Abstract
Compilations of domain–domain interactions based on solved structures suggest there are distinct domain pairs that are used repeatedly in different protein contexts to mediate protein–protein interactions. However, not all protein pairs with the corresponding domains that can potentially mediate interaction do interact, even when they are colocalized and coexpressed. It is conceivable that there are structural and sequence features, below the domain level, that play a role in determining the potential of domains to mediate protein–protein interactions. Here, we discover such features by comparing domains that, on the one hand, mediate homodimerization of proteins and, on the other, occur in different proteins that are documented as monomers. Intriguingly, this comparison uncovered surface loops that can be considered as determinants of the interactions. There are enabling loops, which mediate the domain interactions, and disabling loops that prevent the interactions. The presence of the enabling/disabling loops is consistent with the fulfillment/prevention of the interaction and is highly preserved in evolution. This suggests that, along with the preservation of structural elements that enable interaction, evolution maintains elements intended to prevent unwanted interactions. The enabling and disabling loops discovered in this study have implications in prediction of protein–protein interactions, by pointing to the protein regions that determine the interaction. Our results extend the hierarchy of attributes that collectively establish the modularity of domain-mediated protein–protein interactions.
Keywords: modularity, oligomeric state, protein–protein interaction
The function of most proteins depends on their interaction with other proteins. Various lines of evidence support the notion that many protein–protein interactions (PPIs) are mediated by protein domains. Previous studies showed statistically significant overrepresentation of certain domain pairs in large datasets of experimentally determined PPIs (1–4). Subsequently, lists of thousands of structurally based domain pairs that are used repeatedly for mediating the interactions in various protein contexts were published (5, 6). These domain pairs were recently shown to be evolutionarily conserved in PPIs across species and thus can be considered as the building blocks of the organisms' PPI networks (7, 8).
Obviously, not all protein pairs that contain domains with an interaction potential do interact, even when they are coexpressed and colocalized. This implies there are additional features that determine when an interaction will be fulfilled via these domains. It suggests also that PPI modularity is achieved by a hierarchy of features. Domain pairs are at the top of this hierarchy, and they are followed by more delicate intradomain features. Indeed, several groups have characterized intradomain features associated with domain–domain interactions. For example, Littler and Hubbard (9) studied domain interactions within proteins and characterized the interfaces used for interaction by examining domain orientation and sequence conservation. Another study typified the interaction interfaces (10), whereas others determined clusters of spatially close residues along the interface that play a role in domain–domain interaction (11, 12).
An appealing approach for unraveling such intradomain features would be to compare different instances of the same domain pair, where in some instances the domains mediate protein–protein interaction, and in other instances they do not. To this end, two sets of data per domain pair are needed. The first is a positive dataset including verified PPIs mediated by a certain domain pair. The second is a negative dataset including protein pairs with the corresponding domains, which are known to be coexpressed and colocalized but are documented as noninteracting. By identifying subtle differences in the sequence and/or structure of the corresponding domains in the two datasets, intradomain features that are responsible for fulfilling a domain–domain interaction may be identified. However, although generating a positive dataset per domain pair is relatively easy, it is rather difficult to compile protein pairs documented as noninteracting. We managed to overcome this difficulty by studying domains that mediate homo-oligomerization, and that are found also in proteins documented as monomers.
Homomultimers and monomers that include the same domains provide ideal positive and negative sets for such an analysis, especially when the interactions in the homomultimers are mediated by the same domain in the different subunits (self-interacting domain). In both cases, the proteins including the domain are coexpressed and colocalized; however, in the case of homomultimers, the interaction is fulfilled, and in the case of monomers, it is not fulfilled. We compiled a dataset of crystallographically solved monomers, homodimers, and homotetramers of the same domain and compared them by structure and sequence alignment. Of note, the proteins containing the domains in the homomultimeric and monomeric instances are different. Thus, we do not look for a conformational change of a protein that functions once as a monomer and once as a dimer, but we look rather at the inherent sequence and structure differences between two proteins of the same domain, each documented to function in a different oligomeric state. We show that in a substantial fraction of the monomer/homodimer/homotetramer instances, the potential for self-interaction is determined by two types of loops, which either prevent or mediate the interaction. These loops determine the interaction potential of domains and define an additional subtle level of modularity in the PPI network, below the domain level.
Results
Enabling/Disabling Loops Determine the Fate of Interaction.
We compiled a list of 1,670 domains shown crystallographically to mediate homodimerization and homotetramerization via self-interactions (see Methods). Next, we used this catalog to create a dataset of 50 self-interacting domains, each contained in at least two single-domain proteins differing in their oligomeric state. One is a monomer, and the other is a homomultimer. Both proteins were shown experimentally to function in their corresponding oligomeric state. In our data, homodimers and homotetramers represent the domain instances that mediate interaction, and monomers represent the domain instances in which interaction does not occur. Therefore, in the discussion below, the oligomeric state of a protein is used to indicate whether an interaction was fulfilled.
