Abstract
The phosphodiesterases (PDEs) are metal ion-dependent enzymes that regulate cellular signaling by metabolic inactivation of the ubiquitous second messengers cAMP and cGMP. In this role, the PDEs are involved in many biological and metabolic processes and are proven targets of successful drugs for the treatments of a wide range of diseases. However, because of the rapidity of the hydrolysis reaction, an experimental knowledge of the enzymatic mechanisms of the PDEs at the atomic level is still lacking. Here, we report the structures of reaction intermediates accumulated at the reaction steady state in PDE9/crystal and preserved by freeze-trapping. These structures reveal the catalytic process of a PDE and explain the substrate specificity of PDE9 in an actual reaction and the cation requirements of PDEs in general.
Keywords: crystallography, enzyme mechanism, reaction intermediates, freeze-trapping
The phosphodiesterases (PDEs) are a superfamily of enzymes that metabolically inactivate the ubiquitous intracellular messengers cAMP and cGMP. This function involves the PDEs in a broad range of important cellular functions, such as immune response, memory, and vision (1–4). The human genome encodes for 21 PDEs that are categorized into 11 families (2). Alternative splicing results in the generation of >60 identified isoforms. These enzymes share a conserved catalytic domain of approximately 300 aa that is located in the C-terminal region of the protein. The N-terminal regions, which vary among different PDEs, serve regulatory functions including autoinhibition of the catalytic domains or control of subcellular localization (2). The PDEs have different substrate preferences: PDE 4, -7, and -8 preferentially hydrolyze cAMP; PDE5, -6, and -9 are cGMP specific. PDE1, -2, -3, -10, and -11 can hydrolyze both cyclic nucleotides (2). The different substrate preferences, combined with different expression profiles, cellular compartmentalization, and regulation, allow the PDEs to play a very versatile role in cell signal transduction (5).
It is becoming increasingly clear that the physiological role of PDEs is the temporal and spatial control of cyclic nucleotide signaling, not simply inactivation (1, 2, 6). The clearest example of this control is in the fast action of PDE6 required for the temporal resolution of human vision (4). However, a complete understanding of the catalytic mechanism of these enzymes that accounts for the substrate specificity and reaction kinetics at the atomic level is lacking. Crystal structures of the catalytic domains of PDE1B, -2, -3, -4, -5, -7, -9, and -10, by themselves or in complex with inhibitors, substrates, or products, have been reported (7–19). Generally, the active sites of PDEs can be divided into 2 parts: a nucleotide recognition pocket and a hydrolysis center (7, 19). Each PDE has its unique nucleotide recognition pocket, resulting in different substrate specificity and inhibition profiles (7–19). Based on the high-resolution crystal structures of the apo PDE1B and product-bound PDE4B, -4D, and -5A, a so-called “glutamine switch mechanism” was proposed using hydrogen-bond patterns specific for nucleotide bases to explain their substrate selectivity (19). On the other hand, the structures of the hydrolysis center in all PDEs are highly similar, and a universal hydrolysis reaction mechanism is expected. However, because the reactions catalyzed by PDEs progress rapidly in solution, efforts to obtain substrate-bound co-crystal structures of native PDEs have been unsuccessful to date. Thus, a direct observation of substrate binding is lacking. Lack of this information also has resulted in controversy regarding the determinants of substrate specificity (18).
Here, we report on crystallographic studies of PDE9, a cGMP-specific PDE that shed light on these issues. Different species were captured during the hydrolytic process within the crystal lattice of the active PDE by taking advantage of PDE9's high affinity for cGMP (≈170 nM), low affinity for the nonselective PDE inhibitor 3-isobutyl-1-methylxanthine (IBMX) (20–23), and the absence of product inhibition even at high concentrations. Specifically, we prepared crystals of PDE9 catalytic domain in complex with IBMX using a published protocol (16). These crystals then were soaked for different durations with substrate cGMP in the presence of different metal ions to vary the level of enzymatic activity. The complex of PDE9 with substrate, product, and putative reaction intermediates were captured by freeze-trapping (24). From these crystal structures, we deduce a putative reaction scheme.
Results
PDE9 Kinetics.
The kinetics of PDE9 in solution were characterized under steady-state and single-turnover conditions. In steady-state reactions, the wild type PDE9 catalytic domain used for crystallization was found initially to be fully active without divalent metals, because of the incorporation of metal ions during expression. This enzymatic activity gradually dies within an hour when exposed to ≈18 mM Zn2+. The protein also can be inactivated completely by incubation with 10 mM EDTA for 30 min, which strips the original metals. Inclusion of 8 mM MnCl2 or 8 mM MgCl2 or 4 mM MnCl2 plus 4 mM MgCl2 in the reaction buffer can restore fully the activity of PDE9 inactivated by EDTA. After reactivation, both substrate affinity (Km values of 51, 43.5, and 126 nM, respectively) and the maximum velocities (Vmax values of 1.2, 0.91, and 2.0 nmol·min−1·nmol−1 protein, respectively) were similar to initial values before metal stripping (Km of 80 ± 20 nM and Vmax of 1.3 ± 0.2 nmol·min−1·nmol−1 protein in the presence of 8 mM MgCl2). In contrast, when the reaction buffer contained 2 mM EDTA or 8 mM ZnCl2, the protein remained inactive. Similar divalent cation requirements for other PDEs, including the inhibitory effects of Zn2+, have been reported (25).
In reactions that were run with 4,000 nM cGMP, inclusion of 20 μM 5′-GMP reduced the initial reaction rate by only ≈9%, indicating that 5′-GMP does not compete well with cGMP for binding at the active site of PDE9. The lack of product inhibition in the presence of high concentrations of product also was reported for PDE10 (18).
