Abstract
Introduction
Many genes are differentially expressed between androgen-dependent and androgen-independent prostate cancer (CaP). Differential expression analysis and subtractive hybridization previously identified nine genes expressed in intact mice bearing CWR22 tumors and castrated mice bearing recurrent CWR22 tumors but not in regressed tumors. The objectives of this study were to develop an immunostaining method to dual-label foci of proliferating tumor cells [the origin of castration-recurrent CaP (CR-CaP)], to determine which of the nine candidate proteins were differentially expressed in proliferating versus nonproliferating cells at the onset of growth after castration, and to test preclinical findings using clinical specimens of androgen-stimulated benign prostate (AS-BP) and CaP (AS-CaP) and CR-CaP.
Methods
Paraffin-embedded, bromodeoxyuridine-injected CWR22 tumors were hydrated, antigen-retrieved using high heat and high pressure, labeled for each of the nine antigens of interest, visualized using peroxidase, and counterstained with hematoxylin. Mean optical density was calculated for proliferating and nonproliferating areas using automated (nuclear staining) or manual (cytoplasmic staining) image analysis. Prostate tissue microarray sections were immunostained and visually scored.
Results
Immunohistochemistry revealed higher nuclear expression of thioredoxin reductase 1 (TrxR1) in proliferating cells than nonproliferating cells (P < .005). There were no statistical differences between cell types in the expression of other proteins. TrxR1 expression was higher (P < .01) in CR-CaP compared with AS-BP or AS-CaP.
Conclusions
Increased TrxR1 expression in CR-CaP was consistent with increased TrxR1 and BrdU expression at the onset of growth in the CWR22 model. Thioredoxin reductase 1 should be targeted in an attempt to delay or prevent CaP recurrence after castration.
Introduction
Prostate cancer (CaP) remains the most common malignancy and second leading cause of cancer deaths among males in the United States [1]. Prostate cancer is androgen-dependent, and its growth is mediated by an androgen receptor (AR)-regulated gene network. Androgen deprivation therapy causes reduced AR expression [2], apoptosis, decreased cell volume [3], and decline of serum prostate-specific antigen (PSA). However, CaP eventually develops the capacity for recurrent growth in the absence of testicular androgens. Many research investigators are comparing androgen-dependent and castration-recurrent CaP (CR-CaP) in clinical specimens or androgen-sensitive and androgen-independent CaP cell lines using molecular approaches. These experiments may identify many genes whose expression is altered, only a few of which are critical for the development of castration-recurrent growth.
CWR22 is an androgen-dependent human CaP xenograft propagated subcutaneously in nude mice. CWR22 resembles most human CaP; CWR22 secretes PSA, undergoes tumor regression after androgen deprivation therapy, and recurs as a palpable, growing, and ultimately lethal tumor after several months without testicular androgens [4–7]. In an earlier study [8], differential expression analysis was used to identify gene transcripts that were down-regulated after castration but up-regulated in castration-recurrent CWR22 despite the continued absence of testicular androgens. Androgen-regulated transcripts included human kallikrein 2 (hk2), Nkx 3.1, α-tubulin, α-enolase, and insulin-like growth factor binding protein 5 (IGFBP-5). Kim et al. [9] used MIB-1 detection of Ki-67 and automated image analysis in paraffin sections of CWR22 tumors to determine the onset of cellular proliferation after castration. The onset of cellular proliferation on day 90 coincided with an increase in serum PSA. An increase in cellular proliferation was first observed on day 64 after castration. The appearance of proliferating tumors that expressed PSA indicated that these foci might be the precursors of CR-CaP tumors. In a subsequent study [10], subtractive hybridization identified four gene transcripts expressed in intact mice bearing CWR22 tumors and castrated mice bearing recurrent tumors but not in regressed tumors. Northern analysis confirmed temporal association with tumor growth for these candidates: three were proliferation-associated and not androgen-related [tomoregulin, translation elongation factor 1α (EF-1α), Mxi-1] and one was proliferation-associated and androgenregulated [thioredoxin reductase 1 (TrxR1)].
Computer-assisted quantitative image analysis and immunochemistry have enabled in situ protein localization and protein quantification in formalin-fixed, paraffin-embedded tissue. Video image analysis has been used to quantitate AR expression more precisely than visual scoring [11,12]. Mean optical density (MOD) measurements made using automated image analysis successfully quantified the dependence of AR protein levels on serum androgen levels in the CWR22 model [9]. Immunohistochemistry can colocalize proteins of interest and cellular proliferation markers within the same cell.