To find structural attributes that specify an oligomeric state, we carried out a structural superimposition between monomers and homodimer/homotetramer subunits. We compared the interface of the dimer with the monomer's “pseudointerface” (the monomer's surface corresponding to an interface of the dimer's subunit). The superimposition yielded various sequence/structure features that differentiated between the monomeric and multimeric domain instances. In several examples, there was almost no change in the backbone structure of these two protein chains, but there were amino acid differences in the interface region. This, for example, was found by us for the Cytochrome c3 domain and was noted also by Czjzek et al. (13). In some cases, even one residue substitution was found to impose a different oligomeric state for structurally very similar proteins, consistent with previous studies (14, 15). There were also examples in which a single or multiple residue differences between two proteins caused conformational changes that accounted for their difference in the oligomeric state. For other domains, we found published evidence for posttranslational modifications accounting for the difference between the different oligomeric instances (e.g., ref. 16).
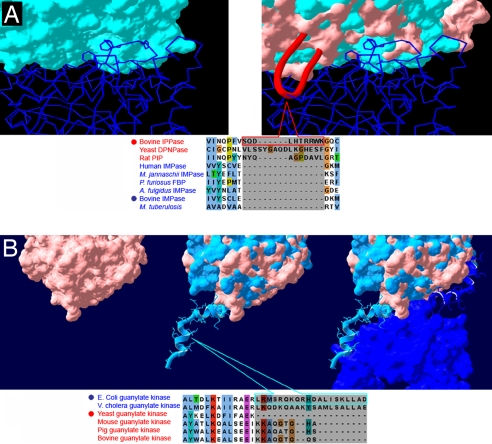
Intriguingly, for 11 of the 50 domains included in our analysis, we could not identify single/multiple amino acid substitutions or posttranslational modifications that could account for the differences between the oligomeric states. We found that the differences in these cases could be attributed to insertions or deletions of protein segments. These were distinct loops on the protein surfaces, which either prevented or enabled interactions. The inhibition or enabling of the interaction is not due to a conformational change caused by the loops, but rather their mere presence determines the interaction, as exemplified in Fig. 1. In Fig. 1A, we show a structural superimposition between one subunit of the homodimeric Inositol monophosphatase and the monomeric Inositol polyphosphate 1-phosphatase. Both are single-domain proteins from bovine, consisting of the Inositol monophosphatase domain. A careful examination of these structures indicated that a segment is looping out in the pseudointerface of the monomer, generating a protrusion that prevents the self-interaction of the domain. This type of loop will hereafter be referred to as a “disabling loop” (shown schematically in Fig. 2A). A structure-based multiple sequence alignment (MSA) of protein sequences sharing this domain (see Methods) revealed that the disabling loop is preserved in proteins documented as monomers but is absent from all homodimers (Fig. 1A Lower).
Fig. 1.
Examples of enabling and disabling loops. (A) Disabling loop. (Upper) Structural superimposition of homodimeric bovine Inositol monophosphatase and the monomeric bovine Inositol polyphosphate 1-phosphatase. On the left, the two subunits of the homodimer are shown, one as a cyan surface and the other as a blue backbone. On the right, the monomer (pink surface) is superimposed onto the cyan homodimer subunit. A red ribbon represents a disabling loop, protruding out of the pseudointerface of the monomer. Clearly, this loop abolishes the self-interaction of the domain. (Lower) A structure-based MSA of the monomer and the homodimer sequences. Sequences whose structures are shown (Upper) are marked with filled circles. Red and blue accessions represent monomers and homodimers, respectively. The disabling loop is represented by the shaded block in the MSA. (B) Enabling loop. (Upper) Structural superimposition of homodimeric Escherichia coli Guanylate kinase and the monomeric Saccharomyces cerevisiae Guanylate kinase. On the left, only the monomer is shown (pink surface). In the middle, a superimposition is shown of the monomer and one subunit of the homodimer (cyan surface). Note the C-terminal extension that protrudes out of the homodimer subunit. On the right, the second subunit of the homodimer is added (blue surface), mediating the self-interaction of the domain. (Lower) A structure-based MSA between the monomer and the homodimer along with other Guanylate kinases. Colors of protein names and filled circles are as in A. The enabling loop is represented by a shaded block in the MSA. The correspondence between the presence of the loop and the oligomeric state is evident.
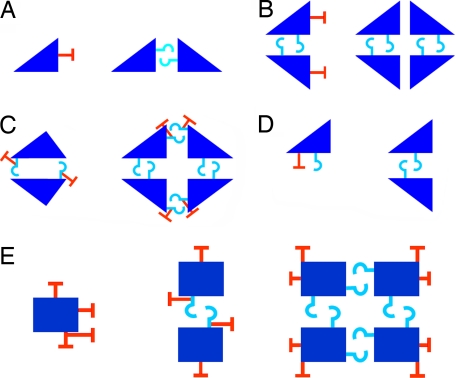
Fig. 2.
