., 1996). Similarly, GRX2 was required for H2O2 resistance during exponential phase growth but not during stationary phase. Stationary phase cells are generally more resistant to conditions of oxidative stress because of the increased synthesis of various antioxidants, including catalase (Izawa et al., 1996), and this may compensate for the lack of GRX2 during this growth phase. Organic peroxides such as tert-butyl hydroperoxide are thought to be detoxified by glutathione-dependent systems, because they are too bulky to be substrates for catalase (Grant and Dawes, 1996), and accordingly, the strain lacking GRX1 and GRX2 was sensitive during both exponential and stationary phases.
The requirement for glutaredoxins in protection against ROS may reflect a specific role in the regulation of a cellular antioxidant(s), or a more general role in protection against oxidants as a result of their disulfide oxidoreductase activity. For example, glutathione peroxidases are antioxidant enzymes that catalyze the breakdown of hydroperoxides using GSH as a reductant (Grant and Dawes, 1996), and in vitro studies have shown that both the glutaredoxin and thioredoxin systems can serve as electron donors for human plasma (selenium-dependent) glutathione peroxidase (Bjornstedt et al., 1994). Thus, the absence of a glutaredoxin may result in a reduced ability to regenerate active glutathione peroxidase after detoxification of ROS. In addition, the antioxidant ascorbic acid, which can detoxify hydrogen peroxide and other forms of ROS, may require glutaredoxin activity for maintenance of its function. Utilization of ascorbic acid results in its conversion to dehydroascorbate, and it is regenerated in a GSH-dependent reaction catalyzed by glutaredoxin and protein disulfide isomerase (Wells et al., 1990; Meister, 1994; Park and Levine, 1996). However, it appears unlikely that Grx1 or Grx2 function in vivo as dehydroascorbate reductases, because the five-carbon analogue erythroascorbic acid is the predominant form detected in yeast (Nick et al., 1986; Kim et al., 1993).
Glutaredoxins catalyze the cleavage of mixed disulfides in the presence of low concentrations of GSH and may therefore protect cells by reducing any mixed disulfides formed during oxidative stress (Chrestensen et al., 1995). In addition, in vitro experiments indicate that glutaredoxins can reactivate a number of oxidized enzymes by reducing the mixed disulfides formed as a result of thiol oxidation (Terada et al., 1992; Terada, 1994; Yoshitake et al., 1994). This repair activity is not restricted to enzymes, because glutaredoxin from human erythrocytes is able to reactivate membrane proteins and to regenerate hemoglobin from the mixed disulfide hemoglobin-S-S-glutathione (Mieyal et al., 1991; Terada et al., 1992). More recently, glutaredoxin has been isolated from the human ocular lens and was found to dethiolate mixed disulfides formed during oxidative stress, preventing loss of lens transparency and cataract formation (Raghavachari and Lou, 1996). Thus, yeast glutaredoxins may function in vivo to reduce mixed disulfides formed as a result of oxidative damage to proteins. Mixed disulfides may be formed by protein S-thiolation when protein sulfydryls are oxidized to from mixed disulfides with low–molecular weight thiols such as GSH (Thomas et al., 1995). No increase in the levels of protein-bound GSH, both under normal growth conditions or after exposure to oxidative stress, was detected in the various glutaredoxin mutants (our unpublished observations). However, differences in protein-GSH conjugates that occur at the subcellular level (e.g., in the vacuole or mitochondrion) would not be detected in our whole-cell extracts, nor would conjugates to other thiols such as cysteine or homocysteine, and this remains the subject of our ongoing investigations.
