Prions are among the most mysterious creatures ever produced in nature. Lacking nucleic acid genomes and composed entirely of proteins, these infectious agents are not eliminated by any traditional sterilization procedures. Prions cause mad cow disease and related disorders in mammals including humans (1). It also appears that proteins with prion properties are widespread in nature and can be found in organisms that are very distant from mammals, for example, in yeast (2, 3). Although yeast prions are likely to be harmful to their hosts (3), they do not kill yeast cells outright and can be propagated through an indefinite number of generations. In this issue of PNAS, Chang et al. (4) use a yeast prion model for deciphering the protein regions that determine prion identities.
The majority of the researchers in the field agree that most prions are self-perpetuating amyloids, fibrous cross-β polymers that reproduce themselves via immobilizing soluble protein of the same sequence and converting it into a unit of polymer, that is, into a prion (2, 3). Then, everything seems simple: a prion is a polymer and a nonprion is a monomer. One major difficulty with this model is that prions generated by proteins of the same sequence are not necessarily the same.
The phenomenon of prion “strains” was first discovered in mammals, where supposedly one and the same protein may produce infectious agents with different biological (incubation period and host specificity) and biochemical (patterns of proteinase resistance) characteristics (5). This phenomenon is well characterized in yeast where strains (or “variants”) can be distinguished by phenotypic patterns, transmissibility in cell generations, and proportion of polymerized versus nonpolymerized protein (6, 7). Usually, “strong” strains are transmitted with higher efficiency and contain more protein in the polymerized state, whereas “weak” strains are transmitted with lower efficiency and contain less protein in the polymerized state. It is known now that major strain characteristics could be maintained through the in vitro stage by the prion protein alone (8, 9). What remains unknown is how they are maintained.
Chang et al. (4) attempted to identify the determinants of prion identity by systematic mutagenesis of the N-proximal region of yeast prion protein Sup35. This region is located within the so-called prion domain (PrD) that controls polymerization capabilities of Sup35. They have generated a set of mutants, each containing a substitution of a specific amino acid residue within this region to proline or glycine, known to destabilize amyloid structures. Mutant proteins were tested for their ability to acquire and maintain the prion state from various preexisting prion strains. The major conclusions are that, first, not the whole N-proximal region is required, and, second, different prion strains require fragments of different lengths. Depending on the strain, the length of the required fragment could vary from 15 to 46 amino acid residues.
The question arises as to whether or not the fragments essential for prion transmission are sufficient for the maintenance of prion strain identity. Although the 15-residue fragment was not enough for such a purpose, fragments as short as 40–50 residues could maintain the strain-specific patterns through the in vitro propagation stage, when fused to unrelated protein (GFP). Such a chimeric protein was able to faithfully transmit these patterns to full-size Sup35 protein upon transfection into the yeast cell. It is truly amazing to see that such a short fragment constituting only 6–7% of total Sup35 protein is all that is needed for “remembering” and reproducing a complex ordered structure and imposing it on the complete Sup35 protein.
Overall, these data complement and further extend the results of other laboratories using different approaches. Shkundina et al. (10) have reported that large fragments of Sup35 PrD are needed to faithfully transmit specific characteristics of weak strains in vivo, compared with strong strains. Toyama et al. (11) have found that a larger piece of Sup35 PrD is protected from hydrogen exchange (and therefore presumably involved in the formation of the axis of an amyloid fiber) in amyloids generated by weak strains in vitro, compared with strong strains. Remarkably, the strain requiring the shortest region for prion conversion in the work by Chang et al. (4) is apparently the strongest one among those studied, if judged by phenotypic characteristics. Therefore, all of these data point in the same direction: the stronger the prion strain is, the shorter is the stretch that it requires for generating amyloid fibers. Size estimate variations between papers could easily be explained by the differences in experimental techniques.
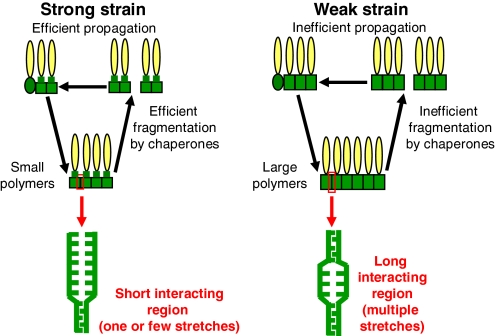
Notably, a larger interacting region would likely lead to a more rigid structure of amyloid fibers, making them more difficult to break. Efficient proliferation of yeast prions in vivo is occurring because of chaperone-mediated breakage of large fibers into the smaller oligomeric seeds, initiating new rounds of polymerization (2). If fibers are more rigid, fewer seeds are produced, resulting in less efficient prion transmission in vivo, fewer proteins in the polymerized state, and a larger polymer size (Fig. 1). All of these characteristics are indeed observed for weak prion strains (7).
Fig. 1.
The length of the interacting region and/or the number of interacting stretches determine strain-specific properties of yeast prions. PrDs are green, the rest of each monomeric unit is yellow. Ellipses correspond to nonprion conformation, and rectangles correspond to prion conformation. Note that according to existing models only PrD acquires prion conformation.
Thus, features that become identifiable only through interaction with the chaperone machinery of the cell are faithfully reproduced in vitro in the absence of any chaperone. Apparently, response to the chaperone action is controlled by the structural organization of an amyloid, which is reproducible both in vivo and in vitro. Indeed, Chang et al. (4) observed that amyloids seeded by different prion strains also exhibit different fiber morphologies in vitro. The ability of the short amino acid stretches to control and reproduce structural parameters of the huge fibers emphasizes the power of structural templating operating at the level of structural assemblies.