Enabling/disabling loops on the interface determine the interaction potential of a domain. A schematic representation of enabling/disabling loops that play a role in assisting/preventing homo-oligomerization. Subunits are depicted as blue triangles or rectangles. Enabling loops are illustrated as cyan hooks, whereas disabling loops are shown as red “T” signs. For the sake of clarity, not all loops are illustrated. (A) Disabling loops in monomers and enabling loops in homodimers. An illustration of the loops in the Dihydropholate reductase domain, which is present in the monomeric Dihydrofolate reductase from Pneumocystis carinii (PDB ID code: 1S3Y, on the left) and in the homodimeric Dihydrofolate reductase from Bacteriophage T4 (PDB ID code 1JUV, on the right). In the homodimer, self-interaction is mediated by an enabling loop, 1JUV [Y96-P110]. In the monomer, the enabling loop is missing and self-interaction is prevented by a disabling loop, 1S3Y [H135-P136]. (B) Disabling loops in homodimers that prevent homotetramerization. An illustration of the Phosphoglycerate mutase domain, which is present in the homodimeric human Bisphosphoglycerate mutase (PDB ID code: 1T8P, on the left) and in the homotetrameric Phosphoglycerate mutase of Mycobacterium tuberculosis (PDB ID code: 1RII, on the right). One interface is common to both structures, where the interaction is mediated by an enabling loop, 1T8P [I126-R153], 1RII [I127-L149]. The second interface, used by the homotetramer (with no enabling loop) becomes disabled in the homodimer, because of a small disabling loop, 1T8P [C145-D146]. (C) Loops that govern two interfaces and are both enabling and disabling. Illustrated are two human proteins that include the Trypsin domain: the homodimeric Granzyme A (PDB ID code: 1ORF, on the left), and the homotetrameric Tryptase beta-2 (PDB ID code: 1A0L, on the right). A single loop, 1ORF [N180-I191], both enables Granzyme A's homodimerization and disables tetramerization. A single loop, 1A0L [K171-V176] both enables one of the tetramerization interactions and disables the Tryptase dimerization interface. (D) Dominant disabling loops and recessive enabling loops. Illustrated are two proteins of the Metallo-beta-lactamase domain, the homodimeric Ribonuclease Z of Bacillus subtilis (PDB ID code: 1Y44, on the right) and the monomeric Beta-lactamase type II from Bacteroides fragilis (PDB ID code 1HLK, on the left). The homodimer has an enabling loop that mediates the self-interaction of the domain, 1Y44 [A10-A16]. When superimposing it with the monomer, it is apparent that the enabling loop, 1HLK [A44-M51], is present in the pseudointerface, but in the same surface, a disabling loop is also present, 1HLK [D74-Q76]. The overall effect is that the disabling loop interferes with the homodimerization potential of the monomer. (E) Multiple prevention of potential interaction interfaces. Illustrated are proteins with the Histidine acid phosphatase domain. This domain has multiple potential self-interacting interfaces, as displayed by the tetrameric 3-phytase B from Aspergillus niger (PDB ID code 1QFX, on the right), the homodimeric Prostatic acid phosphatase from rat (PDB ID code 1RPA, in the center) and the monomeric 4-phytase from E. coli (PDB ID code: 1DKM, on the left). The potential of three interfaces is abolished in the monomeric protein, because of three disabling loops.
We also identified the opposite scenario, where a loop that plays a pivotal role in mediating the self-interaction of a homodimer was absent from the monomer's pseudointerface (exemplified in Fig. 1B Upper). This type of loop will hereafter be referred to as an “enabling loop” (Fig. 2A). A structure-based MSA revealed the preservation of the enabling loop in the homodimers but not in the monomers. In this example of the Guanylate kinase domain, the essentiality of the enabling loop for the stabilization of the protein was experimentally verified (17).
The number of solved protein structures per domain in the group of these 11 domains ranged from 3 to 20 structures (Table 1). In total, there were 73 proteins encompassing this domain set, referred to hereafter as the “core set.” For each domain, we carried out a structure-based MSA of its core set proteins. The positions and lengths of loops were derived from the MSA [Text S1 in supporting information (SI) Appendix], defining for each domain a loop profile that consisted of enabling and/or disabling loops in distinct locations. Accordingly, we determined the characteristic profile for each of the oligomeric states, e.g., an enabling loop and lack of disabling loop for homodimeric domain instances (Text S1 in SI Appendix). As shown in Table 1, in 64 of 73 instances (88%), the profile of enabling/disabling loops of the proteins accounted for their oligomeric state. Twenty-six of the 28 monomers in the core set could be explained by the presence of disabling loops and/or absence of an enabling loop (Table 2). Likewise, 26 of the 28 homodimers and 12 of 17 homotetramers fitted their loop profile (Table 2). The relationship between the oligomeric state and the loop profile was highly statistically significant, with a P value of 10−12 for the monomer–dimer distinction and a P value of 10−6 for the dimer–tetramer distinction (as determined by the χ2 test).
Table 1.