Because Grx2 accounted for the majority of oxidoreductase activity in the cell, its primary function may be to detoxify any mixed disulfides formed as a result of damage caused by ROS. Grx1 could also function in the GSH-dependent disulfide oxidoreductase assay and may therefore be required in addition to Grx2 during certain stress conditions or after the formation of particular mixed disulfide substrates. In agreement with this, the expression of both GRX1 and GRX2 was elevated in response to various stress conditions, including hydrogen peroxide, the superoxide anion, diamide, heat shock, osmotic shock, and stationary phase. Similar levels of induction were seen for GRX1 and GRX2 in response to oxidative stress induced by hydrogen peroxide, menadione, and diamide. However, GRX1 was induced 12- and 6-fold in response to heat and osmotic stress, respectively, compared with the 2-fold inductions seen for GRX2. Increased expression in response to multiple stress conditions is similar to the regulation seen for many stress-responsive genes, including CTT1, DDR2, TPS2, and HSP12 which are controlled by a stress response element (Ruis and Schuller, 1995). In fact, the promoter regions of GRX1 and GRX2 were found to contain putative stress response elements (our unpublished observations), which places them among this family of stress-responsive genes. The expression of other genes forming the glutaredoxin and thioredoxin systems is also induced in response to oxidants, including GSH1, GLR1, and TRX2, which are regulated by the yAP-1 transcriptional activator (Kuge and Jones, 1994; Wu and Moye-Rowley, 1994; Stephen et al., 1995; Grant et al., 1996a,c). Thus, in response to increased ROS, yeast cells can increase the levels of both glutaredoxin and thioredoxin, and this may be one of the primary defenses against oxidative damage to proteins.
Thioredoxin 2 has also been implicated in protection against hydroperoxides, because a trx2 mutant was sensitive to hydrogen peroxide (Kuge and Jones, 1994). In addition, the loss of both TRX1 and TRX2 was found to result in alterations to the cellular GSH redox balance, elevating the levels of oxidized glutathione (Muller, 1996). In contrast, loss of GRX1 or GRX2 did not affect the GSH redox balance, cellular thiol levels, or glutathione reductase activity. It remains to be established how great the functional overlap is between the glutaredoxin and thioredoxin systems, especially given that they appear to be balanced in E. coli (Miranda-Vizuete et al., 1996). However, differences in the function of these two systems have already come to light in yeast, because thioredoxin was found to be essential for sulfur metabolism (Muller, 1991), whereas the glutaredoxin mutants presented here showed wild-type growth rates with sulfate as the sole source of sulfur. This is in contrast to prokaryotic systems, in which both thioredoxin and glutaredoxin serve as hydrogen donors for 3′-phosphoadenosine 5′-phosphosulfate reductase. Overexpression of GRX2 resulted in a slow-growth phenotype, which was not related to problems in sulfur assimilation (our unpublished observations). Recently, it has been demonstrated that glutaredoxin can catalyze both the formation and reduction of mixed disulfides (Ruoppolo et al., 1997), and an increase in either process may account for the slow-growth phenotype. Occasional faster-growing colonies were formed from cells transformed with mcGRX2, and a genetic analysis of these revertants should lead to a better understanding of glutaredoxin function in yeast.
ACKNOWLEDGMENTS
S.L. was sponsored by the Studienstiftung des Deutschen Volkes.