To determine the rate-limiting step of the PDE9 reaction, single-turnover reactions were run in which the amount of enzyme (120 or 240 nM) exceeded that of substrate [3H]cGMP (20 nM) and were quenched at different times by adding a large excess (150 μM) of unlabeled cGMP or 4 N HCl [see supporting information (SI) Appendix]. The data from these studies, combined with data from steady-state kinetic experiments, were used to calculate the various kinetic parameters of the overall reaction. These results indicate that substrate binding, dissociation, and hydrolysis are fast relative to product dissociation, suggesting that this later step is rate limiting (see SI Appendix for detailed reaction kinetics and analysis).
Capturing Guanine Nucleotides in PDE9 Crystals.
In the present study, soaking with guanine nucleotides did not change the overall protein structures or cause any significant conformational changes at the PDE9 active site (Fig. 1). There are two PDE9 molecules in the asymmetric unit, forming a dimer (16). The active site of monomer A is completely exposed to solvent channels. However, the active site of monomer B is occluded by the C-terminal helix of a neighboring monomer A. Because of this difference in crystal contact, guanine nucleotides could displaced IBMX from monomer A completely after brief soaking but could not displace guanine nucleotides from monomer B even after incubation for days (Table S1). Thus, to avoid ambiguities caused by differences in kinetics of inhibitor release, substrate binding, and product release in the two active sites, the complexes described here all represent binding at the monomer A active site.
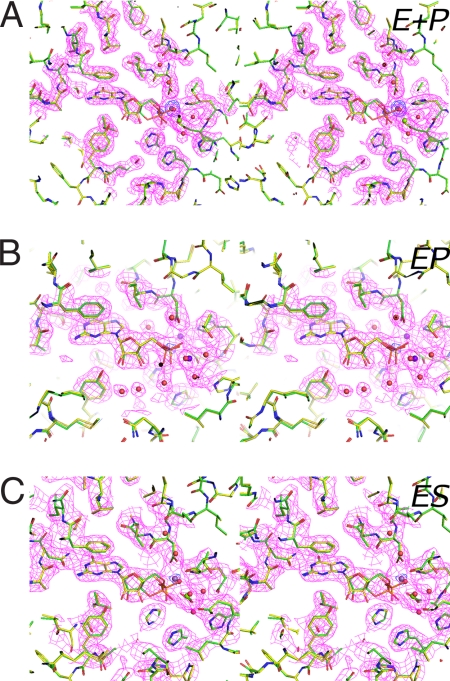
Fig. 1.
Guanine nucleotides captured at monomer A of PDE9 crystals after soaking. The final refined models (protein and ligands in sticks, metal ions, and water in balls) are embedded in the initial 2 Fo−Fc maps calculated before ligands and water molecules were built (pink, 1.2 σ). Figures were prepared using the PyMOL molecular graphics system (DeLano Scientific). Blue, nitrogen; red, oxygen; brown, phosphorus; purple-blue, metal ions. (A) The E+P complex derived from completely hydrolyzed cGMP (1 mM cGMP + 0.5 mM MnCl2, soaked for 2 days). Identical complex was obtained by soaking with 1 mM 5′-GMP + 1 mM −gCl2. The FocGMP −Fo,GMP+no ion electron density map (blue, 10 σ) shows the full occupancy of the first metal (M1) and the precise position of the phosphorus. The electron density for the second metal (M2) is always weaker, even soaked with 20 mM MnCl2. The final model (yellow carbons) was superimposed with the model of the EP complex discussed in Fig. 1B (green carbons). (B) The EP complex captured during reaction in fully active PDE9 crystals (20 mM cGMP + 20 mM MnCl2 + 20 mM MgCl2, from a few seconds to 2 h). The final model (green carbons) was superimposed with the E+P complex model (yellow carbons). The initial Fo−Fc electron density (blue, 15 σ) shows the precise location of the phosphorus. (C) The ES complex captured during reaction in crystals of reduced activity (20 mM cGMP + 2 mM MnCl2, 1 min). The final model (green carbons) was superimposed with the E+P model (yellow carbons). The Fo,cGMP −Fo,GMP+no ion electron density map (blue, 10 σ) indicates significant occupancy of metal ions.
To investigate metal-ion requirements for activity of crystalline PDE9, crystals were soaked for 2 days at 4°C with 1 mM cGMP and different metal ions before flash-cooling to 100 K. A soaking time of 2 days was chosen to ensure the complete hydrolysis of cGMP by crystalline PDE9. Based on unbiased electron densities of captured a PDE9/5′-GMP complex, cGMP was hydrolyzed in crystals in the presence of Mn2+ or Mg2+ (Fig. 1A). The same complex also was obtained by direct soaking crystals with 5′-GMP and Mg2+ (Table S1). Thus, this PDE9/5′-GMP complex is interpreted as the regenerated enzyme/rebound product complex (E+P) formed when the reaction reached the equilibrium after all substrates were completely hydrolyzed. In contrast, in the absence of divalent cations or in the presence of ZnCl2, a PDE9/cGMP complex was captured (Figs. S1 and S2), indicating that the enzyme is inactive under these conditions.
