Abstract
Although pathways for assembly of RNA polymerase (Pol) II transcription preinitiation complexes (PICs) have been well established in vitro, relatively little is known about the dynamic behavior of Pol II general transcription factors in vivo. In vitro, a subset of Pol II factors facilitates reinitiation by remaining very stably bound to the promoter. This behavior contrasts markedly with the highly dynamic behavior of RNA Pol I transcription complexes in vivo, which undergo cycles of disassembly/reassembly at the promoter for each round of transcription. To determine whether the dynamic behavior of the Pol II machinery in vivo is fundamentally different from that of Pol I and whether the static behavior of Pol II factors in vitro fully recapitulates their behavior in vivo, we used fluorescence recovery after photobleaching (FRAP). Surprisingly, we found that all or nearly all of the TATA-binding protein (TBP) population is highly mobile in vivo, displaying FRAP recovery rates of <15 s. These high rates require the activity of the TBP-associated factor Mot1, suggesting that TBP/chromatin interactions are destabilized by active cellular processes. Furthermore, the distinguishable FRAP behavior of TBP and TBP-associated factor 1 indicates that there are populations of these molecules that are independent of one another. The distinct FRAP behavior of most Pol II factors that we tested suggests that transcription complexes assemble via stochastic multistep pathways. Our data indicate that active Pol II PICs can be much more dynamic than previously considered.
Keywords: fluorescence recovery after photobleaching, Mot1, TFIID
Transcription preinitiation complex (PIC) formation involves the assembly of general transcription factors (GTFs) at appropriately remodeled chromatin templates. Regulation of RNA polymerase (Pol) II transcription initiation can occur by many different mechanisms involving facilitated recruitment of PIC components or stimulation of their activities at the promoter. Although the diversity of such regulatory mechanisms is widely appreciated, comparatively little is known about the dynamic behavior of GTFs in vivo vis a vis their interactions with each other and with chromatin. Most nuclear proteins studied to date, including a number of gene-specific transcription factors and chromatin-modifying enzymes, display highly dynamic behavior in vivo (1–3). In many cases, this dynamic behavior is energy-independent (4). Even chromatin structural proteins once thought to be stably bound to DNA display dynamic behavior in vivo (3–9). However, the dynamic behavior of GTFs is largely unexplored. In vitro, a PIC scaffold nucleated by TATA binding protein (TBP) is very stable, a property that facilitates multiple rounds of transcription at an activated promoter (10). A stable TBP association with chromatin is supported by analysis in human cells, which yields a slow fluorescence recovery time for TBP of 20 min (11). In striking contrast, in vivo analysis of RNA Pol I PICs yields a much more dynamic picture, with fluorescence recovery times in the range of seconds to minute (12). These observations support a stochastic model for Pol I PIC assembly, accompanied by dissociation and reassembly for each round of transcription (12, 13). It is not known why Pol I and Pol II PICS should exhibit strikingly different dynamic behaviors. Nor is much understood in either of these systems about what factors regulate the residence times of PIC components on chromatin.
To further investigate Pol II PIC stability in vivo, we used yeast as a model system, allowing us to replace endogenous PIC components with fluorescently tagged versions expressed at endogenous levels. Ready availability of yeast mutants also enabled us to investigate how different factors impact PIC stability. Given the complexity of regulatory mechanisms that affect different aspects of PIC assembly, we hypothesized that PIC disassembly, as manifested in GTF dynamic behavior, is likewise regulated in different ways at different promoters. Relative PIC stability may contribute to promoter-specific transcriptional noise, allowing optimization of transcriptional responses to meet different physiological requirements (14, 15). Measurements of Pol II GTF mobility provide constraints on models defining pathways of PIC assembly and disassembly in vivo and provide general insight into the strengths of chromatin interaction and what factors facilitate global redistribution of GTFs within the nucleus. Identification of significant populations of long-lived, chromatin-bound GTFs, for example, would support the widespread existence of stable PICs. Remarkably, although TBP is rate-limiting for transcription in vivo, we find that nearly all of the TBP is highly mobile in yeast nuclei, and that this mobility is regulated by the cellular factor Mot1. These results suggest that TBP-containing complexes, even some of those formed at active promoters, can undergo rapid cycles of assembly and disassembly.