REFERENCES
- Aslund F, Ehn B, Miranda-Vizuete A, Pueyo C, Holmgren A. Two additional gluterdoxins exist in Escherichia coli: glutardoxin 3 as a hydrogen donor for ribonucleotide reductase in a thioredoxin/glutaredoxin 1 double mutant. Proc Natl Acad Sci USA. 1994;91:9813–9817. doi: 10.1073/pnas.91.21.9813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudin A, Ozier-Kalogeropoulos O, Danouel A, Lacroute F, Cullin C. A simple and efficient method for direct gene deletion in Saccharomyces cerevisiae. Nucleic Acids Res. 1993;21:3329–3330. doi: 10.1093/nar/21.14.3329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berben G, Dumont J, Gilliquet V, Bolle P-A, Hillger F. The YDp plasmids: a uniform set of vectors bearing versatile gene disruption cassettes for Saccharomyces cerevisiae. Yeast. 1991;7:475–477. doi: 10.1002/yea.320070506. [DOI] [PubMed] [Google Scholar]
- Bjornstedt M, Xue J, Huang W, Akesson B, Holmgren A. The thioredoxin and glutaredoxin systems are efficient electron donors to human plasma glutathione peroxidase. J Biol Chem. 1994;269:29382–29384. [PubMed] [Google Scholar]
- Casalone E, Di Ilio C, Federici G, Polsinelli M. Glutathione and glutathione metabolizing enzymes in yeasts. Ant van Leeuw. 1988;54:367–375. doi: 10.1007/BF00393527. [DOI] [PubMed] [Google Scholar]
- Chrestensen CA, Eckman CB, Starke DW, Mieyal JJ. Cloning, expression and characterization of human thioltransferase (glutaredoxin) in E. coli. FEBS Lett. 1995;374:25–28. doi: 10.1016/0014-5793(95)01066-n. [DOI] [PubMed] [Google Scholar]
- Christianson TW, Sikorski RS, Dante M, Shero JH, Hieter P. Multifunctional yeast high-copy-number shuttle vectors. Gene. 1992;110:119–122. doi: 10.1016/0378-1119(92)90454-w. [DOI] [PubMed] [Google Scholar]
- Flattery-O’Brien J, Collinson LP, Dawes IW. Saccharomyces cerevisiae has an inducible response to menadione which differs to that to hydrogen peroxide. J Gen Microbiol. 1993;139:501–507. doi: 10.1099/00221287-139-3-501. [DOI] [PubMed] [Google Scholar]
- Gan Z-R. Yeast thioredoxin genes. J Biol Chem. 1991;266:1692–1696. [PubMed] [Google Scholar]
- Gan Z-R. Cloning and sequencing of a gene encoding yeast thioltransferase. Biochem Biophys Res Commun. 1992;187:949–955. doi: 10.1016/0006-291x(92)91289-3. [DOI] [PubMed] [Google Scholar]
- Gan Z-R, Polokoff MA, Jacobs JW, Sardana MK. Complete amino acid sequence of yeast thioltransferase (glutaredoxin) Biochem Biophys Res Comun. 1990;168:944–951. doi: 10.1016/0006-291x(90)91120-h. [DOI] [PubMed] [Google Scholar]
- Grant CM, Collinson LP, Roe J-H, Dawes IW. Yeast glutathione reductase is required for protection against oxidative stress and is a target gene for yAP-1 transcriptional regulation. Mol Microbiol. 1996a;21:171–179. doi: 10.1046/j.1365-2958.1996.6351340.x. [DOI] [PubMed] [Google Scholar]
- Grant CM, Dawes IW. Synthesis and role of glutathione in protection against oxidative stress in yeast. Redox Rep. 1996;2:223–229. doi: 10.1080/13510002.1996.11747054. [DOI] [PubMed] [Google Scholar]
- Grant CM, MacIver FH, Dawes IW. Glutathione is an essential metabolite required for resistance to oxidative stress in the yeast Saccharomyces cerevisiae. Curr Genet. 1996b;29:511–515. doi: 10.1007/BF02426954. [DOI] [PubMed] [Google Scholar]
- Grant CM, MacIver FM, Dawes IW. Stationary phase induction of GLR1 expression is mediated by the yAP-1 transcriptional protein in Saccharomyces cerevisiae. Mol Microbiol. 1996c;22:739–746. doi: 10.1046/j.1365-2958.1996.d01-1727.x. [DOI] [PubMed] [Google Scholar]