The objective of this study was to develop an immunostaining protocol to sequentially dual-label (peroxidase and fluorescence) CWR22 tumors without diminishing the previously labeled protein and to use computer-assisted image analysis to determine which of the nine candidate proteins was differentially expressed in proliferating versus nonproliferating cells 64 days after castration. The protein of interest would then be examined for overexpression in both androgen-stimulated CaP (obtained from radical prostectomy specimens) and CR-CaP (obtained from transurethral resection specimens from men with urinary retention due to CaP recurrence during androgen deprivation therapy).
Materials and Methods
CWR22 Tumors
CWR22 tumors 64 days after castration demonstrated larger foci of proliferation than reported previously [9]. To study the earliest onset of cellular proliferation after castration CWR22 tumors were studied 50 days after castration.
Twenty nude mice were subcutaneously implanted with CWR22 cell suspensions containing 1 million cells bilaterally as described [5,10,13]. Mice bearing castration-recurrent CWR22 tumors were injected with 2.0 mg of bromodeoxyuridine (BrdU) per animal (Roche, Palo Alto, CA) for 4 hours and killed by cervical dislocation. Tumors were resected and immediately formalin-fixed and paraffin-embedded. The Institutional Animal Care and Use Committee of the University of North Carolina at Chapel Hill approved all procedures used.
Immunohistochemistry
Formalin-fixed, paraffin-embedded CWR22 tumors were cut into 6-µm histologic sections. The sections were deparaffinized, rehydrated through an alcohol gradient, and antigen-retrieved using Citra buffer (Biocare, Walnut Creek, CA) for 3 minutes at 120°C [14]. Cooled sections were incubated with a serum block, blocked for endogenous peroxidase using 3% H2O2 for 5 minutes at 37°C, and blocked with avidin biotin for 15 minutes at 37°C (Vector, Burlingame, CA). Sections were incubated with either mouse monoclonal anti-hk2 (Mayo Clinic, Rochester, MN), goat polyclonal anti-Nkx 3.1 (Santa Cruz Biotechnology, Santa Cruz, CA), goat polyclonal anti-IGFBP-5 (Santa Cruz Biotechnology), rat monoclonal anti-α-tubulin (Abcam, Cambridge, UK), rabbit monoclonal anti-non-neuronal enolase (NNE; Accurate Chemical, Westbury, NY), rabbit polyclonal anti-TrxR1 (Upstate, Charlottesville, VA), mouse monoclonal antitomoregulin (Sakamoto Laboratory, Japan), mouse monoclonal anti-EF-1α (Upstate), or mouse monoclonal anti-Mxi-1 (BD Pharmigen, San Jose, CA) at 1:200 for 1 hour at 37°C. Sections were incubated with the appropriate biotinylated secondary IgG (Vector) 1:200 for 15 minutes at 37°C and ABC amplification (Vector) 1:200 for 15 minutes at 37°C. Specific signal was visualized using diaminobenzidine (Vector) 1:200 for 5 minutes at 37°C. Sections were counterstained with hematoxylin. Single-labeled sections were antigen-retrieved a second time using high-pH Tris buffer (Biocare) for 3 minutes at 120°C to expose DNA-incorporated BrdU [15]. Cooled slides were labeled with sheep polyclonal anti-BrdU (Abcam) at 1:200 for 1 hour at 37°, followed by rhodamine-conjugated F(ab′)2 fragment (Jackson ImmunoResearch, West Grove, PA) at 1:200 for 15 minutes at 37°C. A fluorescent nuclear dye (4′,6-diamidino-2-phenylindole) was used for proteins expressed in the cytoplasm.
Tissue Microarray
A high-density tissue microarray (TMA) was constructed using formalin-fixed, paraffin-embedded human prostate specimens. The tissue microarray was constructed from matched pairs of androgen-stimulated benign prostate (AS-BP) and CaP (AS-CaP) from radical prostatectomy specimens from 23 men and CR-CaP from transurethral resection specimens from 22 men using the Beecher Instruments (Silver Spring, MD) manual tissue arrayer. Androgen-stimulated benign prostate obtained by transurethral resection, colon cancer, and mouse liver cores were included as internal controls for standardization. Six-micrometer sections were cut from donor paraffin blocks and stained with hematoxylin and eosin. A urologic pathologist evaluated the tissue sections and identified BP and CaP. The regions of interest were sampled by removing a 1.5-mm tissue core. These cores were implanted into a recipient paraffin block to create a TMA containing a total of 84 tissue cores.