Results and Discussion
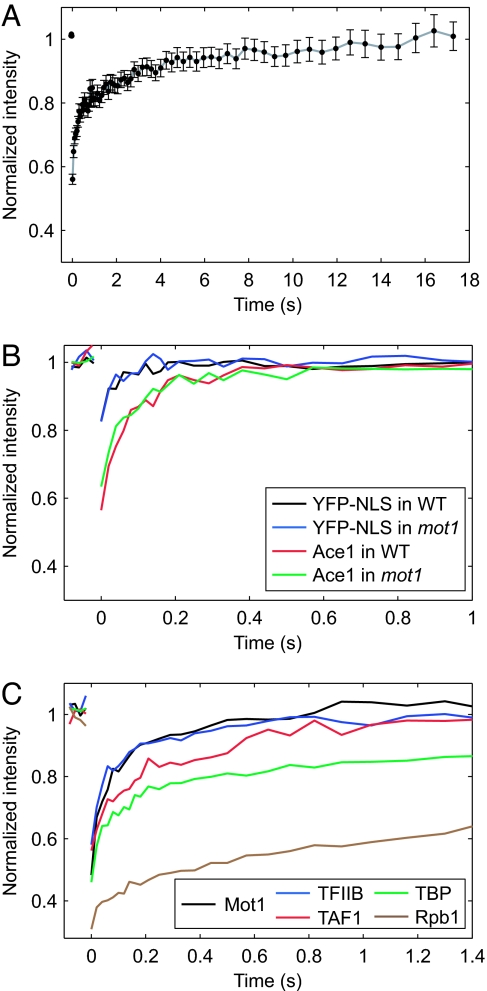
Fluorescence recovery after photobleaching (FRAP) was used to measure the mobilities of GTFs in congenic, homozygous diploid WT or mot1 yeast cells. This approach is technically challenging in yeast (1), but allowed us to express tagged proteins at WT levels, score cell growth phenotypically for the functionality of the tagged proteins, and mutate other regulatory factors to assess their effects on FRAP rates. YFP-tagged GTFs were expressed on chromosomal copies of the genes under the control of their normal promoters. Growth of cells harboring the YFP-tagged alleles was indistinguishable from the untagged parents (data not shown), indicating that the YFP-tagged alleles were fully functional in vivo. The relatively small size of the yeast cell nucleus necessitated measurement of FRAP in a comparatively small bleach spot [see supporting information (SI) Fig. S1 for representative images]. Signal noise resulting from the small size of the bleach spot combined with the relatively faint intranuclear YFP signals was overcome by collection of FRAP data from ≈30–100 individual cells. FRAP normalized intensity was then calculated as the average value ± standard error at each time point after photobleaching. Representative data for TBP-YFP in WT cells are shown in Fig. 1A; for simplicity, subsequent graphs only show the lines derived from the average data.
Fig. 1.
FRAP of TBP and other GTFs. (A) Normalized intensity of FRAP of TBP-YFP in a WT cell. Individual data points correspond to the average intensity at the time point shown, with error bars indicating the SEM between independent measurements. (B) FRAP of YFP-NLS and AceI-GFP in WT and mot1 cells. Cells were imaged and fluorescent signal was calculated before and after photobleaching a portion of the nucleus. Graphs show the recovery kinetics after correcting for background and loss of fluorescence caused by imaging. Level of prebleach fluorescence is normalized to 1; bleach depth is not normalized. Because the same photobleaching conditions were always used, different bleach depths in the FRAP curves reflect the different rates of fluorescent recovery occurring between the end of the photobleach and the first image. For these and all remaining FRAPs, graphs show the average intensity of the bleached spot for the indicated time point. (C) FRAP of PIC components and Mot1-YFP. Relative recovery is shown for components of the Pol II PIC, including TBP-YFP, TAF1-YFP, TFIIB-YFP, and Rbp1-GFP. Rpb1-GFP FRAP was very similar to FRAP of two other Pol II subunits (Fig. S4). Data are represented as described in B.
The results in Fig. 1A show that TBP is highly mobile in yeast nuclei, with ≈90% of the TBP-YFP fluorescence signal recovered in just over 7 s and complete recovery in ≈15 s. The recovery data were biphasic (Fig. S2), indicating that TBP is composed of at least two populations of distinguishable molecules. We cannot rule out the possibility that there is an immobile fraction of TBP, but it is such a tiny proportion of the total that we cannot detect it. For comparison, and in agreement with previously published results (1), mobility of freely diffusible YFP alone was much faster, whereas mobility of the AceI transcription factor was slower than YFP but faster than TBP (Fig. 1 B and C). Mobilities of TFIIB, the RNA Pol II subunit Rpb1, and the TFIID component TBP-associated factor 1 (TAF1) were also measured by FRAP (Fig. 1C). FRAP curves for all of these factors were distinct from one another and from the TBP curve. Therefore, at least some populations of these factors are independent of one another in vivo. As TAF1 is a central component required for TFIID complex assembly (16), the differences in the TAF1 and TBP FRAP curves indicate that most TBP is not stably incorporated into the TFIID complex in vivo.