Structurally based and loop-based annotations of oligomeric states for core set proteins
| Domain name | Annotated monomers | Annotated dimers | Annotated tetramers | Total no. of proteins | Oligomeric state fits the loop profile | Oligomeric state does not fit the loop profile |
|---|---|---|---|---|---|---|
| Trypsin | 4 | 1 | 1 | 6 | 5 | 1 |
| tRNA binding | 1 | 3 | - | 4 | 4 | 0 |
| Protease inhibitor | 1 | 2 | - | 3 | 3 | 0 |
| Guanylate kinase | 2 | 1 | - | 3 | 3 | 0 |
| Carbonyl reductase | 1 | 7 | 12 | 20 | 16 | 4 |
| Dihydrofolate reductase | 7 | 2 | - | 9 | 8 | 1 |
| PfkB carbohydrate kinase | 1 | 3 | - | 4 | 4 | 0 |
| Phosphoglycerate mutase | 1 | 2 | 2 | 5 | 4 | 1 |
| Histidine acid phophatase | 2 | 2 | 1 | 5 | 4 | 1 |
| Inositol monophosphatase | 3 | 4 | - | 7 | 7 | 0 |
| Metallo-beta-lactamase | 5 | 1 | 1 | 7 | 6 | 1 |
| Total | 28 | 28 | 17 | 73 | 64 | 9 |
Loop profiles for each domain and each protein in the core set are detailed in Table S3 in SI Appendix.
Table 2.
Consistency of loop profile-based and structure-based oligomeric state annotation in the “core set”
| Structure-based annotation |
||||
|---|---|---|---|---|
| Monomer | Dimer | Tetramer | ||
| Loop profile-based prediction* | Monomer | 26 | 1 | 0 |
| Dimer | 2 | 26 | 5 | |
| Tetramer | 0 | 1 | 12 | |
*Loop profile of each protein was determined by a structure-based MSA with the appropriate core set proteins (Text S1 and Table S3 in SI Appendix). Statistical significance for the distinction between two oligomeric states by the loop profiles was computed by the χ2 test. Monomers/dimers: P = 2.7·10−12; dimers/tetramers: P = 3.2·10−6.
In 7 of the 11 domains, more than one interface was found to be capable of mediating self-interaction, adding another level of complexity. For some of these domains, disabling loops were found to prevent not only homodimerization of monomers but also tetramerization of dimers via another potential interface (Fig. 2B). In other cases, a special type of loop was identified, which dominates over two (or even three) interfaces of the domain, for example, a loop that simultaneously enables an interaction in one interface and disables an interaction in another (Fig. 2C). Also, in 10 cases, one interface included both a disabling and an enabling loop. In eight of these cases (80%), the disabling loop dominated the enabling loop, which as a result did not fulfill its interaction potential (Fig. 2D). In the other two cases, the disabling loop adapted a conformation that did not disable the interaction. Impressively, in five of seven domains with multiple interfaces the monomers exhibited disabling loops in almost all interfaces to prevent possible interactions (Fig. 2E).
The implications of the enabling and disabling loops are not limited to homodimers but can be extended to heterodimers as well. We gathered experimentally verified heterodimers, in which the two proteins include the same domain (of the core set domains) (see Methods). We filtered this list to include only single-domain proteins, resulting in six heterodimers that involve nine proteins (Table S2 in SI Appendix). The sequences of the domains in the two proteins are similar but not identical. Thus, this dataset allows us to test whether the domain loop profiles in these heterodimers are those that allow self-interaction (Table S2 in SI Appendix). Indeed, we determined the loop profile of each of these proteins and found that seven of nine (78%) of the domains included enabling loops and did not contain disabling loops. This further supports the role of the enabling and disabling loops in determining the interaction potential of a domain.
Implications of Enabling/Disabling Loops in Prediction.
We next attempted to examine whether the loop profile of a domain can be used to predict its self-interaction. For this purpose, we compiled another set of 105 proteins containing these 11 domains, for which Swiss-Prot (18) provides oligomeric state annotation based on experimental evidence (see Methods). The loop profile of each protein sequence in this set was determined by its alignment with the MSA of corresponding proteins in the core set. Based on the loop profile of each protein, we predicted its oligomeric state (see Methods). Indeed, for 83 of the 105 proteins (79%), the prediction was accurate. These results are highly statistically significant (Table 3 and Table S3 in SI Appendix) and suggest that loop considerations may serve as good predictors of oligomeric state while also indicating crucial elements of the interaction. Notably, predicting the oligomeric state by running BLAST and adopting the oligomeric state annotation of the closest homologue yielded comparable prediction accuracy (83%). Of note, the overlap between the correct BLAST predictions and correct loop profile-based predictions is very high, 91%. This observation can be used to increase the reliability of a prediction, as described below.
Table 3.