Because crystalline PDE9 retains activity, we were able to capture the hydrolytic reactions in crystals using a freeze-trapping method (24), and from these snapshots we could deduce a reaction scheme. Reactions were initiated by transferring one PDE9 crystal into a 5-μl soaking solution containing 20 mM cGMP with Mn2+ or Mg2+ or both. After a period of time ranging from a few seconds to up to 2 h, the reactions were stopped by flash-cooling in liquid nitrogen. X-ray crystallographic data were collected at 100 K on these frozen crystals (Table S1). Within these short reaction times, cGMP still was abundant in the soaking solution, because under optimal conditions, the amount of active PDE9 (1 × 1013 molecules) in a typical crystal used for soaking (≈150 × 150 × 150 μm3) could hydrolyze only ≈ 2% of the total amount of cGMP in the soaking solution (1.2 × 1015 in 6 × 1016 molecules). Thus, the reactions were at a kinetic steady state when crystals were flash-cooled
Under conditions in which the enzyme is fully active (20 mM cGMP + 20 mM MnCl2 + 20 mM MgCl2), cGMP was cleaved in a few seconds, and a PDE9/5′-GMP complex was captured (Fig. 1B) that differed from the putative E+P complex described previously. We interpret this PDE9/5′-GMP complex as the enzyme/product (EP) complex of the hydrolysis reaction, a species existed right after the cleavage of the P-O3′ bond. This interpretation is consistent with the conclusions from the kinetic studies that cGMP hydrolysis is fast and that 5′-GMP dissociation is the rate-limiting step of the reaction. Significantly, in this EP complex the ribose and phosphates had well defined electron densities (Fig. 1B); in contrast, in the E+P complex, the ribose and phosphate had weaker electron densities that consistently were much less defined (Fig. 1A). Judged by the similar average B factors of the guanine bases and their protein partners, the guanine nucleotides should have full occupancies in both complexes. Similarly, the ribose and phosphates in the EP complexes also should have full occupancies. On the other hand, to best fit the electron densities, the ribose and phosphates in the E+P complexes had to be modeled with lower occupancy at PDE9 active site (Fig. S3). These observations indicate that the EP and E+P complexes are distinct species and that enzyme-ligand interactions are weaker in the latter.
Based on observations that Mn2+ or Mg2+ activates PDE9 in a concentration-dependent fashion (20, 21), we also soaked PDE9 in Mn2+ at less than fully activating concentrations. In crystals soaked for 1 min with 20 mM cGMP plus 2 mM MnCl2, the unbiased electron density maps indicated that cGMP clearly was not hydrolyzed (Fig. 1C). Because crystalline PDE9 was active in the presence of Mn2+ (Fig. 1A) and the observed PDE9/cGMP complex under this condition has a conformation different from those observed in inactive crystals (Fig. 1C and Figs. S1 and S2), we interpret this complex as being the enzyme/substrate complex (ES) of the hydrolytic reaction.
Recognition of cGMP by Active PDE9.
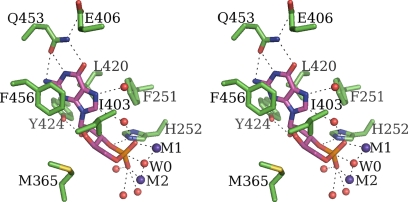
PDE9 is highly specific for the catalysis of cGMP compared with cAMP (Km of ≈170 nM vs. 250 μM, respectively; refs. 22–25). The basis for this specificity is apparent from our crystal structures. In all nucleotide species captured, the guanine base adopts an extended anti-conformation relative to the ribose and locates at almost identical position in the nucleotide recognition pocket of PDE9 (Fig. 2). Like all PDEs, the PDE9 nucleotide recognition pocket consists of a “hydrophobic clamp” (7–19) formed by the side chains of Phe-456 and Leu-420 (7–19). One side of the guanine base of bound nucleotide stacks with the aromatic side chain of Phe-456, and the other side contacts the side chain of Leu-420 (Fig. 2). The base also makes additional hydrophobic contacts with the side chains of Phe-251 and Ile-403 (Fig. 2). All these hydrophobic residues are either invariant (Phe-456) or semiconserved across the PDE family (7–19). At the edge of the nucleotide recognition pocket, almost co-planar with the base, is the side chain of the invariant Gln-453, the putative key residue in substrate selectivity of PDEs (19). In PDE9 the Oε and Nε atoms of Gln-453 form a pair of hydrogen bonds with N1 and O6, respectively, of the guanine base (Fig. 2). This hydrogen-bond pattern obviously is specific toward guanine base and is key for recognition of cGMP by PDE9.
Fig. 2.
Stereo diagram of the putative ES complex model illustrates the recognition of cGMP (magenta carbons) by active PDE9 (green carbons). Hydrogen bonds are shown by dashed lines.
Several additional contacts outside the recognition pocket contribute to binding. The C1′, O4′, C4′, and C5′ side of the nucleotide ribose contacts the side chains of Phe-456, Met-365, and Ile-403. Met-365 is hydrophobic in all PDEs (Met/Ile/Leu). Except for these contacts, the O2′ and O3′ side of the ribose and the phosphates are exposed to a hydrophilic environment, with the O2′ forming a hydrogen bond with Tyr-424 Oη and O3′ and the phosphates interacting with the hydrolysis center of PDE9 (Fig. 2).
Hydrolysis Center of PDE9.