Immunohistochemistry
A 6-µm histologic section was cut from the TMA and was deparaffinized, rehydrated through an alcohol gradient, and antigen-retrieved using a Citra buffer (Biocare) for 3 minutes at 120°C. The cooled section was incubated with a serum block, blocked for endogenous peroxidase using 0.03% H2O2 for 5 minutes at 37°C followed by a block of avidin biotin for 15 minutes at 37°C (Vector). The section was incubated with anti-TrxR1 1:200 for 1 hour at 37°C [10]. The section was incubated with biotinylated antimouse IgG (Vector) 1:200 for 15 minutes at 37°C and ABC amplification (Vector) 1:200 for 15 minutes at 37°C. Specific signal was visualized using diaminobenzidine (Vector) 1:200 for 5 minutes at 37°C. The section was counterstained with hematoxylin.
Image Acquisition and Analysis
The image acquisition system consisted of a Leica DMRA2 microscope (Leica Microsystems Inc, Bannockburn, IL) with a motorized stage controller (Ludl Electronic Products Ltd, Hawthorne, NY), a three-chip CCD camera with controller (Hamamatsu, Bridgewater, NJ), and a FlashPoint 3D image grabber card (Integral Technologies, Indianapolis, IN) in a Pentium 4-based personal computer. Image Pro Plus 4.5 (MediaCybernetics, Carlsbad, CA) software was used to capture and store the images. Images with 24-bit color depth and spatial resolution of 640 x 480 pixels were stored as uncompressed TIFF files.
Nuclear immunostaining
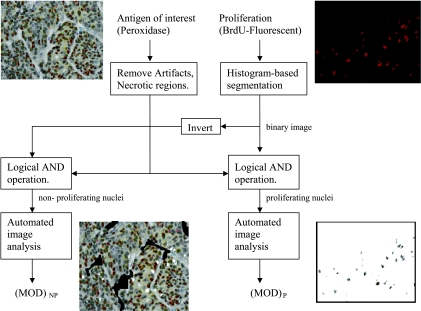
A flowchart illustrated the image analysis steps involved when the protein of interest was expressed in nuclei (TrxR1, Nkx 3.1, IGFBP-5, and tomoregulin; Figure 1). The BrdU-immunostained fluorescent image and the peroxidase image (immunostained with protein of interest) were captured from the same location in the specimen. Stromal areas and regions of artifact were removed from the peroxidase image. The fluorescent image was used to create nuclear masks of proliferating nuclear areas and all nuclear areas. Histogram-based segmentation (Image Pro Plus 4.5) was used to create a binary mask. The threshold was selected to make nuclear boundaries visible. The mask was added to the peroxidase image to create a proliferation image. The mask from proliferating regions was inverted and added to the mask from all nuclear areas before addition to peroxidase image to create a nonproliferation image. Mean optical density was calculated for proliferating and nonproliferating images using automated nuclear image analysis [12]. The parameters for image analysis were determined using a macro in Image Pro Plus 4.5. The image background was set at white (grayscale level = 255) for MOD calculations.
Figure 1.
Flowchart illustrating computer-assisted image analysis for images where protein of interest is expressed in nuclei. NP indicates nonproliferating; P, proliferating.
Cytoplasmic immunostaining
Automated nuclear analysis software could not be used when the protein of interest was expressed in the cytoplasm (hk2, α-tubulin, NNE, EF-1α, Mxi-1). The analysis was performed manually by visually selecting cytoplasmic areas adjacent to the identified nuclear regions in proliferating and nonproliferating images. Mean optical density was calculated from those regions using Image Pro Plus 4.5.
Visual scoring
Epithelial and stromal regions from AS-BP, ASCaP, and CR-CaP cores on prostate TMA sections were visually scored from 0 (staining same as background) to 4 (intense staining).
