Sequences of DNA provide documentary evidence of the evolutionary past undreamed of by pioneers such as Darwin and Wallace, but their potential as sources of evolutionary information is still far from being realized. A major hindrance to progress has been confusion regarding the role of positive (Darwinian) selection, i.e., natural selection favoring adaptive mutations. In particular, problems have arisen from the widespread use of certain poorly conceived statistical methods to test for positive selection (1, 2). Thousands of papers are published every year claiming evidence of adaptive evolution on the basis of computational analyses alone, with no evidence whatsoever regarding the phenotypic effects of allegedly adaptive mutations. But it would be a mistake to dismiss Yokoyama et al.'s (3) study, in this issue of PNAS, of the evolution of visual pigments in vertebrates as more of the same. For, unlike all too many recent papers in the field, this study is solidly grounded in biology.
Evolution of Vertebrate Rhodopsins
Visual pigments represent a particularly well chosen system for understanding the molecular basis of adaptive evolution. The wavelength of maximal absorption (λmax) is an easily measured phenotype directly related to the functional utility of a visual protein in dif ferent environments. Amino acid replacements that change the spectral sensitivity of visual pigments thus provide examples of molecular changes having direct ecological relevance.
Yokoyama et al. (3) concentrated on the rhodopsins, the visual pigments used by vertebrates for dim-light vision. When active at twilight, vertebrates living in shallow water or on land encounter light that falls mainly in the range between 400 and 500 nm. Correspondingly, fishes that inhabit shallow water share with terrestrial amphibians, birds, and mammals rodopsins with λmax values ≈500 nm. By contrast, Yokoyama et al. found that deep-sea fishes tend to have rhodopsins with λmax values ≈480 nm, corresponding to the available light in their environment. Exceptions to this trend were seen in deep-sea fishes that migrate upward in the water column at night; these species' rhodopsins showed λmaxs ≈490 nm, intermediate between those of deep- and shallow-water fishes. A deep-sea fish known as the shining loosejaw (Aristostomias scintillans) provides another interesting exception. This species produces red bioluminescence from suborbital light organs, and its rhodopsin has λmax ≈700 nm, in the red end of the light spectrum.
Yokoyama et al. (3) used phylogenetic methods to reconstruct the relationships among vertebrate rhodopsin genes and to predict the sequences ancestral to present-day rhodopsins. Moreover, combining informatics with laboratory work, they engineered 11 of these predicted ancestral rhodopsins and determined their λmax values. Thus, it was possible for them to infer which changes were responsible for the evolution of novel sensitivities in vertebrate rhodopsins.
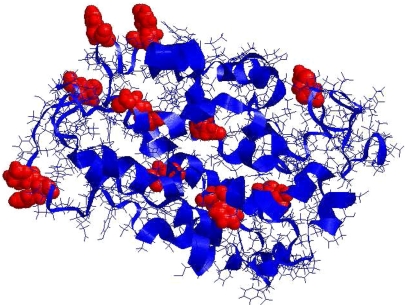
Their most remarkable finding was that 15 amino acid replacements at just 12 amino acid sites (Fig. 1) could account for the evolution of λmax values of most contemporary vertebrate rhodopsins (3). Moreover, certain of these functionally significant amino acid changes occurred multiple times over the course of evolution. For example, the replacement of alanine by serine at site 292 occurred nine times, and the replacement of aspartic acid by asparagine at position 83 occurred seven times (3). In addition, similar functional effects could be obtained in different ways. For example, they discovered three different combinations of mutations, involving entirely different residues, each of which has the effect of lowering λmax by 14–20 nm (3).
Fig. 1.
Structure of bovine rhodopsin, showing (space-filled, red) the 12 amino acid sites at which changes affecting λmax have occurred in the evolution of vertebrates (3).