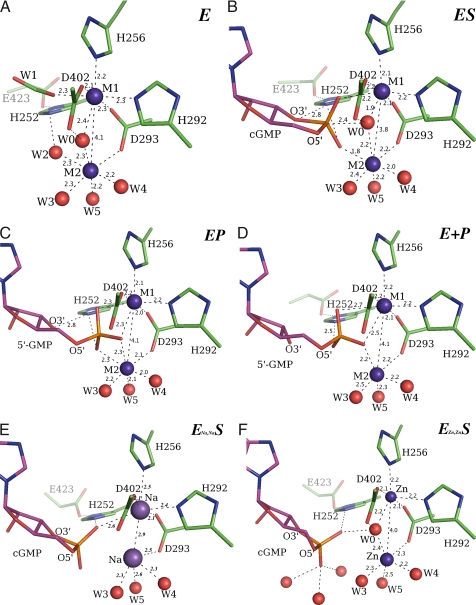
The hydrolysis centers of PDEs contain two divalent cations, denoted M1 and M2, respectively (Fig. 3). Even with the high concentrations used, the only divalent ions found are at the hydrolysis centers of PDE9. M1 and M2 are coordinated by protein side chains that are strictly conserved across PDEs (7–19). M1 has four coordination partners, Asp-293, Asp-402, His-292, and His-256. M2 has only one, Oδ of Asp-293. Proximal to M1 and M2 are the strictly conserved His-252 and its hydrogen-bond partner Glu-423. Modeling and theoretic calculations suggest that His-252 is the general acid of PDE hydrolysis, donating a proton to the leaving O3′ during reaction (7, 10, 26; see later discussion).
Fig. 3.
Structures of PDE9 hydrolysis centers. Distances within hydrogen bond lengths are shown by dashed lines. Some key distances are given also. For clarity, only protein side chains are shown. (A) An example of unliganded, active PDE9 hydrolysis center (active site B of crystals soaked with 1 mM cGMP + 0.5 mM MnCl2). In this case, both M1 and M2 are Mn2+ ions. Similarly, M1/M2 can have Mn2+-Mg2+, Zn2+-Mg2+, or Mg2+-Mg2+ configurations. When M1 is Mn2+, W1 is a formate ion from solvent. W1 is a water with partial occupancy when M1 is Zn2+ or Mg2+. (B). cGMP (magenta carbons) interacting with the active PDE9 hydrolysis center (green carbons) in the putative ES complex. In the ES and EP complex, only H252 Nε2-O3′ interaction has ideal geometry for hydrogen bond. (C) 5′-GMP interacting with the active PDE9 hydrolysis center in the putative EP complex. (D) 5′-GMP interacting with the active PDE9 hydrolysis center in the putative E+P complex. (E) cGMP bound at the PDE9 hydrolysis center inactivated by Na+ ions occupying both the M1 and M2 sites. (F) Bound cGMP at the inactive PDE9 hydrolysis center. Zn2+ ions occupy both M1 and M2 sites.
In addition to the protein side chains, both M1 and M2 have coordinating waters. These waters complete the coordination configurations of the metal ions in a manner dependent on the identity of the metal ions (Fig. 3). All these water molecules are held in place by conserved protein side chains. In the absence of bound nucleotide, W0 bridges M1 and M2, and W1 and W2 coordinate with M1 and M2, respectively (Fig. 3A). At nucleotide-bound hydrolysis centers, W0, W1, and W2 are differentially displaced (Fig. 3 B–F).
The results described in the previous paragraphs indicate that soaking different metal ions in the PDE9 protein conferred distinct catalytic properties that resulted in different ligands being captured at the active site. The chemical identities of M1 and M2 in the protein sample used for crystallization could not be determined directly by anomalous signals at the various wavelengths attempted (1.0, 1.54, and 1.7 Å). M2 is expected to have lower anomalous signals than M1 because it is bound more loosely (Fig. 3) and its electron density is at the level of a Mg2+ even when 20 mM Mn2+ or 2 mM Zn2+ was used (Fig. 1 and Fig. S2). Nonetheless, the available evidence suggests that initially after purification of PDE9, M1 and M2 are Zn2+ and Mg2+, respectively. The identity of M1 as Zn2+ in this initial PDE9 sample was confirmed indirectly by x-ray fluorescence signal at the zinc absorption edge. The coordination geometry and the height of electron density of M2 are consistent with those of Mg2+. Both Zn2+ and Mg2+ in the original PDE9 sample can be exchanged completely by cations in the soaking solution. For example, soaking with cGMP plus Mn2+ diminished zinc x-ray fluorescence signals but at the same time resulted in M1 electron density at the level of a full occupancy Mn2+ (typical Fo, ion−Fo, no ion electron density peaks > 25σ, Fig. 1A).
Soaking PDE9 crystals with Mn2+, Mg2+, or both cations resulted in active enzyme. Soaking with Mn2+ alone resulted in a full occupancy at the M1 site, but electron density for M2 remained at the level of a full occupancy Mg2+ ion (Fig. 1A). On the other hand, soaking with only Mg2+ resulted in full occupancy at both sites. Soaking with both Mn2+ and Mg2+ resulted in a full occupancy by Mn2+ at M1 and full occupancy by Mg2+ at M2. Thus, Mn2+ (or Zn2+) seems to be the preferred ion for the occupation of the M1 site, whereas Mg2+ seems to be the preferred ion for the occupation of M2. These binding preferences are consistent with the results of the kinetic assay, which indicated that the mixture of Mn2+ and Mg2+ reactivated metal-depleted PDE9 better than either ion alone. Taken together, these data indicate that the hydrolysis center of active PDE9 can have Zn2+-Mg2+, Mn2+-Mg2+, Mn2+-Mn2+ or Mg2+-Mg2+, and crystal structures of each of these configurations were observed (Fig. 3 B–D).
Soaking without divalent cations or with Zn2+ resulted in inactive PDE9. After soaking without divalent cations, both M1 and M2 had significantly weaker electron densities and shifted positions significantly (Fig. S1), indicating that they probably are Na+ ions. The significant longer coordination distances are typical for Na+ (Fig. 3E) (27). Soaking with Zn2+ must result in an inactive Zn2+-Zn2+ center (Fig. S2 and Fig. 3F), because the putative Zn2+-Mg2+ hydrolysis center is active, and Zn2+ has been reported to bind PDEs tightly, inhibiting PDEs competitively against Mn2+ or Mg2+ at concentrations >1 μM (25).