Statistical Analysis
Student's paired t tests were performed on the MOD data sets from proliferating versus nonproliferating nuclei or cytoplasm to determine whether the differences between image pairs was statistically significant. A comparison of means (Student's t test) was performed to determine whether the difference in visual scores from epithelia or stroma among different tissue types was statistically significant.
Results
Bromodeoxyuridine labeling required DNA denaturing with high-pH buffer that did not degrade the peroxidase label, but BrdU labeling interfered with peroxidase immunostaining when performed first. Super heating, high-pressure antigen retrieval was used twice on the same slide at different pH. Antiproteins were optimized using highheat, high-pressure antigen retrieval and neutral pH Citra buffer.
The expression of four nuclear and five cytoplasmic proteins were quantified in castration-recurrent CWR22 tumors 50 days after castration (Table 1). Thioredoxin reductase 1 was expressed at higher levels in proliferating cells than in nonproliferating cells (P < .005). The expression of hk2, Nkx 3.1, IGFBP-5, α-tubulin, NNE, tomoregulin, EF-1α, and Mxi-1 was similar between proliferating and nonproliferating cells.
Table 1.
Expression of Nine Proteins in Castration Recurrent CWR22 Tumors.
| Proteins | Subcellular Localization | Dual Method Using Quantitative Image Analysis | |
|---|---|---|---|
| MOD of High/Low Proliferation | P* | ||
| hk2 | C | 0.212/0.213 | .473 |
| Nkx 3.1 | N | 0.891/0.913 | .149 |
| IGFBP-5 | N | 0.937/0.938 | .821 |
| α-tubulin | C | 0.410/0.425 | .094 |
| NNE | C | 0.366/0.356 | .065 |
| TrxR1 | N | 1.034/0.963 | .003 |
| Tomoregulin | N | 1.226/1.235 | .685 |
| EF-1α | C | 0.208/0.209 | .056 |
| Mxi-1 | C | 0.193/0.192 | .179 |
P value in bold indicates significance.
C indicates cytoplasmic; N, nuclear.
Student's t test for paired samples computed for 95% confidence interval.
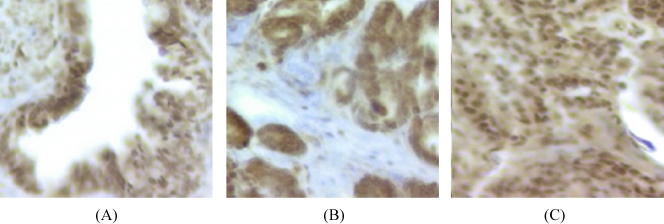
A total of 25 cores (9 benign prostatic hyperplasia, 8 AS-CaP, and eight recurrent CaP) were found unsuitable for analysis on the prostate TMA. Qualitative visual assessment demonstrated that TrxR1 was expressed at higher levels in CR-CaP than in AS-BP and AS-CaP (Figure 2). Visual scoring revealed similar levels of immunostaining in epithelia and stroma of AS-BP and AS-CaP tissue specimens (Table 2). Castration-recurrent CaP exhibited higher levels of immunostaining in both epithelia and stroma.
Figure 2.
TrxR1 immunostaining in (A) AS-BP, (B) AS-CaP, and (C) CR-CaP tissue specimens.
Table 2.
Comparison of TrxR1 Expression in AS-BP, AS-CaP, and CR-CaP in Prostate TMA.
| Epithelia | Stroma | P | |
|---|---|---|---|
| AS-BP | 2.88 ± 0.50 | 1.69 ± 0.48 | >.05 |
| AS-CaP | 2.59 ± 0.62 | 1.35 ± 0.49 | >.05 |
| CR-CaP | 3.47 ± 1.07 | 2.18 ± 1.13 | <.01 |
Student's t test computed at 95% confidence interval.