The Failure of Codon-Based Methods
Point mutations in protein-coding regions are of two types: nonsynonymous (those that change the amino acid) and synonymous (those that do not change the amino acid). Evolutionary biologists have long reasoned that comparison of the rate of fixation of these two types of mutations (adjusted for opportunity because nonsynonymous mutations are about three times as likely as synonymous mutations as a result of the properties of the genetic code) can provide information regarding the past action of natural selection. In most comparisons between related genes, the number of synonymous substitutions per synonymous site (dS) exceeds the number of nonsynonymous substitutions per nonsynonymous site (dN). This pattern is explained by the fact that most nonsynonymous mutations are deleterious because they disrupt protein structure, whereas synonymous mutations are selectively neutral or nearly so. Thus natural selection (so-called “purifying selection”) acts to eliminate a majority of nonsynonymous mutations, lowering dN relative to dS.
In a small number of cases, however, natural selection acts to favor repeated nonsynonymous changes in a limited set of codons, leading to a pattern whereby dN exceeds dS (4). The classic example involves the vertebrate major histocompatibility complex (MHC), where a coevolutionary race with pathogens drives repeated changes in the residues that bind peptides presented to host immune cells (5). The MHC and a few similar cases provide examples where positive selection favoring repeated amino acid changes has reversed the usual pattern, leading to dN > dS (4).
The so-called “codon-based” methods of testing for positive selection are derived from an unwarranted generalization of the MHC case. These methods typically use a likelihood ratio test to decide between two models of sequence evolution, one of which includes a category of codons with dN > dS. As a follow-up, Bayesian methods can be used to identify individual codons with dN > dS, and the latter are considered to be subject to positive selection. These methods thus assume that the existence of even a single codon with dN > dS implies positive selection. But this assumption is demonstrably false, because codons with dN > dS are likely to occur by chance even under strong purifying selection because of the stochastic nature of the mutational process (2, 6). Yet, despite their shaky foundations, numerous publications have used these methods as the basis for claims of positive selection at the molecular level.
The data of Yokoyama et al. (3) provided an ideal opportunity to test whether codon-based methods can indeed be used to identify adaptive evolution. Because those authors knew the amino acid replacements that have led to adaptive changes in rhosopsins, they could test whether these same residues are identified by the codon-based methods. In fact, the results showed that the codon-based methods were 100% off-target. When Bayesian methods were applied to a set of closely related rhodopsin sequences, eight sites were identified as “positively selected.” Yet not one of these sites was among the 12 sites known to be involved in adaptive changes in rhodopsin sensitivity. Moreover, amino acid changes at these sites were shown experimentally to have no effect on λmax and thus almost certainly to lack any adaptive significance (3).
These results support the theoretical prediction that, because of the faulty logic in their underlying assumptions, codon-based focus mainly on statistical artifacts rather than true cases of positive selection (2). An additional finding of Yokoyama et al. (3) further supported this interpretation. When more distantly related genes were added to the data set, the Bayesian methods no longer identified any sites as positively selected. Such anomalous results are not unexpected because individual codons with dN > dS are more likely to occur by chance alone when sequences are closely related, because of the random nature of the mutational process (2, 6).
Non-Darwinian Mechanisms
Yokoyama et al. (3) were able to identify the molecular basis of functionally significant changes in dim-light vision of vertebrates. But their approach could not necessarily determine the population processes that originally gave rise to those changes; in particular, there was no direct evidence that natural selection was actually involved in fixing adaptive changes. Contrary to a widespread impression, natural selection does not leave any unambiguous “signature” on the genome, certainly not one that is still detectable after tens or hundreds of millions of years. To biologists schooled in Neo-Darwinian thought processes, it is virtually axiomatic that any adaptive change must have been fixed as a result of natural selection. But it is important to remember that reality can be more complicated than simplistic textbook scenarios.
Combining informatics with laboratory work, they engineered 11 ancestral rhodopsins.