Proposed Reaction Mechanism of PDE9.
At the hydrolysis centers of active PDE9, both M1 and M2 have distorted octahedral coordination and similar M1-M2 distances (3.8–4.0 Å) (Fig. 3 A–C). Quantum computations on an active Zn2+-Mg2+ hydrolysis center indicated that at physiological pH W0 can partially dissociate into a hydroxide ion, and, as a hydroxide ion, W0 was suggested to be the nucleophile of the hydrolysis reaction of PDEs (26, 28). Presumably, W0 also can be a partial hydroxide ion at Mn2+-Mg2+, Mn2+-Mn2+, or Mg2+-Mg2+ hydrolysis centers. In the absence of bound nucleotide, W1 was found to be either a negative ion from solvent (COO−) when M1 is Mn2+ or a water with 50% occupancy when it is Zn2+ or Mg2+, but W2 is always a water with full occupancy (Fig. 3A). In the ES complex, the axial and equatorial phosphate oxygen of cGMP coordinates with M1 and M2, respectively, displacing both W1 and W2 (Fig. 3B). The bridging hydroxide W0 is 2.4Å away from the positively charged phosphorus, and the W0-P-O3′ angle is 174°, indicating that W0 is in fact the nucleophile of the hydrolysis reaction. As the reaction proceeds to the EP complex, the phosphorus center inverts, the O3′-P bond breaks, and W0 becomes part of the newly formed 5′-GMP (Fig. 3C). During the transition from ES to EP, only O3′ and phosphorous shifted visibly. In both the ES complex and the EP complex, His-252 Nε2 interacts with O3′ and 2 phosphate oxygens, but the geometry is ideal only for a Nε2-O3′ hydrogen bond (Fig. 3 B and C). As mentioned previously, in the E+P complex only 50% of the PDE9 hydrolysis center was occupied by ribose and phosphates; the other 50% was unliganded. In the E+P complex His-252 Nε2 no longer forms a hydrogen bond with O3′ and, instead, forms a hydrogen bond with one of the phosphate oxygens (Fig. 3D). The E+P complex also loses at least 2 phosphate-metal coordination bonds observed in the EP complex (compare Fig. 3 C and D). In all cGMP complexes studied here, the ribose has 3′-endo-4′-exo puckering, generally found in cyclic nucleotides (29). It retained 3′-endo-4′-exo puckering in the EP complex but changed to 3′-endo puckering in the E+P complex. This 3′-endo puckering is typical in nucleosides and nucleotides (29).
The hydrolytic mechanism of PDE9 proposed here explains the metal-ion requirements for activity. In this mechanism, the metal-bridging nucleophile W0 must be a hydroxide ion, and substrate phosphate must be activated by both M1 and M2 (Fig. 3 B and C). At the cGMP-bound Na+-Na+ hydrolysis center, W0 is missing, and the phosphate apparently cannot be activated by Na+ ions (Fig. 3E). At the cGMP-bound Zn2+-Zn2+ hydrolysis center, the phosphate does not coordinate with either M1 or M2, and W0 is within hydrogen-bond distances of both Asp-402 Oδ2 and cGMP phosphate axial oxygen (Fig. 3F). A theoretic calculation based on a Zn2+-Mg2+ center indicates that as a hydroxide ion W0 donates its single proton to Asp-402 Oδ2 to form a hydrogen bond (26); thus the observation that W0 forms hydrogen bonds with both Asp-402 Oδ2 and cGMP phosphate suggests that W0 may not dissociate easily into hydroxide ion at a Zn2+-Zn2+ hydrolysis center.
Effect of His-252 Mutagenesis.
The data described in the previous sections suggest that His-252 has a role as the general acid in the reaction. To verify this proposed role, we compared the activity in solution of the catalytic domain of wild type PDE9 enzyme and an enzyme with specific point mutations. The wild type enzyme had a Km of 80 ± 20 nM and Vmax of 1.3 ± 0.2 nmol·min−1·nmol−1 protein. Mutating His-252 to Ala completely inactivated the enzyme. His-296 is another strictly conserved residue side chain that forms a π-π stacking interaction with the His-252 side chain. Substituting Ala for His-296 had no effect on the affinity of the enzyme for substrate (Km of 80 ± 20 nM). However, the His-296/Ala substitution reduced Vmax to 0.28 ± 0.08 nmol·min−1·nmol−1 protein. These results are consistent with a critical role of His-252 as the putative general acid. His-296 may play a supporting role in the reaction by anchoring the orientation of His-252 and stabilizing its protonation state.
Discussion
Using a freeze-trapping technique, we report the capture of ligand/enzyme complexes of a PDE that spans the entire reaction path during the hydrolysis of the phosphor-ester bond of a cyclic nucleotide. The structures obtained also shed further light on the substrate specificity of PDE9 in an actual reaction and on the cation requirements of PDEs in general. These data reveal the putative mechanism of catalysis for this important class of enzymes.