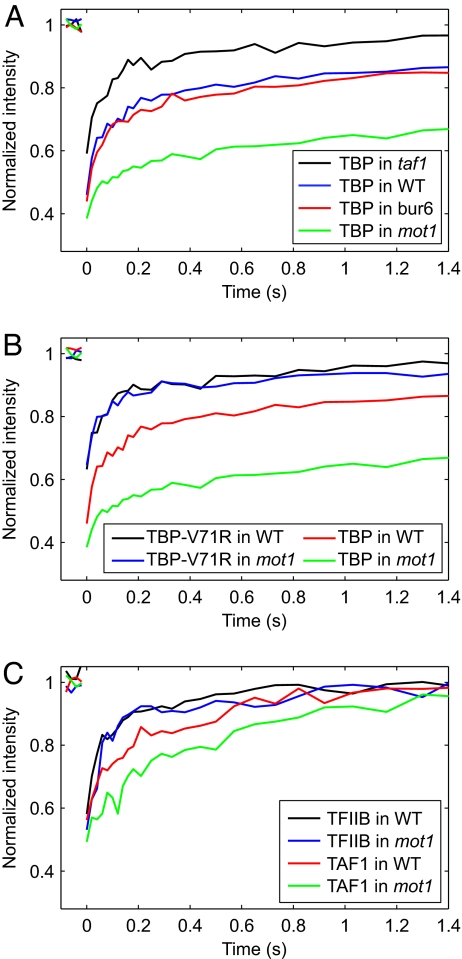
Fluorescence recovery rate depends on the size of the diffusing complex (i.e., the diffusion coefficient) and the extent to which the mobility of the diffusing complex is retarded by association with immobile nuclear constituents (9, 17). Polypeptide size cannot explain these results because the much larger TAF1 and Mot1 proteins display markedly faster recovery rates than the rather small TBP (Fig. 1C). Additionally, TFIIB and Mot1 have very different native molecular weights but were the most highly mobile transcription factors tested. The dependence of diffusion rate on molecular volume means that an 8-fold increase in mass would be required for a modest 2-fold decrease in the rate of free diffusion. Appreciable effects on diffusion-limited recovery rates based on size alone would require all of these proteins to be constituents of much larger complexes than are physiologically plausible (4). Thus, a significant effect on GTF mobility is likely association with chromatin, which is essentially immobile over the time course of these experiments. Consistent with this idea, Rpb1 displayed slow recovery, as expected, if it were assembled into Pol II molecules that were engaged in transcription. The highly mobile behavior of TFIIB is consistent with previous results (11) and suggests that rather than being stably associated with chromatin its binding is rate limiting and/or it is rapidly released after the onset of initiation (10, 18–20). To determine the extent to which chromatin binding retards TBP mobility, FRAP was performed in a strain expressing the DNA binding-defective TBP mutant V71R (21). As shown in Fig. 2B, the V71R mutation increased TBP mobility, supporting the suggestion that most TBP molecules are chromatin-associated in vivo, and that most chromatin binding by TBP is short-lived. The level of TBP V71R was a small fraction of the WT TBP level (Fig. S3), indicating that the faster mobility of the mutant TBP was not attributable to being overexpressed.
Fig. 2.
FRAP of GTFs in WT and mutant cells. (A) FRAP of TBP in mutant cells. TBP-YFP recovery was visualized in mot1, bur6, and taf1 cells to determine the effects on mobility when essential regulators of TBP binding are disrupted. The TBP-YFP curve in WT cells is replotted from Fig. 1C for comparison. (B) FRAP of TBP that is defective for DNA binding. TBP V71R-YFP was visualized in WT and mot1 cells to determine the effect of chromatin binding on TBP dynamics. WT TBP-YFP curves are replotted from A for comparison. (C) FRAP of GTFs in mot1 cells. TFIIB-YFP and TAF1-YFP were visualized in mot1 cells to determine the specificity of Mot1's function on TBP. Curves of TFIIB-YFP and Taf1-YFP in WT cells are replotted from Fig. 1C for comparison.