As well as natural selection, nonselective (or “non-Darwinian”) mechanisms may play a role in the origin of adaptive phenotypes. The most important non-Darwinian process is chance fluctuation in gene frequency or genetic drift, which can lead to the fixation of selectively neutral mutations (those with no effect on fitness) or sometimes even of slightly deleterious mutations. Kimura (7) coined the term “Dykhuizen-Hartl effect” to describe an originally neutral mutation that later becomes adaptive in a changed environment, including a changed biochemical environment resulting from other amino acid replacements in the same protein (8).
A process of this sort seems likely to have played a role in giving rise to adaptive changes in the spectral sensitivity of at least some vertebrate rhodopsins. Yokoyama et al. (3) found that functional changes in rhodopsins sometimes required a combination of amino acid replacements at several different sites. For example, a rhodopsin from the Japanese conger eel with λmax ≈480 nm achieved this sensitivity through the interaction of three different amino acid replacements (at sites 195, 195, and 292). There does not seem to be any way that natural selection could favor an amino acid replacement that would be of adaptive value only if two other replacements were to occur as well. Rather, it seems more plausible to hypothesize that two of the three amino acid changes were selectively neutral and were individually fixed by genetic drift. When those two changes had been fixed, natural selection would then favor the third change, because, in the context of the two previous changes, it yielded an adaptive phenotype.
Setting a New Standard
In recent years the literature of evolutionary biology has been glutted with extravagant claims of positive selection on the basis of computational analyses alone, including both codon-based methods and other questionable methods such as the McDonald-Kreitman test (1, 9). This vast outpouring of pseudo-Darwinian hype has been genuinely harmful to the credibility of evolutionary biology as a science. It is to be hoped that the work of Yokoyama et al. (3) will help put an end to these distressing tendencies. By incorporating experimental evidence regarding the phenotypic effects of reconstructed evolutionary changes, this study sets a new standard for studies of adaptive evolution at the molecular level. In addition, by providing evidence that non-Darwinian and Darwinian processes are likely to be involved in the evolution of adaptive phenotypes, it points the way toward a new, more realistic appreciation of the evolutionary process.
Footnotes
The author declares no conflict of interest.
See companion article on page 13480.
References
- 1.Hughes AL. Looking for Darwin in all the wrong places: The misguided quest for positive selection at the nucleotide sequence level. Heredity. 2007;99:364–373. doi: 10.1038/sj.hdy.6801031. [DOI] [PubMed] [Google Scholar]
- 2.Hughes AL, Friedman R. Codon-based tests of positive selection, branch lengths, and the evolution of mammalian immune system genes. Immunogenetics. 2008;60:495–506. doi: 10.1007/s00251-008-0304-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yokoyama S, Tada T, Zhang H, Britt L. Elucidation of phenotypic adaptations: Molecular analyses of dim-light vision proteins in vertebrates. Proc Natl Acad Sci USA. 2008;105:13480–13485. doi: 10.1073/pnas.0802426105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hughes AL. Adaptive Evolution of Genes and Genomes. New York: Oxford Univ Press; 1999. [Google Scholar]
- 5.Hughes AL, Nei M. Pattern of nucleotide substitution at MHC class I loci reveals overdominant selection. Nature. 1988;335:167–170. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 6.Hughes AL, Friedman R. Variation in the pattern of synonymous and nonsynonymous difference between two fungal genomes. Mol Biol Evol. 2005;22:1320–1324. doi: 10.1093/molbev/msi120. [DOI] [PubMed] [Google Scholar]
- 7.Kimura M. The Neutral Theory of Molecular Evolution. Cambridge, UK: Cambridge Univ Press; 1983. [Google Scholar]
- 8.Dykhuizen D, Hartl DL. Selective neutrality of 6PGD allozymes in E. coli and the effects of genetic background. Genetics. 1980;96:801–817. doi: 10.1093/genetics/96.4.801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hughes AL, Friedman R, Rivailler P, French JO. Synonymous and nonsynonymous polymorphism vs. divergence in bacteria. Mol Biol Evol. 2008 doi: 10.1093/molbev/msn166. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]