The theoretical basis for the utility of the freeze-trapping technique is that at cryotemperature (100 K in our case), the reaction rates of many enzyme reactions drop so dramatically that intermediate species resulting from unstable reactions can be preserved long enough for mono-wavelength x-ray data collection (30, 31). In general, this approach does not perturb the reactions being studied and has been used to preserve many intrinsically unstable reaction intermediates (24, 30, 31). Several factors specific to PDE9 contributed to the success of this approach. PDE9 was crystallized with the low-affinity inhibitor IBMX, which can be displaced quickly and completely by cGMP, which has high binding affinity. The turnover rate of PDE9 is sufficiently slow that substrate remained abundant at reasonably achievable short reaction times. Thus, in the short soaking times used, the total amount of cGMP in the soaking solution exceeded that of enzyme in the crystal, and the substrate concentration remained far above the experimentally determined Km of 80 nM. Under these conditions, the reaction in PDE9 crystals should reach a kinetic steady state at which reaction intermediates should accumulate at the rate-limiting step (31, 32). These intermediate reaction/enzyme complexes then were captured by rapid freezing.
It is not surprising that the EP complex was captured in crystals of fully active PDE9 at steady state, because kinetic studies indicate that product dissociation is the rate-limiting step of the reaction. This EP complex clearly is different from the E+P complex formed by rebinding of dissociated product. The greater mobility of ribose and phosphate suggests that at equilibrium 5′-GMP interacts with PDE9 more weakly in the E+P complex than in the EP complex. Indeed, compared with the EP complex, the E+P complex loses the ribose-His-252 hydrogen bond and two phosphate-metal coordination bonds. These interactions may contribute to the stabilization of the EP complex and increase the energy barrier to its conversion to E+P. The existence of two different product complexes explains why product does not compete well with substrate for binding at the PDE9 active center and why product dissociation is the rate-limiting step of the reaction.
Because PDE9 activity requires activating metals such as Mn2+ or Mg2+, PDE9/cGMP complexes can be obtained in crystals inactivated either by stripping the activating metals or replacing the activating metals with the inhibitory Zn2+. Incubating with EDTA or guanine nucleotides strips both M1 and M2 of divalent cations that are replaced by Na+, resulting in inactive enzyme. Zn2+ alone also could be incorporated into both metal sites and would result in inactive enzyme.
Significantly, a putative ES complex of the reaction also was captured in crystals with reduced activity (20 mM cGMP + 2 mM MnCl2, 1 min) (Fig. 1C). The unusual high substrate/metal ratio used in these experiments and the strong phosphate–metal interactions during reaction may decrease M1 and M2's occupancies partially, as evidenced by slightly lower electron density for M1 and M2 (Fig. 1C), and may reduce enzyme activity. A decrease in metal occupancies may change the kinetics both by reducing the hydrolysis rate and by increasing the product release rate.
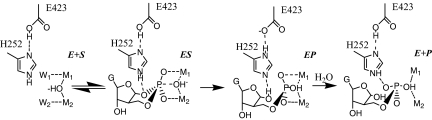
The structures of the ES, EP, and E+P complexes of PDE9 provide snapshots of the PDE9 hydrolysis reaction from which a putative reaction path can be constructed (Fig. 4), with the protonation states of the His-252–Glu-423 pair constructed based on theoretic calculations (26). In this proposed reaction scheme, both metal ions have catalytic roles, acting as Lewis acids, whereas the His-252 side chain acts as a general acid. Apparently, the proton relay between His-252 and Glu-423 is important in neutralizing the accumulated negative charge of O3′ during the transition from ES to EP (Fig. 4). To regenerate the His-252–Glu-423 hydrogen-bond network, His-252 needs to break the hydrogen bond with O3′ and abstract a proton from solvent (E+P in Fig. 4).
Fig. 4.
Schematic illustration of the proposed reaction pathway for PDE9. E, apo enzyme. ES, enzyme/substrate. EP, enzyme/product complex. E+P, the regenerated enzyme/rebound product complexes. Dashed lines represent hydrogen/coordination bonds.
Finally, the data presented here explain the substrate specificity of PDE9. The hydrogen bonds observed between the Gln-453 side chain and the guanine base are specific for guanine base (Fig. 2). The orientation of the Gln-453 side chain is fixed by a hydrogen bond between its Nε atom and Oε of Glu-406 (Fig. 2). The occurrence of Glu-406 and its interaction with Gln-453 is unique to PDE9. The side chain of Glu-406 is held in place in turn by hydrogen bonds with Oη of Tyr-490 and Oγ of Ser-496. The negatively charged Glu-406 side chain may polarize the side chain amide of Gln-453, making it a much better partner for accepting and donating hydrogen bonds. This unique polarization makes the hydrogen bond pattern with Gln-453 a feature more pronounced in PDE9 than in other PDEs, accounting for the high specificity of PDE9 toward cGMP over cAMP.
Materials and Methods
Protein Expression, Purification, and Crystallization.
The protocols for protein production and crystallization were adopted from the report of the co-crystal structure of the PDE9 catalytic domain with IBMX (16). Crystals were confirmed to have the same space group and similar diffraction qualities when tested in the x-ray beam.
PDE9 mutants of H252/A, H296/A, and H252/A/H296/A were generated by PCR mutagenesis for kinetic studies. The mutant proteins were expressed and purified in the same manner as the wild-type protein.
Ligand Soaking and Structure Determination.
The soaking experiments were carried out at 4°C by transferring several PDE9/IBMX co-crystals (≈150 × 150 × 150 μm) to a 5-μl soaking solution drop containing 4.2 M sodium formate (pH = 7.9), 5 mM DTT, and a combination of cGMP or 5′-GMP with metal ions (Table S1). All ligands and metals were added from 100-mM stocks in H2O. Crystals were soaked for different times and then were flash-cooled in liquid nitrogen before data collection. X-ray crystallographic data were collected at 100K, either in house with a Raxis-4++ detector or with an HTC detector mounted on a FRE x-ray generator (Rigaku) or at synchrotron source (IMCA-CAT). Data were processed with program suite HKL2000 (33), and further calculations were done with CCP4 program package (34). After rigid body and one round of geometry restraint maximum-likelihood refinement with program REFMAC (35) on the initial PDE9/IBMX complex model (PDB ID code 1TBM, with water molecules and ligands removed), ligands were recognized and built into electron density with graphic program O (36). Cross-validation on refinement was maintained by using the same set of reference reflections used in the original refinement (37). Soaking conditions, data collection, and refinement statistics are listed in Table S1.