- Grant CM, MacIver FH, Dawes IW. Glutathione synthetase is dispensable for growth under both normal and oxidative stress conditions in the yeast Saccharomyces cerevisiae due to an accumulation of the dipeptide gamma-glutamylcysteine. Mol Biol Cell. 1997;8:1699–1707. doi: 10.1091/mbc.8.9.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halliwell B. Reactive oxygen species in living systems: source, biochemistry, and role in human disease. Am J Med. 1991;91:3C–14S–3C–38S. doi: 10.1016/0002-9343(91)90279-7. [DOI] [PubMed] [Google Scholar]
- Harder J, Follmann H. Identification of a free radical and oxygen dependence of ribonucleotide reductase in yeast. Free Radical Res Commun. 1990;10:281–286. doi: 10.3109/10715769009149896. [DOI] [PubMed] [Google Scholar]
- Hassan HM, Fridovich I. Intracellular production of superoxide radical and of hydrogen peroxide by redox active compounds. Arch Biochem Biophys. 1979;196:385–395. doi: 10.1016/0003-9861(79)90289-3. [DOI] [PubMed] [Google Scholar]
- Holmgren A. Hydrogen donor system for Escherichia coli ribonucleotide diphosphate reductase dependent upon glutathione. Proc Natl Acad Sci USA. 1976;73:2275–2279. doi: 10.1073/pnas.73.7.2275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmgren A. Glutathione-dependent synthesis of deoxyribonucleotides. J Biol Chem. 1979;254:3664–3671. [PubMed] [Google Scholar]
- Holmgren A. Thioredoxin and glutaredoxin systems. J Biol Chem. 1989;264:13963–13966. [PubMed] [Google Scholar]
- Holmgren A. Glutaredoxin: structure and function. In: Vina J, editor. Glutathione: Metabolism and Physiological Functions. Boca Raton, FL: CRC Press; 1990. pp. 146–154. [Google Scholar]
- Holmgren A, Aslund F. Glutaredoxin. Methods Enzymol. 1995;252:283–292. doi: 10.1016/0076-6879(95)52031-7. [DOI] [PubMed] [Google Scholar]
- Hopper S, Johnson RS, Vath JE, Biemann K. Glutaredoxin from rabbit bone marrow. J Biol Chem. 1989;264:20438–20447. [PubMed] [Google Scholar]
- Izawa S, Inoue Y, Kimura A. Importance of catalase in the adaptive response to hydrogen peroxide: analysis of acatalasaemic Saccharomyces cerevisiae. Biochem J. 1996;320:61–67. doi: 10.1042/bj3200061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamieson DJ. Saccharomyces cerevisiae has distinct adaptive responses to both hydrogen peroxide and menadione. J Bacteriol. 1992;174:6678–6681. doi: 10.1128/jb.174.20.6678-6681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HS, Seib PA, Chung OK. d-Erythroascorbic acid in baker’s yeast and effects on wheat dough. J Food Sci. 1993;58:845–862. [Google Scholar]
- Kosower NS, Kosower EM. Diamide: an oxidant probe for thiols. Methods Enzymol. 1995;251:123–133. doi: 10.1016/0076-6879(95)51116-4. [DOI] [PubMed] [Google Scholar]
- Kuge S, Jones N. YAP1 dependent activation of TRX2 is essential for the response of Saccharomyces cerevisiae to oxidative stress by hydroperoxides. EMBO J. 1994;13:655–664. doi: 10.1002/j.1460-2075.1994.tb06304.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lammers M, Follmann H. Deoxyribonucleotide biosynthesis in yeast (Saccharomyces cerevisiae). A ribonucleotide reductase system of sufficient activity for DNA synthesis. Eur J Biochem. 1984;140:281–287. doi: 10.1111/j.1432-1033.1984.tb08099.x. [DOI] [PubMed] [Google Scholar]