The interpretation of rapid redistribution of TBP among chromatin sites in vivo is at odds with the longevity of the TBP–DNA complex measured in vitro, but could be explained by the actions of transcriptional regulatory factors that directly interact with TBP and thereby regulate transcription on a global scale. An alternative, not mutually exclusive, possibility is that most TBP in vivo is nonspecifically bound to low-affinity DNA sites. The NC2 complex, a heterodimer of Bur6 and Ydr1 (22), promotes relocation of TBP along the DNA contour, allowing TBP to move toward or away from a TATA box (23). Mobility of TBP was not much different in bur6 cells compared with WT cells (Fig. 2A), indicating that Bur6 does not contribute to the TBP recovery rate. In contrast, TBP mobility was substantially increased in taf1 cells compared with WT (Fig. 2A), consistent with a role for the Taf1-containing TFIID complex in stabilizing TBP binding to promoters. In striking contrast, TBP mobility was drastically decreased in mot1 cells compared with WT cells (Fig. 2A). Mot1-mediated regulation of TBP's dynamic behavior was specific for TBP, as mobilities of YFP alone, AceI, and TFIIB were indistinguishable in WT and mot1 cells (Figs. 1B and 2C). These results are fully consistent with Mot1's biochemical activity as a TBP–DNA dissociating enzyme (24, 25), and indicate that Mot1 plays a unique and critical role in establishing the large-scale dynamic behavior of TBP in vivo. Although other factors such as proteasomes, chromatin remodelers, and chaperones have also been implicated in destabilizing nuclear protein interactions with chromatin (1, 26–28), the Mot1 in vitro and in vivo data combine to provide perhaps the clearest mechanistic explanation for how dynamic behavior is generated in vivo for a protein with high intrinsic affinity for chromatin.
We found that Mot1 also had some effect on the recovery rate of TAF1 (Fig. 2C), which could be evidence that TAF1 is sequestered by transcriptionally inactive TBP-containing complexes more stably bound to chromatin in mot1 cells. The altered mobility of TAF1 in mot1 cells is also consistent with the observation that many Mot1-activated genes are TAF1-dependent, and although TFIID and Mot1 are not apparently physically associated, ChIP experiments suggest that a defective form of TFIID is assembled at promoters when Mot1 function is impaired (25).
Fig. 3 summarizes all of the FRAP results by displaying the mobility of each factor on the basis of its recovery rate with associated error, calculated as described in Materials and Methods. This analysis provides quantitative support for the differences in factor mobility described above. We also performed pairwise comparisons by using an extra sum-of-squares F test to determine which curves were significantly different from one another with >99% confidence (P < 0.01). Notably, TBP, TFIIB, TAF1, and Rpb1 have significantly different recovery rates (Fig. 3A), indicating that they are not stably associated with one another. The recovery of TBP is drastically faster when DNA-binding activity is lost (Fig. 3B), consistent with the suggestion that association with chromatin limits TBP mobility. Quantitative analysis showed that the TBP recovery rate is markedly reduced in mot1 cells. The TAF1 recovery rate is also significantly reduced in mot1 cells, but to a much smaller extent (Fig. 3B). These effects are specific, because recovery rates for the other proteins are not distinguishably different in WT versus mot1 cells (Fig. 3C), nor was there a significant effect on TBP mobility in bur6 cells (Fig. 3B), despite the fact that MOT1 and BUR6 have overlapping functions (29–31). These comparisons were performed by using single-component models to fit the complete FRAP curves. However, in some cases, notably TBP and Pol II, two-component models yielded much better fits. These better fits did not alter the conclusions shown in Fig. 3 (see Fig. S2 and Fig. S4).
Fig. 3.
Dot plots to quantify FRAP comparisons. The recovery rate and standard error for each FRAP curve were plotted to highlight key differences. Significant differences in pairwise comparisons are indicated by brackets with closed diamonds; open diamonds denote no significant difference. The x axis is a logarithmic scale showing the values of the recovery rate for each curve, calculated as described in Materials and Methods.
With a TBP FRAP rate of 15 s, these results demonstrate extraordinarily dynamic behavior for TBP in vivo. Estimates for the abundance of TBP in vivo vary somewhat, but there appear to be ≈20,000 molecules of TBP per haploid yeast cell (32). About a third of these molecules would be required for expression of all of the annotated genes, but transcription is much more complex than this, with close to 90% of the entire yeast genome transcribed (33). If all active PICs were long-lived (i.e., with lifetimes longer than a few seconds), we would expect to see a significant immobile fraction in the FRAP of TBP. The absence of a measurable immobile fraction suggests that the PICs of at least some, and perhaps many, active promoters are rapidly assembled and disassembled rather than stable. There may be very stable TBP-containing complexes at some active promoters, but the proportion of these in comparison to the total TBP pool is too small to be detectable. The rate of PIC disassembly may vary greatly from promoter to promoter, just as the kinetic and thermodynamic parameters of the assembly reaction vary from promoter to promoter. Active PIC disassembly by Mot1 (and probably other factors) may be facilitated by, or contribute to, the inefficiency of Pol II initiation observed in vivo (34). PIC instability at active promoters may also be critical for integration of transcriptional responses with other regulatory events and may provide a mechanism for ensuring the timely termination of transcription at deactivated genes. In contrast to most DNA binding proteins, TBP binds in vitro with high stability to a variety of DNA sequences (35–37); similarly stable and promiscuous binding in vivo could affect start site utilization on a global scale and deplete the pool of free TBP. We propose that Mot1-mediated rapid recycling of TBP from chromatin is essential to limit spurious PIC assembly and to provide an adequate pool of free TBP for timely transcriptional regulation. Hence an active process limits the residency times, thereby reducing them from what is expected based on in vitro binding constants. Consistent with this notion, results indicate that TBP is limiting for transcription in both yeast (38) and mammalian cells (39, 40).