Discussion
To make direct comparison of protein expression in proliferating versus nonproliferating cells, a total of 7 years were invested to evaluate different approaches. The goal was an immunostaining protocol that allowed dual labeling of either Ki-67 or BrdU (primary label) and other proteins of interest (second label). Recent publications described the unique ability of high-heat, high-pressure antigen retrieval methods to yield good immunostaining results [14,16]. The effect of repeated high-pressure antigen retrieval using differing pH buffers on immunostaining quality was concerning, but the second high-pH retrieval produced no noticeable loss of intensity of peroxidase immunostaining. High-pressure antigen retrieval was reported to increase endogenous peroxidase activity and nonspecific staining [17] and nonspecific background increased slightly after two sequences of high-pressure retrieval. Therefore, peroxidase was labeled before the second antigen retrieval, which made the hydrogen peroxide blocking step sufficient to quench this activity. A hybrid peroxidase/fluorescent dual-labeling protocol was developed to derive robust images for image analysis. Cell proliferation was determined using fluorescent-labeled BrdU. The proliferating cells were identified using fluorescence that did not interfere with image analysis for the peroxidase-labeled primary. The method was demonstrated for simultaneous assessment of proliferation and protein expression.
Thioredoxin (Trx), and thioredoxin reductase (TrxR) comprise the thioredoxin system, which exists in nearly all living cells. The system is crucial for the maintenance of reduced intracellular redox environment, cellular growth, defense against oxidative stress, and control of apoptosis [18]. The function of Trx as a disulfide reductase in mammalian cells depends on the activity of the selenoenzyme, TrxR [19], which catalyzes the nicotinamide adenine dinucleotide phosphate-dependent reduction of active disulfide sites of Trx to a di-thiol. Humans have three isoforms of TrxRs, namely TrxR1, TrxR2, and TrxR3. Although Trx is predominantly cytosolic, it quickly translocates into the nucleus in response to NF-κB activation by UVB irradiation or tumor necrosis factor α treatment [20]. In this study, TrxR1 expression was predominantly nuclear in CWR22 tumors 50 days after castration. The reasons for nuclear expression of TrxR1 in CWR22 tumors require further study.
Thioredoxin increases the DNA-binding activity of number of transcription factors, including AP-1, AP-2, nuclear receptors, including glucocorticoid and estrogen receptors [21]. A variety of potent TrxR inhibitors has been shown to alter the cancer-related properties of tumors and malignant cells. For example, 1,2-[bis (2-benzysoselenazolone-3(2H)-ketone)]ethane, reversed the phenotype of five human leukemia cell lines [22]. Yoo et al. [23] used TrxR1 knockdown in a mouse cancer cell line driven by the oncogene k-ras to demonstrate the need for TrxR1 expression for cancer growth. Reduction of TrxR activity in human hepatocellular carcinoma cells by transfection of TrxR antisense RNA inhibited cell growth [24]. Thioredoxin expression was also found to be associated with lymph node status and prognosis in early operable non-small cell lung cancer [25]. Baker et al. [26] found Trx-binding protein, a possible tumor-suppressor gene, when induced in response to hypoxia in pancreatic cancer cells resulted in increased apoptosis and increased sensitivity to platinum anticancer therapy. Increased Trx expression in human colorectal cancer was found to be associated with decreased patient survival [27]. Thioredoxin reductase 1 has therefore been implicated as a potential target for cancer therapy [28].
Among nine proteins differentially expressed in androgen-stimulated versus castration-recurrent CWRtumors, only TrxR1 was differentially expressed in proliferation versus nonproliferating cells at the onset of recurrent growth 50 days after castration. Rigorous testing was made possible by the development of a dual immunostaining method and quantitative image analysis. The CWR22 model was used to associate TrxR1 with the earliest possible onset of castration-recurrent growth. Increase in TrxR1 expression in human CR-CaP specimens is consistent with increased TrxR1 and BrdU expression in the CWR22 model. Thioredoxin reductase 1 should be targeted to delay or prevent CaP recurrence after castration.
Footnotes
Supported by Department of Defense Prostate Cancer Research Program grant DAMD 01-1-0340, National Cancer Institute (NCI) grant PO1-CA-77739 and, in part, by the NCI Cancer Center Support Grant to Roswell Park Cancer Institute CA016156.