- Meister A. Glutathione-ascorbic acid antioxidant system in animals. J Biol Chem. 1994;269:9397–9400. [PubMed] [Google Scholar]
- Mieyal JJ, Starke DW, Gravina SA, Dothey C, Chung JS. Thioltransferase in human red blood cells: purification and properties. Biochemistry. 1991;30:6088–6097. doi: 10.1021/bi00239a002. [DOI] [PubMed] [Google Scholar]
- Minakuchi K, Yabushita T, Masumura T, Ichihara K, Tanaka K. Cloning and sequence analysis of a cDNA encoding rice glutaredoxin. FEBS Lett. 1994;337:157–160. doi: 10.1016/0014-5793(94)80264-5. [DOI] [PubMed] [Google Scholar]
- Miranda-Vizuete A, Rodriguez-Ariza A, Toribio F, Holmgren A, Lopez-Barea J, Pueyo C. The levels of ribonucleotide reductase, thioredoxin, glutaredoxin 1, and GSH are balanced in Escherichia coli K12. J Biol Chem. 1996;271:19099–19103. doi: 10.1074/jbc.271.32.19099. [DOI] [PubMed] [Google Scholar]
- Muller EGD. Thioredoxin deficiency in yeast prolongs S phase and shortens the G1 interval of the cell cycle. J Biol Chem. 1991;266:9194–9202. [PubMed] [Google Scholar]
- Muller EGD. Deoxyribonucleotides are maintained at normal levels in a yeast thioredoxin mutant defective in DNA synthesis. J Biol Chem. 1994;269:24466–24471. [PubMed] [Google Scholar]
- Muller EGD. A redox-dependent function of thioredoxin is necessary to sustain a rapid rate of DNA synthesis in yeast. Arch Biochem Biophys. 1995;318:356–361. doi: 10.1006/abbi.1995.1240. [DOI] [PubMed] [Google Scholar]
- Muller EGD. A glutathione reductase mutant of yeast accumulates high levels of oxidized glutathione and requires thioredoxin for growth. Mol Biol Cell. 1996;7:1805–1813. doi: 10.1091/mbc.7.11.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nick JA, Leung CT, Loewus FA. Isolation and identification of erythroascorbic acid in Saccharomyces cerevisiae and Lypomyces starkeyi. Plant Sci. 1986;46:181–187. [Google Scholar]
- Park JB, Levine M. Purification, cloning and expression of dehydrascorbic acid-reducing activity from human neutrophils: identification as glutaredoxin. Biochem J. 1996;315:931–938. doi: 10.1042/bj3150931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavachari N, Lou MF. Evidence for the presence of thioltransferase in the lens. Exp Eye Res. 1996;63:433–441. doi: 10.1006/exer.1996.0133. [DOI] [PubMed] [Google Scholar]
- Rice-Evans CA, Diplock AT, Symons MCR. In: Techniques in Free Radical Research. Burdon RH, Van Knippenberg PH, editors. Vol. 22. Amsterdam: Elsevier; 1991. pp. 227–230. [Google Scholar]
- Rose M, Botstein D. Construction and use of gene fusions to lacZ (β-galactosidase) which are expressed in yeast. Methods Enzymol. 1983;101:167–180. doi: 10.1016/0076-6879(83)01012-5. [DOI] [PubMed] [Google Scholar]
- Ruis H, Schuller C. Stress signaling in yeast. Bioessays. 1995;17:959–965. doi: 10.1002/bies.950171109. [DOI] [PubMed] [Google Scholar]
- Ruoppolo M, Lundstrom-Ljung J, Talamo F, Pucci P, Marino G. Effect of glutaredoxin and protein disulfide isomerase on the glutathione-dependent folding of ribonuclease A. Biochemistry. 1997;36:12259–12267. doi: 10.1021/bi970851s. [DOI] [PubMed] [Google Scholar]
- Schmitt ME, Brown TA, Trumpower BL. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990;18:3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Lawrence CW. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1974. [Google Scholar]