At first glance a surprising feature of our results is that TBP FRAP dynamics in live yeast cells (Fig. 1C) are much faster than in mammalian cells (11). However, what appears to be conserved between yeast and mammals are the relative mobilities of PIC components. TFIIB in both systems exhibited the fastest recoveries followed by TBP and then Pol II (Fig. 1C) (11, 41). Furthermore, within each system, TBP and Pol II recoveries operated on comparable time scales: at least 20 min in mammalian cells and <1 min in yeast cells. This may reflect, at least in part, the high number of promoter-proximal stalled Pol II complexes in mammalian cells (42). Another possibility is that this dramatic shift in timing reflects the fact that the average yeast gene is at least an order of magnitude shorter than its mammalian counterpart. If rates of Pol elongation are roughly comparable, then the time to transcribe a typical yeast gene should be significantly less than for a typical mammalian gene. This elongation time is thought to be associated with recovery of the slower second component of mammalian Pol II FRAP curves (41, 43). We found that yeast Pol II FRAPs were also well fit by a model containing a slower second component (Fig. S4). However, this slower component recovered much faster in yeast than in mammalian cells, a result explicable in terms of the time to transcribe the average gene in the two organisms. Another interesting difference in the Pol II FRAPs was the fraction of molecules in the slower second component. This was much larger in yeast (60–80%; Fig. S4) than in mammalian cells (25%), raising the possibility that a higher fraction of nuclear Pol II may be engaged in transcription in yeast.
Combining the available yeast and mammalian in vivo data, we are led to the conclusion that the time to transcribe a gene may influence the residence times of other PIC components. Exactly how such coordination might be achieved is unknown, but increasing evidence for physical communication between the 5′ and 3′ ends of genes (44, 45) provides a plausible framework. Regardless of the explicit mechanism of coordination, the in vivo data suggest that TBP association with chromatin could be variable and gene-dependent, rather than consistently “stable” or “unstable.” Viewed in this context, it becomes apparent that the in vitro measurements of TBP that have spawned the stable TBP scaffold model are not likely to capture all of the relevant features present in vivo. These include not only a role for Mot1, but also potentially other factors involved in the possible coordination of TBP residency and Pol II elongation times.
Materials and Methods
Yeast Strains and Growth Conditions.
Homozygous diploid strains expressing C-terminal YFP fusions were constructed by homologous recombination of PCR-generated DNA fragments using standard yeast genetic approaches as described (46). Tagging vectors were obtained from EUROSCARF. Diploid strains were better suited than haploids for FRAP experiments because their nuclei are larger. TAF1-YFP and TBP-V71R-YFP experiments were conducted in ade5Δ strains to reduce autofluorescence. TBP-V71R-YFP was expressed in haploid cells from a low-copy plasmid under control of the GPD promoter derived from pTSK274 (47). For a complete list of strains used in this study, see Table S1. To prepare cells for FRAP, strains were grown overnight in 3 ml of complete synthetic medium-histidine at 28°C. Cultures were diluted 1:30 into 3 ml of prewarmed synthetic media and grown for 3–4 h before imaging. Five-hundred-microliter aliquots were centrifuged briefly in glass tubes, and 2 μl of concentrated cell suspension was dropped onto Lab-Tek II chambers. Agarose slabs were used as cover slips to extend the life of the sample.
Cell Imaging and FRAP.
FRAP experiments were carried out on a Zeiss 510 confocal microscope with a 100 × 1.3 NA oil immersion objective. Cells were imaged with a 488-nm laser line at low laser intensity (0.60–0.75%) to reduce bleaching caused by imaging. Bleaching was performed with the 488-nm line from a 40-mW argon laser operation at 60–75% laser power. A 7-ms bleach pulse was used, producing an incomplete photobleach in an approximately circular area of 0.7 μm in diameter. Fluorescence recovery was monitored every 20 ms. All of the experiments were repeated at least twice, and the bleach conditions were identical for all experiments. The total cell number analyzed for each experiment was routinely >30. For each data set shown, separate FRAPs were performed and then averaged to generate a single FRAP curve.