Evaluation of oligomeric state prediction by the loop profiles for the “test set” proteins
| Experiment-based annotation |
||||
|---|---|---|---|---|
| Monomer | Dimer | Tetramer | ||
| Loop profile-based prediction* | Monomer | 9 | 3 | 0 |
| Dimer | 5 | 63 | 8 | |
| Tetramer | 0 | 6 | 11 | |
*Loop profile of each protein was determined by structure-based MSA with the appropriate core set proteins (Text S1 and Table S3 in SI Appendix). Statistical significance for the distinction between two oligomeric states by the loop profiles was computed by the χ2 test. Monomers/dimers: P = 1.5·10−6; dimers/tetramers: P = 5·10−6.
There were an additional 202 proteins in Swiss-Prot containing these domains that had oligomeric state annotation. However, these annotations were not based on experimental evidence but were based on sequence similarity to proteins with known oligomeric state. This annotation method is analogous to the above-mentioned prediction by BLAST. Based on our results above, it is obvious that not all of these annotations are correct, and that intersecting them with the loop profile-based predictions will increase their reliability. For 146 of the 202 proteins (≈72%), the loop profile was consistent with the annotation, supporting their putative oligomeric state. Most of the remaining 56 proteins are probably misannotated in Swiss-Prot, as we demonstrated above that inconsistent correct predictions are rather rare.
Discussion
Intradomain Features Play a Role in Molecular Recognition.
The widely accepted conjecture that domain–domain interactions are important building blocks of protein–protein interactions, as shown by us and others (7, 8), raises the question of specificity and prevention of cross-reactivity between interacting domains. In general, amino acid substitutions between different domain instances contribute to specificity (19). One illuminating example is provided by coiled-coil proteins, which share many sequence and structural characteristics among them. There, specificity in interaction is gained by charged and polar residues at the interface between coiled-coil strands (20). Here, we used sequence and structural comparisons between monomeric and homodimeric proteins sharing the same domain to find elements discriminating between interacting and noninteracting domains. In addition to amino acid substitution per se, we showed that in 22% of the studied domains, self-interaction potential is dictated by enabling or disabling loops whose presence or absence is highly evolutionarily preserved. These are salient domain segments that are present on the surface of homodimers and absent from monomers, or vice versa, thereby determining whether a domain instance will self-interact to make a higher-order homomultimer (or heterodimer). It is conceivable that such enabling and disabling loops are used also in determining the specificity of interactions between two different domains. This conjecture is also supported by Chan et al. (21), who recently demonstrated a possible role for insertions and deletions in the regulation and modification of PPIs.
Our analysis indicated that, although there are domains in principle capable of interaction with identical domains, the actual realization of such an interaction depends on their loop profiles. An interaction might be fulfilled when a disabling loop is not present, and/or an enabling loop is present. Likewise, an interaction might be prevented because of the presence of a disabling loop, the absence of an enabling loop, or the presence of both an enabling and a disabling loop. Regarding the latter, we found that, in most cases, the disabling loop dominates the enabling loop.
Insights into Enabling and Disabling Loops.
The disabling and enabling loops have on average the same length of 8–9 aa, but the standard deviation of the disabling loops is much larger (10.2 compared with 6 aa). Because the disabling loop protrudes out of the interface and prevents the interaction, its size is less critical than that of an enabling loop, which mediates the self-interaction. We also identified longer loops of 14 residues on average, which are found at the junction of two interfaces, concurrently enabling interaction in one interface and disabling interaction in another. These loops are found in 5 of the 11 domains and are of special interest, because they simultaneously control the interaction of two interfaces and thus can be regarded as novel hotspots in proteins.
One prominent feature identified in our study is that in approximately half of the domains (6 of 11 domains), the enabling/disabling loops are proximal to the boundaries of the domain. The example of the Guanylate kinase domain described above (Fig. 1B) demonstrates such a scenario, where the 20 C-terminal residues that make up the enabling loop are close to the boundary of the domain. Although these findings may simply suggest that, in some of these cases, the Pfam domain boundaries should be readjusted, they may also have more far-reaching implications. It is possible that, in analogy to genes that have proximal regulatory elements, proteins are built of domains and proximal short segments that determine their interactions with other proteins.
Functional Implications.
The function of a protein is tightly associated with its interacting partners. This holds also for self-interactions, where the function is associated with the oligomeric state. Hence, it is expected that the features responsible for the interaction would be preserved in evolution. Indeed, loop profiles are highly preserved in evolution (Fig. 1 and Text S1 in SI Appendix), supporting the structural and functional importance of enabling/disabling loops. In some cases, the differences in the loop profile may reflect adaptation to specific environments. For example, dimeric proteins with the Dehydropholate reductase domain have an enabling loop that is absent from the monomeric proteins. Thermodynamic measurements show that a homodimer, found in a thermophile, is more stable than its monomeric relative, found in a mesophile (22). This indicates that the formation of a specific oligomeric state is advantageous in a specific environment.
One of our intriguing findings is the prevention of interaction in all possible interfaces. We showed that monomers exhibit disabling loops in almost all pseudointerfaces that may mediate an interaction, implying that it is crucial to maintain these proteins in their monomeric state. This may result for various reasons, such as the capability of monomers to diffuse much faster than proteins of higher oligometric state (23).