- Spyrou G, Holmgren A. Deoxyribonucleoside triphosphate pools and growth of glutathione-depleted 3T6 mouse fibroblasts. Biochem Biophys Res Commun. 1996;220:42–46. doi: 10.1006/bbrc.1996.0353. [DOI] [PubMed] [Google Scholar]
- Stephen DWS, Rivers SL, Jamieson DJ. The role of the YAP1 and YAP2 genes in the regulation of the adaptive oxidative stress responses of Saccharomyces cerevisiae. Mol Microbiol. 1995;16:415–423. doi: 10.1111/j.1365-2958.1995.tb02407.x. [DOI] [PubMed] [Google Scholar]
- Terada T. Thioltransferase can utilize cystamine the same as glutathione as a reductant during the restoration of cystamine-treated glucose 6-phosphate dehydrogenase activity. Biochem Mol Biol Int. 1994;34:723–727. [PubMed] [Google Scholar]
- Terada T, Oshida T, Nishimura M, Maeda H, Hara T, Hosomi S, Mizoguchi T, Nishihara T. Study on human erythrocyte thioltransferase: comparative characterization with bovine enzyme and its physiological role under oxidative stress. J Biochem. 1992;111:688–692. doi: 10.1093/oxfordjournals.jbchem.a123819. [DOI] [PubMed] [Google Scholar]
- Thomas JA, Poland B, Honzatko R. Protein sulphydryls and their role in the antioxidant function of protein S-thiolation. Arch Biochem Biophys. 1995;319:1–9. doi: 10.1006/abbi.1995.1261. [DOI] [PubMed] [Google Scholar]
- Thompson JD, Higgins GD, Gibson TJ. ClustalW: Improving sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandeputte C, Guizon I, Genestie-Denis I, Vannier B, Lorenzon G. A microtiter plate assay for total glutathione and glutathione disulfide contents in cultured/isolated cells: performance study of a new miniaturized protocol. Cell Biol Toxicol. 1994;10:415–421. doi: 10.1007/BF00755791. [DOI] [PubMed] [Google Scholar]
- Vlamis-Gardikas A, Aslund F, Spyrou G, Bergman T, Holmgren A. Cloning, overexpression, and characterization of glutaredoxin 2, an atypical glutaredoxin from Escherichia coli. J Biol Chem. 1997;272:11236–11243. doi: 10.1074/jbc.272.17.11236. [DOI] [PubMed] [Google Scholar]
- Wells WW, Xu DP, Yang Y, Rocque PA. Mammalian thioltransferase (glutaredoxin) and protein disulfide isomerase have dehydroascorbate reductase activity. J Biol Chem. 1990;265:15361–15364. [PubMed] [Google Scholar]
- Wells WW, Yang Y, Deits TL. Thioltransferases. Adv Enzymol. 1993;66:149–199. doi: 10.1002/9780470123126.ch4. [DOI] [PubMed] [Google Scholar]
- Wenzel TJ, Teunissen AWRH, Steensma HY. PDA1 mRNA: a standard for quantitation of mRNA in Saccharomyces cerevisiae superior to ACT1 mRNA. Nucleic Acids Res. 1995;23:883–884. doi: 10.1093/nar/23.5.883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu A, Moye-Rowley WS. GSH1, which encodes γ-glutamylcysteine synthetase, is a target gene for yAP-1 transcriptional regulation. Mol Cell Biol. 1994;14:5832–5839. doi: 10.1128/mcb.14.9.5832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia T-H, Bushweller JH, Sodano P, Billeter M, Bjornberg O, Holmgren A, Wuthrich K. NMR structure of oxidized Escherichia coli glutaredoxin: comparison with reduced E. coli glutaredoxin and related proteins. Protein Sci. 1992;1:310–321. doi: 10.1002/pro.5560010302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshitake S, Nanri H, Fernando MR, Minakimi S. Possible differences in the regenerative roles played by thioltransferase and thioredoxin of oxidatively damaged proteins. J Biochem. 1994;116:42–46. doi: 10.1093/oxfordjournals.jbchem.a124500. [DOI] [PubMed] [Google Scholar]