Data Processing and Analysis.
Individual FRAP curves were background subtracted and corrected for bleaching caused by imaging (27). About 30–100 separate FRAP experiments were performed and then averaged to generate a single FRAP curve. Images were acquired at equidistant time intervals throughout the measurement. For the analysis, data points were binned so that later time points were evenly distributed on a logarithmic time scale (48). This binning avoids overly weighting the slower phase of the FRAP curve. To determine the rate of recovery binned FRAP curves were fit with a function known to provide a useful empirical fit to many FRAP curves (28):
where θ is the bleach depth and determines the fluorescence intensity at the beginning of the recovery. χ is the recovery rate and determines how fast the recovery takes place: smaller values lead to a slower recovery. I0 and I1 are Bessel functions of the first kind. The Matlab routine nlinfit was used to fit the models to experimental data.
The preceding equation provides a good empirical fit to many one-component FRAP curves. We extended this equation to generate a two-component model that we found provided a much better fit to some of the yeast FRAP curves. The two-component model was:
where φ is the fraction of molecules in the slow component and k is the recovery rate of that component.
To test whether FRAP curves are significantly different, we used the extra sum-of-squares F test (49) to compare the FRAP curves by using the one-component model. This test determines whether two data sets can be described by the same χ (null hypothesis) or if they have to be described with separate χ (alternative hypothesis).
Supplementary Material
Acknowledgments.
We thank Tom Misteli and Sui Huang for discussions and Staton Wade and Woo-sin Park for comments on this manuscript. This work was supported by National Institutes of Health Grant GM55763 (to D.T.A.) and the intramural program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.H.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801901105/DCSupplemental.
References
- 1.Karpova TS, Chen TY, Sprague BL, McNally JG. Dynamic interactions of a transcription factor with DNA are accelerated by a chromatin remodeler. EMBO Rep. 2004;5:1064–1070. doi: 10.1038/sj.embor.7400281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Phair RD, et al. Global nature of dynamic protein–chromatin interactions in vivo: Three-dimensional genome scanning and dynamic interaction networks of chromatin proteins. Mol Cell Biol. 2004;24:6393–6402. doi: 10.1128/MCB.24.14.6393-6402.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shav-Tal Y, Darzacq X, Singer RH. Gene expression within a nuclear landscape. EMBO J. 2006;25:3469–3479. doi: 10.1038/sj.emboj.7601226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Misteli T. Protein dynamics: Implications for nuclear architecture and gene expression. Science. 2001;291:843–847. doi: 10.1126/science.291.5505.843. [DOI] [PubMed] [Google Scholar]
- 5.Cheutin T, et al. Maintenance of stable heterochromatin domains by dynamic HP1 binding. Science. 2003;299:721–725. doi: 10.1126/science.1078572. [DOI] [PubMed] [Google Scholar]
- 6.Dion MF, et al. Dynamics of replication-independent histone turnover in budding yeast. Science. 2007;315:1405–1408. doi: 10.1126/science.1134053. [DOI] [PubMed] [Google Scholar]
- 7.Jamai A, Imoberdorf RM, Strubin M. Continuous histone H2B and transcription-dependent histone H3 exchange in yeast cells outside of replication. Mol Cell. 2007;25:345–355. doi: 10.1016/j.molcel.2007.01.019. [DOI] [PubMed] [Google Scholar]
- 8.Mito Y, Henikoff JG, Henikoff S. Histone replacement marks the boundaries of cis-regulatory domains. Science. 2007;315:1408–1411. doi: 10.1126/science.1134004. [DOI] [PubMed] [Google Scholar]
- 9.van Holde KE, Zlatanova J. Scanning chromatin: A new paradigm? J Biol Chem. 2006;281:12197–12200. doi: 10.1074/jbc.R500025200. [DOI] [PubMed] [Google Scholar]
- 10.Yudkovsky N, Ranish JA, Hahn S. A transcription reinitiation intermediate stabilized by activator. Nature. 2000;408:225–229. doi: 10.1038/35041603. [DOI] [PubMed] [Google Scholar]