The preservation of the disabling loops in monomeric proteins to prevent an unwanted interaction may be considered as a result of nature's negative design. Negative design is a term used to describe the outcome of a positive selection process that yields a sequence element that disfavors a particular interaction and/or function. This principle was originally introduced in the context of protein design (24). Negative design was also shown in naturally occurring beta-strands that avoid aggregation (25). Also, there have been recent attempts to identify conservation of residues that abrogate the potential of proteins to interact (26). The finding of disabling loops is an illuminating example of the negative design principle.
Predictive Implications.
According to the presence or absence of enabling/disabling loops in known structures, a unique loop profile was generated for each oligomeric state for all 11 studied domains (Table S3 in SI Appendix). These loop profiles can be used to determine whether a domain can self-interact and hence can be used for predicting the oligomeric state of proteins carrying this domain. As we demonstrated for a set of annotated proteins that were not included in the core set, the accuracy of this prediction is very high (79%, P < 4.98·10−6). As demonstrated in Results, the oligomeric state can be also predicted with comparable accuracy by a BLAST run. Intersection of the two lists of predictions (according to BLAST and the loop profiles) yields highly accurate predictions. Of note, BLAST matches include gaps without any indication of their spatial location. Thus, adding the loop profile considerations not only raises the reliability of the annotation but also pinpoints the location of protein fragments that might mediate the interaction, providing an indicative starting point for studying the relationship among sequence, structure, and function by mutation experiments.
Examination of the wrong predictions for which structures were available revealed in some cases unusual structures of the loops, as for example in a tetramer of the Estradiol 17-beta-dehydrogenase domain. Here, a loop that is predicted as a disabling loop, intended to prevent the tetramerization fails to do so because of sequence and structure changes. This observation leads to the conjecture that, in addition to the mere presence of a loop, delicate sequence considerations may add another level of fine-tuning to the interactions (Fig. 3).
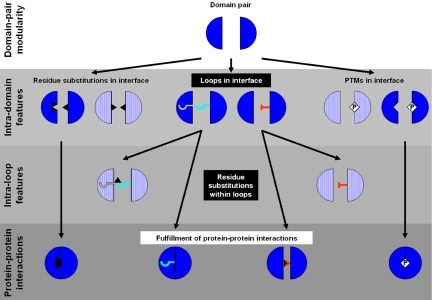
Fig. 3.
PPI modularity is achieved by a hierarchy of features. Domain pairs are at the top level in the hierarchy of features that yield the modularity of PPIs. Intradomain features define additional levels of modularity below the domain level, including enabling and disabling loops. Intra-loop features may define yet an additional level of modularity. Half circles represent potential interaction domains. Filled shapes represent fulfilled interactions, whereas dotted shapes represent interactions that are not fulfilled. Black triangles stand for sequence variations. PTMs, posttranslational modifications.
The predictive potential of our approach is highlighted by the prevalence of single-domain proteins with the core set domains in sequence databases (7,639 proteins in Swiss-Prot and TrEMBL), for all of which we can assign oligomeric state annotation with high confidence. An example of annotations for proteins with the pfkB kinase carbohydrate domain is presented in Table S4 in SI Appendix.
Modularity and Specificity of PPIs.
Modularity can be defined as the use of the same building block in different contexts. Modular arrangements necessitate the presence of additional features that optimize specificity, thereby preventing cross-reactivity. PPIs are modular in the sense that the same domain pairs mediate different interactions. Here, we show that insertions and deletions of loops within domains play an important role in determining their specificity of interaction. Thus, we add a not previously characterized intradomain attribute to the hierarchy of features that determine the interaction potential of domains (Fig. 3).
Although it is easy to understand how disabling loops prevent unwanted interactions, mediating an interaction via enabling loops opens new questions concerning specificity. In the analysis of heterodimers, we showed their domains do not contain disabling loops and contain enabling loops, thus, capable of undergoing self-interaction. However, how is undesired heterodimerization prevented between two different proteins consisting of the same domains and the same enabling loops? One way to prevent cross-reaction is by subtle sequence differences between the loops. This leads to the same conjecture that stemmed from the above discussion regarding the incorrect predictions, that in addition to the mere presence of a loop, there might be an additional level of fine-tuning contributed by delicate sequence considerations. Thus, there seems to be a hierarchy that underlies the PPI network, where domain pairs are at the top of this hierarchy, followed by enabling/disabling loops, and concluding in intra-loop sequence differences that dictate the interactions (Fig. 3).
In this article, we focus on single-domain proteins that undergo self-interaction. Similar analyses can be conducted for multidomain proteins that homodimerize via the same domain, as demonstrated recently by Posy et al. for type I/II Cadherins (27). Furthermore, our analysis of heterodimers that interact via the same domain implies that enabling/disabling loops are not unique for homo-oligomers but seem to be more general and hold for heterodimerization also. Similarly, our analysis can be extended from self-interacting domains to pairs of domains (X,Y) that were shown crystallographically to mediate interaction in various protein contexts. To study these heterodomain interactions, two protein pairs with domains X and Y need to be analyzed: one pair of interacting proteins and the other of proteins documented as noninteracting, even when they are coexpressed and colocalized. Examples for the latter can be gathered from large-scale experiments, looking for proteins with domains X and Y that in all experiments were not documented as interacting or as involved in the same complex. Thus, it would be possible to identify for each structurally derived domain–domain interaction the domain features that allow versatility in their potential to interact.