Kinetic Analysis.
PDE9 activities were assayed using a radioactive Scintillation Proximity Assay (SPA) kit, using [3H]cGMP (Amersham Biosciences) as substrates. For steady-state kinetics, 1.5 nM enzyme in 50-μl reaction buffer (50-mM Tris, pH = 7.5, 8 mM MgCl2) with varying amounts of [3H]cGMP was used. The reactions were run at 20°C for 30 min and then were quenched by adding 25-μl reconstituted SPA beads supplemented with 5 μM unlabeled cGMP. The initial reaction rates were calculated on two repeated measurements of the amount of [3H]5′-GMP generated after 30 min reaction. The Km and Vmax values were determined using equation [S]/V0 = Km/Vmax + [S]/Vmax, with Woolf-Hanes Plot ([S]/V0 vs. [S]) generated at 8 cGMP concentrations ([S]), varying from 800 nM down using a 1:2 dilution scheme. To test divalent cations effects on the activities of the wild-type PDE9, a double-dilution scheme was used. The wild-type PDE9 stock used for crystallization first was diluted ≈1,000-fold into a buffer of 50 mM Tris, pH 7.5, 10 mM EDTA and incubated at 4°C for 30 min to remove the original metals. The protein then was diluted 200-fold to 1.5 nM into 50 μl of buffer of 50 mM Tris, pH 7.5, supplemented with 8 mM ZnCl2, MnCl2, MgCl2 or 4 M MnCl2 plus 4 mM MgCl2, respectively, or with 2 mM EDTA, for activity measurements.
For the single-turnover kinetics, reactions were started by hand mixing 400 μl 480- or 240-nM enzyme in reaction buffer with 400 μl of 40 nM [3H]cGMP. At different reaction times, 50 μl of product reaction solution was removed and added either to 100 μl supplemented with 150 μM unlabeled cGMP or to 100 μl reconstituted SPA beads 4N HCl. For reactions stopped by 4N HCl, the solution was neutralized immediately by a mixture of 100 μl of 4N NaOH and 50 μl 1M Tris, pH 7.5; 50 μl neutralized solution then was added to 100 μl of reconstituted SPA beads for product detection. The amounts of labeled product were measured on two repeated reactions. Reactions also were run with enzyme omitted (background) or added at the same time as the quenching agents (time 0 reactions).
Supplementary Material
Acknowledgments.
Ann Subashi characterized PDE9 samples used for crystallization using isothermal calorimetric methods. We thank members of the Pfizer Groton X-ray Crystallography Group for help with data collection and structure determination. We also thank Jay Pandit and David Nettleton for their comments on the manuscript.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 3DY8, 3DYL, 3DYN, 3DYQ, and 3DYS).
This article contains supporting information online at www.pnas.org/cgi/content/full/0708850105/DCSupplemental.
References
- 1.Beavo JA, Brunton L. Cyclic nucleotide research–still expanding after half a century. Nat Rev Mol Cell Biol. 2002;3:710–718. doi: 10.1038/nrm911. [DOI] [PubMed] [Google Scholar]
- 2.Mehats C, Andersen CB, Filopanti M, Jin SL, Conti M. Cyclic nucleotide phosphodiesterases and their role in endocrine cell signaling. Trends Endocrinol Metab. 2002;13:29–35. doi: 10.1016/s1043-2760(01)00523-9. [DOI] [PubMed] [Google Scholar]
- 3.Monfort P, Munoz MD, Felipo V. Molecular mechanisms of the alterations in NMDA receptor-dependent long-term potentiation in hyperammonemia. Metab Brain Dis. 2005;20:265–274. doi: 10.1007/s11011-005-7905-5. [DOI] [PubMed] [Google Scholar]
- 4.Baylor D. How photons start vision. Proc Natl Acad Sci USA. 1996;93:560–565. doi: 10.1073/pnas.93.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breer H, Boekhoff I, Tareilus E. Rapid kinetics of second messenger formation in olfactory transduction. Nature. 1990;345:65–68. doi: 10.1038/345065a0. [DOI] [PubMed] [Google Scholar]
- 6.Houslay M, Adams D. PDE4 cAMP phosphodiesterases: Modular enzymes that orchestrate signalling cross-talk, desensitization and compartmentalization. Biochem J. 2003;370:1–18. doi: 10.1042/BJ20021698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xu R, et al. Atomic structure of PDE4: Insights into phosphodiesterase mechanism and specificity. Science. 2000;288:1822–1825. doi: 10.1126/science.288.5472.1822. [DOI] [PubMed] [Google Scholar]
- 8.Lee M, Markowitz J, Lee J, Lee H. Crystal structure of phosphodiesterase 4D and inhibitor complex(1) FEBS Lett. 2002;530:53–58. doi: 10.1016/s0014-5793(02)03396-3. [DOI] [PubMed] [Google Scholar]
- 9.Sung B, et al. Structure of the catalytic domain of human phosphodiesterase 5 with bound drug molecules. Nature. 2003;425:98–102. doi: 10.1038/nature01914. [DOI] [PubMed] [Google Scholar]