- 11.Chen D, Hinkley CS, Henry RW, Huang S. TBP dynamics in living human cells: Constitutive association of TBP with mitotic chromosomes. Mol Biol Cell. 2002;13:276–284. doi: 10.1091/mbc.01-10-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dundr M, et al. A kinetic framework for a mammalian RNA polymerase in vivo. Science. 2002;298:1623–1626. doi: 10.1126/science.1076164. [DOI] [PubMed] [Google Scholar]
- 13.Aprikian P, Moorefield B, Reeder RH. New model for the yeast RNA polymerase I transcription cycle. Mol Cell Biol. 2001;21:4847–4855. doi: 10.1128/MCB.21.15.4847-4855.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Blake WJ, et al. Phenotypic consequences of promoter-mediated transcriptional noise. Mol Cell. 2006;24:853–865. doi: 10.1016/j.molcel.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 15.Raser JM, O'Shea EK. Control of stochasticity in eukaryotic gene expression. Science. 2004;304:1811–1814. doi: 10.1126/science.1098641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singh MV, Bland CE, Weil PA. Molecular and genetic characterization of a Taf1p domain essential for yeast TFIID assembly. Mol Cell Biol. 2004;24:4929–2942. doi: 10.1128/MCB.24.11.4929-4942.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sprague BL, Pego RL, Stavreva DA, McNally JG. Analysis of binding reactions by fluorescence recovery after photobleaching. Biophys J. 2004;86:3473–3495. doi: 10.1529/biophysj.103.026765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Roberts SG, Choy B, Walker SS, Lin YS, Green MR. A role for activator-mediated TFIIB recruitment in diverse aspects of transcriptional regulation. Curr Biol. 1995;5:508–516. doi: 10.1016/s0960-9822(95)00103-5. [DOI] [PubMed] [Google Scholar]
- 19.Sandaltzopoulos R, Becker PB. Heat shock factor increases the reinitiation rate from potentiated chromatin templates. Mol Cell Biol. 1998;18:361–367. doi: 10.1128/mcb.18.1.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zawel L, Kuman KP, Reinberg D. Recycling of the general transcription factors during RNA polymerase II transcription. Genes Dev. 1995;9:1479–1490. doi: 10.1101/gad.9.12.1479. [DOI] [PubMed] [Google Scholar]
- 21.Jackson-Fisher AJ, Chitikila C, Mitra M, Pugh BF. A role for TBP dimerization in preventing unregulated gene expression. Mol Cell. 1999;3:717–727. doi: 10.1016/s1097-2765(01)80004-6. [DOI] [PubMed] [Google Scholar]
- 22.Kim S, Cabane K, Hampsey M, Reinberg D. Genetic analysis of the Ydr1–Bur6 repressor complex reveals an intricate balance among transcriptional regulatory proteins in yeast. Mol Cell Biol. 2000;20:2455–2465. doi: 10.1128/mcb.20.7.2455-2465.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schluesche P, Stelzer G, Piaia E, Lamb DC, Meisterernst M. NC2 mobilizes TBP on core promoter TATA boxes. Nat Struct Mol Biol. 2007;14:1196–1201. doi: 10.1038/nsmb1328. [DOI] [PubMed] [Google Scholar]
- 24.Auble DT, et al. Mot1, a global repressor of RNA polymerase II transcription, inhibits TBP binding to DNA by an ATP-dependent mechanism. Genes Dev. 1994;8:1920–1934. doi: 10.1101/gad.8.16.1920. [DOI] [PubMed] [Google Scholar]
- 25.Dasgupta A, Juedes SA, Sprouse RO, Auble DT. Mot1-mediated control of transcription complex assembly and activity. EMBO J. 2005;24:1717–1729. doi: 10.1038/sj.emboj.7600646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Freeman BC, Yamamoto KR. Disassembly of transcriptional regulatory complexes by molecular chaperones. Science. 2002;296:2232–2235. doi: 10.1126/science.1073051. [DOI] [PubMed] [Google Scholar]
- 27.Stavreva DA, McNally JG. Fluorescence recovery after photobleaching (FRAP) methods for visualizing protein dynamics in living mammalian cell nuclei. Methods Enzymol. 2004;375:443–455. doi: 10.1016/s0076-6879(03)75027-7. [DOI] [PubMed] [Google Scholar]