Methods
Generation of the Core Set.
We extracted from the nonredundant database of structurally derived domain–domain interactions of Itzhaki et al. (7) all self-interacting domains, resulting in a database of 1,670 such domains. Using information from the Pfam (28) (Nov 2006 edition) and Protein Data Bank (PDB) (29) databases, we extracted crystallographically solved single-domain proteins containing domains from the above-mentioned list. In cases where a complex structure was not available directly from the PDB database, the PQS server (30) was used as a source for the atomic coordinates. For each self-interacting domain, we verified that there are at least two protein representatives, which differ in their oligomerization state. The first is documented as an experimentally verified monomer, and the second is an experimentally verified homodimer or a homotetramer [oligomeric state annotation was obtained from Swiss-Prot (18)]. This filtration procedure yielded a list of 50 such domains.
Finding Enabling/Disabling Loops.
Each monomer was superimposed on a homodimer subunit (homodimers were superimposed on homotetramers), and enabling/disabling loops were manually assigned for each structure after a visual inspection (31). The 11 domains with defined enabling/disabling loops were cross-checked with a structure-based MSA that was constructed for all domain representatives using the EXPRESSO server (32). This server uses structural templates and alignments to create a more precise MSA (relative to sequence-based-only methods). The minimal length of a loop was set to two residues.
Analysis of the Test Set.
To assess the predictive power of the loop profile, we generated a dataset of additional proteins containing these 11 domains, for which oligomeric state annotations were available. These annotations were based on Swiss-Prot or BRENDA (33). The proteins were divided into two sets based on the annotation reliability: One set included proteins whose oligomeric state annotation was based on experimental evidence, whereas the other included proteins annotated “by similarity” (annotation is transferred from another annotated protein of similar sequence). The oligomeric state of a test protein was based on the compatibility of its loop profile to either the loop profile of monomer or multimer. Each test protein of a certain domain was aligned to the structure-based MSA of the core proteins of the same domain. According to this alignment, the presence or absence of enabling/disabling loops was determined. These enabling/disabling loops defined the loop profile of the test protein. If this loop profile fitted a loop profile of a monomer, it was predicted as a monomer. If it fitted the loop profile of a dimer or tetramer, it was predicted accordingly. This alignment was carried out by using the EXPRESSO server with manual adjustments.
Analysis of Heterodimers.
We created a list of heterodimers using binary interactions documented in the BioGrid (34) and IntAct (35) PPI databases (Table S2 in SI Appendix). These dimers were mapped to the Pfam database (28) and were then filtered to include only interactions between single-domain proteins of the 11 core set domains. The interaction potential of each protein (six interactions, nine proteins) was assigned according to the loop profile of its corresponding domain.
Supplementary Material
Acknowledgments.
We thank Ora Furman, Yael Altuvia, Ruth Hershberg, and Pinchas Akiva for valuable discussions and useful comments on the manuscript. This work was supported by European Union grant 3D-repertoire, contract number LSHG-CT-2005-512028.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801207105/DCSupplemental.
References
- 1.Sprinzak E, Margalit H. Correlated sequence-signatures as markers of protein-protein interaction. J Mol Biol. 2001;311:681–692. doi: 10.1006/jmbi.2001.4920. [DOI] [PubMed] [Google Scholar]
- 2.Ng SK, Zhang Z, Tan SH. Integrative approach for computationally inferring protein domain interactions. Bioinformatics. 2003;19:923–929. doi: 10.1093/bioinformatics/btg118. [DOI] [PubMed] [Google Scholar]
- 3.Deng M, Mehta S, Sun F, Chen T. Inferring domain-domain interactions from protein-protein interactions. Genome Res. 2002;12:1540–1548. doi: 10.1101/gr.153002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gomez SM, Noble WS, Rzhetsky A. Learning to predict protein-protein interactions from protein sequences. Bioinformatics. 2003;19:1875–1881. doi: 10.1093/bioinformatics/btg352. [DOI] [PubMed] [Google Scholar]
- 5.Finn RD, Marshall M, Bateman A. iPfam: Visualization of protein-protein interactions in PDB at domain and amino acid resolutions. Bioinformatics. 2005;21:410–412. doi: 10.1093/bioinformatics/bti011. [DOI] [PubMed] [Google Scholar]