However, it would be premature to conclude that the mystery of prion strains is solved. First of all, one should be cautious when interpreting mutagenesis data. As proline or glycine substitutions can also destabilize adjacent structures, it is not guaranteed that every residue confined to the required region would necessarily have a crucial impact on its own. Amazingly, this could mean that actual determinants of prion identity are even shorter than those uncovered by Chang et al. (4), whereas longer regions identified by their approach could simply be composed of two or several structural elements each. Indeed, even mutagenesis data suggest that for one of the strains, the required region is composed of two “arms” with less important residues in between. Tools with higher power are needed to resolve this issue.
Prions generated by proteins of the same sequence are not necessarily the same.
Recently, Tessier and Lindquist (12) reported that most PrD-derived peptides, which are capable of binding the PrD-containing piece of Sup35 and promoting its conversion into an amyloid in vitro, include a fragment between amino acid residues 9 and 20. The role of this fragment in the determination of strain-specific binding properties was also proposed. This fragment is also present in all regions required by various strains in the work by Chang et al. (4) and includes “amyloid stretch,” a vague hexapeptide consensus identified in the vast majority of proteins capable of amyloid formation in vitro (13). Higher probability of generation of the short amyloidogenic stretches by various sequences with the same amino acid composition may explain why derivatives of the yeast prion proteins with reshuffled PrDs usually maintain prion-forming capabilities (14). Differences between prion strains formed by proteins of the same sequence could be explained by different numbers of potential amyloid stretches involved in amyloid formation in each case (Fig. 1).
Are we, then, close to deciphering the code of structural templating? Possibly, but not certainly. Even if we are on the right track, it will take the courage and persistence of investigators to identify the other sequences.
Acknowledgments.
Work on yeast prions in my laboratory is supported by National Institutes of Health Grant R01GM58763 and National Science Foundation Grant MCB 0614772.
Footnotes
The author declares no conflict of interest.
See companion article on page 13345.
References
- 1.Prusiner SB. Prions. Proc Natl Acad Sci USA. 1998;95:13363–13383. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chernoff YO. Amyloidogenic domains, prions, and structural inheritance: Rudiments of early life or recent acquisition? Curr Opin Chem Biol. 2004;8:665–671. doi: 10.1016/j.cbpa.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 3.Wickner RB, Edskes HK, Shewmaker F, Nakayashiki T. Prions of fungi: Inherited structures and biological roles. Nat Rev Microbiol. 2007;5:611–618. doi: 10.1038/nrmicro1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang H-Y, Lin J-Y, Lee H-C, Wang H-L, King C-Y. Strain-specific sequences required for yeast [PSI+] prion propagation. Proc Natl Acad Sci USA. 2008;105:13345–13350. doi: 10.1073/pnas.0802215105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Morales R, Abid K, Soto C. The prion strain phenomenon: Molecular basis and unprecedented features. Biochim Biophys Acta. 2007;1772:681–691. doi: 10.1016/j.bbadis.2006.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Derkatch IL, Chernoff YO, Kushnirov VV, Inge-Vechtomov SG, Liebman SW. Genesis and variability of [PSI] prion factors in Saccharomyces cerevisiae. Genetics. 1996;144:1375–1386. doi: 10.1093/genetics/144.4.1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tanaka M, Collins SR, Toyama BH, Weissman JS. The physical basis of how prion conformations determine strain phenotypes. Nature. 2006;442:585–589. doi: 10.1038/nature04922. [DOI] [PubMed] [Google Scholar]
- 8.King CY, Diaz-Avalos R. Protein-only transmission of three yeast prion strains. Nature. 2004;428:319–323. doi: 10.1038/nature02391. [DOI] [PubMed] [Google Scholar]
- 9.Tanaka M, Chien P, Naber N, Cooke R, Weissman JS. Conformational variations in an infectious protein determine prion strain differences. Nature. 2004;428:323–328. doi: 10.1038/nature02392. [DOI] [PubMed] [Google Scholar]
- 10.Shkundina IS, Kushnirov VV, Tuite MF, Ter-Avanesyan MD. The role of the N-terminal oligopep-tide repeats of the yeast Sup35 prion protein in propagation and transmission of prion variants. Genetics. 2006;172:827–835. doi: 10.1534/genetics.105.048660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Toyama BH, Kelly MJ, Gross JD, Weissman JS. The structural basis of yeast prion strain variants. Nature. 2007;449:233–237. doi: 10.1038/nature06108. [DOI] [PubMed] [Google Scholar]
- 12.Tessier PM, Lindquist S. Prion recognition elements govern nucleation, strain specificity, and species barriers. Nature. 2007;447:556–561. doi: 10.1038/nature05848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pastor MT, Esteras-Chopo A, Serrano L. Hacking the code of amyloid formation: The amyloid stretch hypothesis. Prion. 2007;1:9–14. doi: 10.4161/pri.1.1.4100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ross ED, Edskes HK, Terry MJ, Wickner RB. Primary sequence independence for prion formation. Proc Natl Acad Sci USA. 2005;102:12825–12830. doi: 10.1073/pnas.0506136102. [DOI] [PMC free article] [PubMed] [Google Scholar]