References
- 1.Jemal A, Siegal R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics 2008. CA Cancer J Clin. 2008;58:71–96. doi: 10.3322/CA.2007.0010. [DOI] [PubMed] [Google Scholar]
- 2.Prins GS, Birch L. Immunocytochemical analysis of androgen receptor along the ducts of the separate rat prostate lobes after androgen withdrawal and replacement. Endocrinology. 1993;132:169–178. doi: 10.1210/endo.132.1.8419121. [DOI] [PubMed] [Google Scholar]
- 3.Kyprianou N, Isaacs JT. Activation of programmed cell death in the rat ventral prostate after castration. Endocrinology. 1988;122:552–562. doi: 10.1210/endo-122-2-552. [DOI] [PubMed] [Google Scholar]
- 4.Pretlow TG, Wolman SR, Micale MA, Pelley RJ, Kursh ED, Resnick MI, Bodner DR, Jacobberger JW, Delmoro CM, Giaconia JM. Xenografts of primary human prostatic carcinoma. J Natl Cancer Inst. 1993;85:394–398. doi: 10.1093/jnci/85.5.394. [DOI] [PubMed] [Google Scholar]
- 5.Wainstein MA, He F, Robinson D, Kung H-J, Schwartz S, Giaconia JM, Edgehouse NL, Pretlow TP, Bodner DR, Kursh ED, et al. CWR22: androgen-dependent xenograft model derived from a primary human prostatic carcinoma. Cancer Res. 1994;54:6049–6052. [PubMed] [Google Scholar]
- 6.Nagabhushan M, Miller CM, Pretlow TP, Giaconia JM, Edgehouse NL, Schwartz S, Kung HJ, de Vere White RW, Gumerlock PH, Resnick MI, et al. CWR22: the first human prostate cancer xenograft with strongly androgen-dependent and relapsed strains both in vivo and in soft agar. Cancer Res. 1996;56:3042–3046. [PubMed] [Google Scholar]
- 7.Tan J-A, Sharief Y, Hamil KG, Gregory CW, Zang D-Y, Sar M, Gumerlock PH, de Vere White RW, Pretlow TG, Harris SE, et al. Dehydroepiandrosterone activates mutant androgen receptors expressed in the androgen-dependent human prostate cancer xenograft CWR22 and LNCaP cells. Mol Endocrinol. 1997;11:450–459. doi: 10.1210/mend.11.4.9906. [DOI] [PubMed] [Google Scholar]
- 8.Gregory CW, Hamil KG, Kim D, Hall SH, Pretlow TG, Mohler JL, French FS. Androgen receptor expression in androgen-independent prostate cancer is associated with increased expression of androgen-regulated genes. Cancer Res. 1998;58:5718–5724. [PubMed] [Google Scholar]
- 9.Kim D, Gregory CW, French FS, Smith GJ, Mohler JL. Androgen receptor expression and cellular proliferation during transition from androgen-dependent to recurrent growth after castration in the CWR22 prostate cancer xenograft. Am J Pathol. 2002;160:219–226. doi: 10.1016/S0002-9440(10)64365-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mohler JL, Morris TL, Ford OH, III, Alvey RF, Sakamoto C, Gregory CW. Identification of differentially expressed genes associated with androgenindependent growth of prostate cancer. Prostate. 2002;51:247–255. doi: 10.1002/pros.10086. [DOI] [PubMed] [Google Scholar]
- 11.Prins GS, Sklarew RJ, Pertschuk LP. Image analysis of androgen receptor immunostaining in prostate cancer accurately predicts response to hormonal therapy. J Urol. 1998;159:641–649. [PubMed] [Google Scholar]
- 12.Kim D, Gregory CW, Smith GJ, Mohler JL. Immunohistochemical quantification of androgen receptor expression using color video image analysis. Cytometry. 1999;35:2–10. doi: 10.1002/(sici)1097-0320(19990101)35:1<2::aid-cyto2>3.3.co;2-p. [DOI] [PubMed] [Google Scholar]
- 13.Gregory CW, Johnson RT, Jr, Presnell SC, Mohler JL, French FS. Androgen receptor regulation of G1 cyclin and cyclin-dependent kinase function in the CWR22 human prostate cancer xenograft. J Androl. 2001;22:537–548. [PubMed] [Google Scholar]
- 14.Leong AS, Lee ES, Yin H, Kear M, Haffajee Z, Pepperall D. Super-heating antigen retrieval. Appl Immunohistochem Mol Morphol. 2002;10:263–268. doi: 10.1097/00129039-200209000-00014. [DOI] [PubMed] [Google Scholar]