- 6.Stein A, Russell RB, Aloy P. 3did: Interacting protein domains of known three-dimensional structure. Nucleic Acids Res. 2005;33:D413–D417. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Itzhaki Z, Akiva E, Altuvia Y, Margalit H. Evolutionary conservation of domain-domain interactions. Genome Biol. 2006;7:R125. doi: 10.1186/gb-2006-7-12-r125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schuster-Bockler B, Bateman A Reuse of structural domain-domain interactions in protein networks. BMC Bioinformatics. 2007;8:259. doi: 10.1186/1471-2105-8-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Littler SJ, Hubbard SJ. Conservation of orientation and sequence in protein domain-domain interactions. J Mol Biol. 2005;345:1265–1279. doi: 10.1016/j.jmb.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 10.Kim WK, Henschel A, Winter C, Schroeder M. The many faces of protein-protein interactions: A compendium of interface geometry. PLoS Comput Biol. 2006;2:e124. doi: 10.1371/journal.pcbi.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Reichmann D, et al. The modular architecture of protein-protein binding interfaces. Proc Natl Acad Sci USA. 2005;102:57–62. doi: 10.1073/pnas.0407280102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.del Sol A, Carbonell P. The modular organization of domain structures: insights into protein-protein binding. PLoS Comput Biol. 2007;3:e239. doi: 10.1371/journal.pcbi.0030239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Czjzek M, Guerlesquin F, Bruschi M, Haser R. Crystal structure of a dimeric octaheme cytochrome c3 (M-13. (r) 26,000) from Desulfovibrio desulfuricans Norway. Structure (London) 1996;4:395–404. doi: 10.1016/s0969-2126(96)00045-7. [DOI] [PubMed] [Google Scholar]
- 14.Andres AM, et al. Positive selection in MAOA gene is human exclusive: Determination of the putative amino acid change selected in the human lineage. Hum Genet. 2004;115:377–386. doi: 10.1007/s00439-004-1179-6. [DOI] [PubMed] [Google Scholar]
- 15.De Colibus L, et al. Three-dimensional structure of human monoamine oxidase A (MAO A): Relation to the structures of rat MAO A and human MAO B. Proc Natl Acad Sci USA. 2005;102:12684–12689. doi: 10.1073/pnas.0505975102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woodcock JM, Murphy J, Stomski FC, Berndt MC, Lopez AF. The dimeric versus monomeric status of 14–3-3zeta is controlled by phosphorylation of Ser58 at the dimer interface. J Biol Chem. 2003;278:36323–36327. doi: 10.1074/jbc.M304689200. [DOI] [PubMed] [Google Scholar]
- 17.Hible G, et al. Calorimetric and crystallographic analysis of the oligomeric structure of Escherichia coli GMP kinase. J Mol Biol. 2005;352:1044–1059. doi: 10.1016/j.jmb.2005.07.042. [DOI] [PubMed] [Google Scholar]
- 18.Bairoch A, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 20.Newman JR, Keating AE. Comprehensive identification of human bZIP interactions with coiled-coil arrays. Science. 2003;300:2097–2101. doi: 10.1126/science.1084648. [DOI] [PubMed] [Google Scholar]
- 21.Chan SK, Hsing M, Hormozdiari F, Cherkasov A Relationship between insertion/deletion (indel) frequency of proteins and essentiality. BMC Bioinformatics. 2007;8:227. doi: 10.1186/1471-2105-8-227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dams T, et al. The crystal structure of dihydrofolate reductase from Thermotoga maritima: Molecular features of thermostability. J Mol Biol. 2000;297:659–672. doi: 10.1006/jmbi.2000.3570. [DOI] [PubMed] [Google Scholar]
- 23.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 24.Hecht MH, Richardson JS, Richardson DC, Ogden RC. De novo design, expression, and characterization of Felix: A four-helix bundle protein of native-like sequence. Science. 1990;249:884–891. doi: 10.1126/science.2392678. [DOI] [PubMed] [Google Scholar]
- 25.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim S, Tsai J, Kagiampakis I, LiWang P, Vannucci M. Detecting protein dissimilarities in multiple alignments using Bayesian variable selection. Bioinformatics. 2007;23:245–246. doi: 10.1093/bioinformatics/btl566. [DOI] [PubMed] [Google Scholar]
- 27.Posy S, Shapiro L, Honig B. Sequence and structural determinants of strand swapping in cadherin domains: do all cadherins bind through the same adhesive interface? J Mol Biol. 2008;378:952–966. doi: 10.1016/j.jmb.2008.02.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bateman A, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Berman HM, et al. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Henrick K, Thornton JM. PQS: A protein quaternary structure file server. Trends Biochem Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 31.Guex N, Peitsch MC. SWISS-MODEL and the Swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 32.Poirot O, Suhre K, Abergel C, O'Toole E, Notredame C. 3DCoffee@igs: A web server for combining sequences and structures into a multiple sequence alignment. Nucleic Acids Res. 2004;32:W37–40. doi: 10.1093/nar/gkh382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schomburg I, et al. BRENDA, the enzyme database: Updates and major new developments. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Breitkreutz BJ, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerrien S, et al. IntAct-open source resource for molecular interaction data. Nucleic Acids Res. 2007;35:D561–D565. doi: 10.1093/nar/gkl958. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.