Abstract
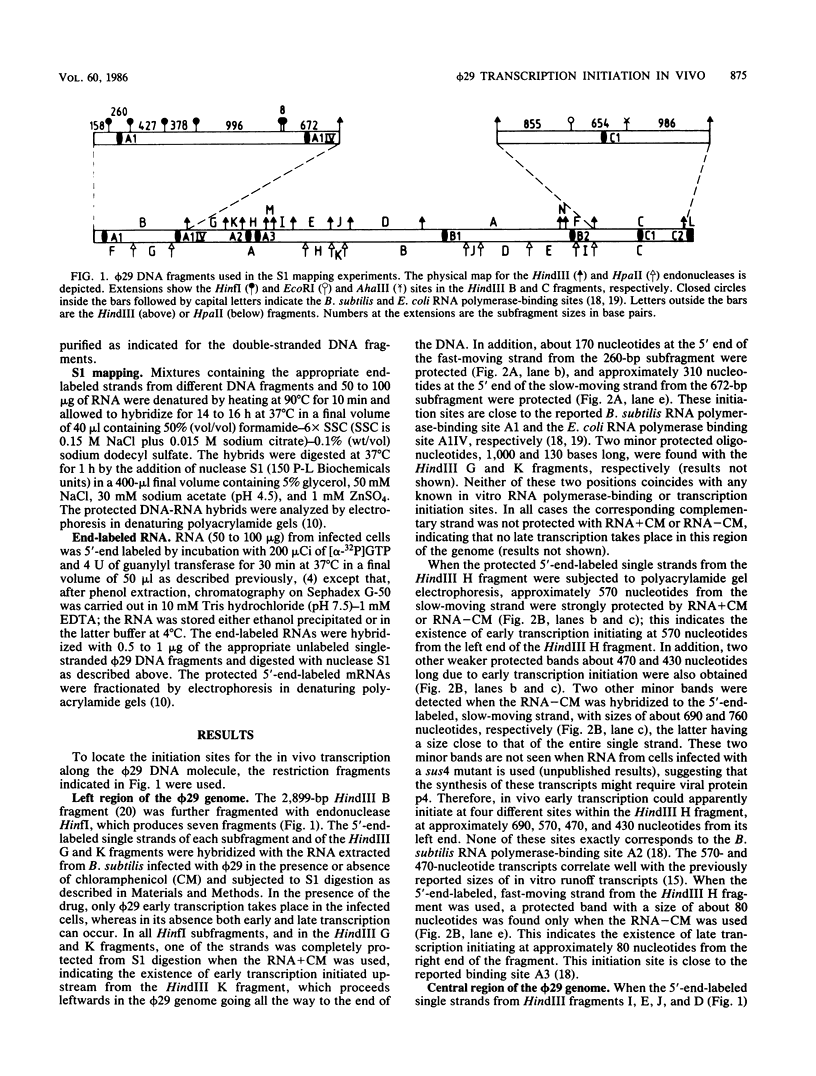
The initiation sites of the RNA transcripts synthesized in vivo in Bacillus subtilis infected with bacteriophage phi 29 have been mapped by S1 protection experiments. Nine transcription initiation sites were localized along the entire phi 29 genome, close to previously reported B. subtilis and Escherichia coli RNA polymerase-binding sites. Eight of these sites corresponded to early transcription and only one corresponded to late transcription. By using 5'-end-labeled RNA, four of the early sites and the late one were shown to be the main sites where initiation of transcription occurs in vivo in the phi 29 genome.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Blanco L., García J. A., Salas M. Cloning and expression of gene 2, required for the protein-primed initiation of the Bacillus subtilis phage phi 29 DNA replication. Gene. 1984 Jul-Aug;29(1-2):33–40. doi: 10.1016/0378-1119(84)90163-x. [DOI] [PubMed] [Google Scholar]
- Carrascosa J. L., Camacho A., Moreno F., Jiménez F., Mellado R. P., Viñuela E., Salas M. Bacillus subtilis phage phi29. Characterization of gene products and functions. Eur J Biochem. 1976 Jul 1;66(2):229–241. doi: 10.1111/j.1432-1033.1976.tb10512.x. [DOI] [PubMed] [Google Scholar]
- Christie G. E., Calendar R. Bacteriophage P2 late promoters. Transcription initiation sites for two late mRNAs. J Mol Biol. 1983 Jul 15;167(4):773–790. doi: 10.1016/s0022-2836(83)80110-7. [DOI] [PubMed] [Google Scholar]
- Coleman J., Green P. J., Inouye M. The use of RNAs complementary to specific mRNAs to regulate the expression of individual bacterial genes. Cell. 1984 Jun;37(2):429–436. doi: 10.1016/0092-8674(84)90373-8. [DOI] [PubMed] [Google Scholar]
- Coleman J., Hirashima A., Inokuchi Y., Green P. J., Inouye M. A novel immune system against bacteriophage infection using complementary RNA (micRNA). Nature. 1985 Jun 13;315(6020):601–603. doi: 10.1038/315601a0. [DOI] [PubMed] [Google Scholar]
- Escarmís C., Salas M. Nucleotide sequence at the termini of the DNA of Bacillus subtilis phage phi 29. Proc Natl Acad Sci U S A. 1981 Mar;78(3):1446–1450. doi: 10.1073/pnas.78.3.1446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escarmís C., Salas M. Nucleotide sequence of the early genes 3 and 4 of bacteriophage phi 29. Nucleic Acids Res. 1982 Oct 11;10(19):5785–5798. doi: 10.1093/nar/10.19.5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvey K. J., Yoshikawa H., Ito J. The complete sequence of the Bacillus phage phi 29 right early region. Gene. 1985;40(2-3):301–309. doi: 10.1016/0378-1119(85)90053-8. [DOI] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65(1):499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- Mellado R. P., Barthelemy I., Salas M. In vitro transcription of bacteriophage phi 29 DNA. Correlation between in vitro and in vivo promoters. Nucleic Acids Res. 1986 Jun 25;14(12):4731–4741. doi: 10.1093/nar/14.12.