- 28.Stavreva DA, Müller WG, Hager GL, Smith CL, McNally JG. Rapid GR exchange at a promoter: Coupling to transcription and regulation by chaperones and proteasomes. Mol Cell Biol. 2004;24:2682–2697. doi: 10.1128/MCB.24.7.2682-2697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dasgupta A, Darst RP, Martin KJ, Afshari CA, Auble DT. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proc Natl Acad Sci USA. 2002;99:2666–2671. doi: 10.1073/pnas.052397899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Geisberg JV, Holstege FC, Young RA, Struhl K. Yeast NC2 associates with the RNA polymerase II preinitiation complex and selectively affects transcription in vivo. Mol Cell Biol. 2001;21:2736–2742. doi: 10.1128/MCB.21.8.2736-2742.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prelich G. Saccharomyces cerevisiae BUR6 encodes a DRAP1/NC2α homolog that has both positive and negative roles in transcription in vivo. Mol Cell Biol. 1997;17:2057–2065. doi: 10.1128/mcb.17.4.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Borggrefe T, Davis R, Bareket-Samish A, Kornberg RD. Quantitation of the RNA polymerase II transcription machinery in yeast. J Biol Chem. 2001;276:47150–47153. doi: 10.1074/jbc.M109581200. [DOI] [PubMed] [Google Scholar]
- 33.David L, et al. A high-resolution map of transcription in the yeast genome. Proc Natl Acad Sci USA. 2006;103:5320–5325. doi: 10.1073/pnas.0601091103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darzacq X, et al. In vivo dynamics of RNA polymerase II transcription. Nat Struct Mol Biol. 2007;14:796–806. doi: 10.1038/nsmb1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoopes BC, LeBlanc JF, Hawley DK. Kinetic analysis of yeast TFIID-TATA box complex formation suggests a multistep pathway. J Biol Chem. 1992;267:11539–11547. [PubMed] [Google Scholar]
- 36.Patikoglou GA, et al. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Petri V, Hsieh M, Brenowitz M. Thermodynamic and kinetic characterization of the binding of the TATA binding protein to the adenovirus E4 promoter. Biochemistry. 1995;34:9977–9984. doi: 10.1021/bi00031a020. [DOI] [PubMed] [Google Scholar]
- 38.Patterson GH, Schroeder SC, Bai Y, Weil PA, Piston DW. Quantitative imaging of TATA-binding protein in living yeast cells. Yeast. 1997;14:813–825. doi: 10.1002/(SICI)1097-0061(19980630)14:9<813::AID-YEA280>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 39.Bush SD, Richard P, Manley JL. Variations in intracellular levels of TATA binding protein can affect specific genes by different mechanisms. Mol Cell Biol. 2008;28:83–92. doi: 10.1128/MCB.00809-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Colgan J, Manley JL. TFIID can be rate limiting in vivo for TATA-containing, but not TATA-lacking, RNA polymerase II promoters. Genes Dev. 1992;6:304–315. doi: 10.1101/gad.6.2.304. [DOI] [PubMed] [Google Scholar]
- 41.Kimura H, Sugaya K, Cook PR. The transcription cycle of RNA polymerase II in living cells. J Cell Biol. 2002;159:777–782. doi: 10.1083/jcb.200206019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wu JQ, Snyder M. RNA polymerase II stalling: Loading at the start prepares genes for a sprint. Genome Biol. 2008;9:220. doi: 10.1186/gb-2008-9-5-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dundr M, et al. In vivo kinetics of Cajal body components. J Cell Biol. 2004;164:831–842. doi: 10.1083/jcb.200311121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perkins KJ, Lusic M, Mitar I, Giacca M, Proudfoot NJ. Transcription-dependent gene looping of the HIV-1 provirus is dictated by recognition of pre-mRNA processing signals. Mol Cell. 2008;29:56–68. doi: 10.1016/j.molcel.2007.11.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Singh BN, Hampsey M. A transcription-independent role for TFIIB in gene looping. Mol Cell. 2007;27:806–816. doi: 10.1016/j.molcel.2007.07.013. [DOI] [PubMed] [Google Scholar]
- 46.Sheff MA, Thorn KS. Optimized cassettes for fluorescent protein tagging in Saccharomyces cerevisiae. Yeast. 2004;21:661–670. doi: 10.1002/yea.1130. [DOI] [PubMed] [Google Scholar]
- 47.Karpova TS, et al. Concurrent fast and slow cycling of a transcriptional activator at an endogenous promoter. Science. 2008;319:466–469. doi: 10.1126/science.1150559. [DOI] [PubMed] [Google Scholar]
- 48.Waharte F, Brown CM, Coscoy S, Coudrier E, Amblard F. A two-photon FRAP analysis of the cytoskeleton dynamics in the microvilli of intestinal cells. Biophys J. 2005;88:1467–1478. doi: 10.1529/biophysj.104.049619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Motulsky H, Christopoulos A. Fitting Models to Biological Data Using Linear and Nonlinear Regression: A Practical Guide to Curve Fitting. New York: Oxford Univ Press; 2004. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.