- 15.Shi SR, Cote RJ, Wu L, Liu C, Datar R, Shi Y, Liu D, Lim H, Taylor CR. DNA extraction from archival formalin-fixed, paraffin-embedded tissue sections based on the antigen retrieval principle: heating under the influence of pH. J Histochem Cytochem. 2002;50:1005–1011. doi: 10.1177/002215540205000802. [DOI] [PubMed] [Google Scholar]
- 16.Shi SR, Cote RJ, Taylor CR. Antigen retrieval techniques: current perspectives. J Histochem Cytochem. 2001;49:931–937. doi: 10.1177/002215540104900801. [DOI] [PubMed] [Google Scholar]
- 17.Kim SH, Jung KC, Shin YK, Lee KM, Park YS, Choi YL, Oh KI, Kim MK, Chung DH, Son HG, et al. The enhanced reactivity of endogenous biotin-like molecules by antigen retrieval procedures and signal amplification with tyramine. Histochem J. 2002;34:97–103. doi: 10.1023/a:1020954611464. [DOI] [PubMed] [Google Scholar]
- 18.Arner ESJ, Holmgren A. The thioredoxin system in cancer. Semin Cancer Biol. 2006;16:420–426. doi: 10.1016/j.semcancer.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 19.Arner ES, Holmgren A. Physiological functions of thioredoxin and thioredoxin reductase. Eur J Biochem. 2000;267:6102–6109. doi: 10.1046/j.1432-1327.2000.01701.x. [DOI] [PubMed] [Google Scholar]
- 20.Hirota K, Murata M, Sachi Y, Nakamura H, Takeuchi JKM, Yodoi J. Distinct roles of thioredoxin in the cytoplasm and in nucleus. A two-step mechanism of redox regulation of transcription factor NF-kappaB. J Biol Chem. 1999;274:27891–27897. doi: 10.1074/jbc.274.39.27891. [DOI] [PubMed] [Google Scholar]
- 21.Nishinaka Y, Nakamura H, Masutani H, Yodoi J. Redox control of cellular function by thioredoxin: A new therapeutic direction in host defense. Arch Immunol Ther Exp. 2001;49:285–292. [PubMed] [Google Scholar]
- 22.Zhao F, Yan J, Deng S, Lan L, He F, Kuang B, Zeng H. A thioredoxin reductase inhibitor induces growth inhibition and apoptosis in five cultured human carcinoma cell lines. Cancer Lett. 2006;236:46–53. doi: 10.1016/j.canlet.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 23.Yoo MH, Xu XM, Carlson BA, Patterson AD, Gladyshev VN, Hatfield DL. Targeting thioredoxin reductase 1 reduction in cancer cells inhibits selfsuficient growth and DNA replication. PLoS ONE. 2007;2:e1112. doi: 10.1371/journal.pone.0001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gan L, Yang XL, Liu Q, Xu HB. Inhibitory effects of thioredoxin reductase antisense RNA on the growth of human hepatocellular carcinoma cells. J Cell Biochem. 2005;96:653–664. doi: 10.1002/jcb.20585. [DOI] [PubMed] [Google Scholar]
- 25.Kakolyris S, Giatromanolaki A, Koukourakis M, Powis G, Souglakos J, Sivridis E, Georgoulias V, Gatter KC, Harris AL. Thioredoxin expression is associated with lymph node status and prognosis in early operable non-small cell lung cancer. Clin Cancer Res. 2001;7:3087–3091. [PubMed] [Google Scholar]
- 26.Baker AF, Koh MY, Williams RR, James B, Wang H, Tate WR, Gallegos A, Von Hoff DD, Han H, Powis G. Identification of thioredoxin-interacting protein 1 as a hypoxia-inducible factor 1alpha-induced gene in pancreatic cancer. Pancreas. 2008;36:178–186. doi: 10.1097/MPA.0b013e31815929fe. [DOI] [PubMed] [Google Scholar]
- 27.Raffel J, Bhattacharyya AK, Gallegos A, Cui H, Einspahr JG, Alberts DS. Increased expression of thioredoxin-1 in human colorectal cancer is associated with decreased patient survival. J Lab Clin Med. 2003;142:46–51. doi: 10.1016/S0022-2143(03)00068-4. [DOI] [PubMed] [Google Scholar]
- 28.Biaglow JE, Miller RA. The thioredoxin reductase/thioredoxin system: novel redox targets for cancer therapy. Cancer Biol Ther. 2005;4(1):6–13. doi: 10.4161/cbt.4.1.1434. [DOI] [PubMed] [Google Scholar]