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado R. P., Delius H., Klein B., Murray K. Transcription of sea urchin histone genes in Escherichia coli. Nucleic Acids Res. 1981 Aug 25;9(16):3889–3906. doi: 10.1093/nar/9.16.3889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado R. P., Moreno F., Viñuela E., Salas M., Reilly B. E., Anderson D. L. Genetic analysis of bacteriophage phi 29 of Bacillus subtilis: integration and mapping of reference mutants of two collections. J Virol. 1976 Aug;19(2):495–500. doi: 10.1128/jvi.19.2.495-500.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellado R. P., Salas M. High level synthesis in Escherichia coli of the Bacillus subtilis phage phi 29 proteins p3 and p4 under the control of phage lambda PL promoter. Nucleic Acids Res. 1982 Oct 11;10(19):5773–5784. doi: 10.1093/nar/10.19.5773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray C. L., Rabinowitz J. C. Nucleotide sequences of transcription and translation initiation regions in Bacillus phage phi 29 early genes. J Biol Chem. 1982 Jan 25;257(2):1053–1062. [PubMed] [Google Scholar]
- Salas M. A new mechanism for the initiation of replication of phi 29 and adenovirus DNA: priming by the terminal protein. Curr Top Microbiol Immunol. 1984;109:89–106. doi: 10.1007/978-3-642-69460-8_4. [DOI] [PubMed] [Google Scholar]
- Schachtele C. F., De Sain C. V., Anderson D. L. Transcription during the development of bacteriophage phi29: definition of "early" and "late" phi29 ribonucleic acid. J Virol. 1973 Jan;11(1):9–16. doi: 10.1128/jvi.11.1.9-16.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sogo J. M., Inciarte M. R., Corral J., Viñuela E., Salas M. RNA polymerase binding sites and transcription map of the DNA of Bacillus subtilis phage phi29. J Mol Biol. 1979 Feb 5;127(4):411–436. doi: 10.1016/0022-2836(79)90230-4. [DOI] [PubMed] [Google Scholar]
- Sogo J. M., Lozano M., Salas M. In vitro transcription of the Bacillus subtilis phage phi 29 DNA by Bacillus subtilis and Escherichia coli RNA polymerases. Nucleic Acids Res. 1984 Feb 24;12(4):1943–1960. doi: 10.1093/nar/12.4.1943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshikawa H., Ito J. Nucleotide sequence of the major early region of bacteriophage phi 29. Gene. 1982 Mar;17(3):323–335. doi: 10.1016/0378-1119(82)90149-4. [DOI] [PubMed] [Google Scholar]