- 10.Huai Q, Colicelli J, Ke H. The crystal structure of AMP-bound PDE4 suggests a mechanism for phosphodiesterase catalysis. Biochemistry. 2003;42:13220–13226. doi: 10.1021/bi034653e. [DOI] [PubMed] [Google Scholar]
- 11.Huai Q, et al. Three-dimensional structures of PDE4D in complex with roliprams and implication on inhibitor selectivity. Structure. 2003;11:865–873. doi: 10.1016/s0969-2126(03)00123-0. [DOI] [PubMed] [Google Scholar]
- 12.Huai Q, Liu Y, Francis S, Corbin JD, Ke H. Crystal structures of phosphodiesterases 4 and 5 in complex with inhibitor 3-isobutyl-1-methylxanthine suggest a conformation determinant of inhibitor selectivity. J Biol Chem. 2004;279:13095–13101. doi: 10.1074/jbc.M311556200. [DOI] [PubMed] [Google Scholar]
- 13.Xu RX, et al. Crystal structures of the catalytic domain of phosphodiesterase 4B complexed with AMP, 8-Br-AMP, and rolipram. J Mol Biol. 2004;337:355–365. doi: 10.1016/j.jmb.2004.01.040. [DOI] [PubMed] [Google Scholar]
- 14.Iffland A, et al. Structural determinants for inhibitor specificity and selectivity in PDE2A using the wheat germ in vitro translation system. Biochemistry. 2005;44:8312–8325. doi: 10.1021/bi047313h. [DOI] [PubMed] [Google Scholar]
- 15.Scapin G, et al. Crystal structure of human phosphodiesterase 3B: Atomic basis for substrate and inhibitor specificity. Biochemistry. 2004;43:6091–6100. doi: 10.1021/bi049868i. [DOI] [PubMed] [Google Scholar]
- 16.Huai Q, et al. Crystal structure of phosphodiesterase 9 shows orientation variation of inhibitor 3-isobutyl-1-methylxanthine binding. Proc Natl Acad Sci USA. 2004;101:9624–9629. doi: 10.1073/pnas.0401120101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang H, Liu Y, Chen Y, Robinson H, Ke H. Multiple elements jointly determine inhibitor selectivity of cyclic nucleotide phosphodiesterases 4 and 7. J Biol Chem. 2005;280:30949–30955. doi: 10.1074/jbc.M504398200. [DOI] [PubMed] [Google Scholar]
- 18.Wang H, et al. Structural insight into substrate specificity of phosphodiesterase 10. Proc Natl Acad Sci USA. 2007;104:5782–5787. doi: 10.1073/pnas.0700279104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang KY, et al. A glutamine switch mechanism for nucleotide selectivity by phosphodiesterases. Mol Cell. 2004;15:279–286. doi: 10.1016/j.molcel.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 20.Fisher D, Smith J, Pillar J, St Denis S, Cheng J. Isolation and characterization of PDE9A, a novel human cGMP-specific phosphodiesterase. J Biol Chem. 1998;273:15559–15564. doi: 10.1074/jbc.273.25.15559. [DOI] [PubMed] [Google Scholar]
- 21.Wang P, Wu P, Egan RW, Billah MM. Identification and characterization of a new human type 9 cGMP-specific phosphodiesterase splice variant (PDE9A5). Differential tissue distribution and subcellular localization of PDE9A variants. Gene. 2003;314:15–27. doi: 10.1016/s0378-1119(03)00733-9. [DOI] [PubMed] [Google Scholar]
- 22.Soderling S, Bayuga S, Beavo J. Identification and characterization of a novel family of cyclic nucleotide phosphodiesterases. J Biol Chem. 1998;273:15553–15558. doi: 10.1074/jbc.273.25.15553. [DOI] [PubMed] [Google Scholar]
- 23.Guipponi M, et al. Identification and characterization of a novel cyclic nucleotide phosphodiesterase gene (PDE9A) that maps to 21q22.3: Alternative splicing of mRNA transcripts, genomic structure and sequence. Hum Genet. 1998;103:386–392. doi: 10.1007/s004390050838. [DOI] [PubMed] [Google Scholar]
- 24.Petsko GA, Ringe D. Observation of unstable species in enzyme-catalyzed transformations using protein crystallography. Curr Opin Chem Biol. 2000;4:89–94. doi: 10.1016/s1367-5931(99)00057-5. [DOI] [PubMed] [Google Scholar]
- 25.Omburo G, Brickus T, Ghazaleh F, Colman R. Divalent metal cation requirement and possible classification of cGMP-inhibited phosphodiesterase as a metallohydrolase. Arch Biochem Biophys. 1995;323:1–5. doi: 10.1006/abbi.1995.0001. [DOI] [PubMed] [Google Scholar]
- 26.Salter AE, Wierzbicki A . The mechanism of cyclic nucleotide hydrolysis in the Phosphodiesterase catalytic site. J Phys Chem. 2007;111:4547–4552. doi: 10.1021/jp066582+. [DOI] [PubMed] [Google Scholar]
- 27.Harding M. Metal-ligand geometry relevant to proteins and in proteins: Sodium and potassium. Acta Cryst Allogr. 2002;D58:872–874. doi: 10.1107/s0907444902003712. [DOI] [PubMed] [Google Scholar]
- 28.Zhan C-G, Zheng F. First computational evidence for a catalytic bridging hydroxide ion in a phosphodiesterase active site. J Am Chem Soc. 2001;123:2835–2838. doi: 10.1021/ja005529a. [DOI] [PubMed] [Google Scholar]
- 29.Sundaralingam M. Structure and conformation of nucleosides and nucleotides and their analogs as determined by x-ray diffraction. Ann NY Acad Sci. 1975;255:3–42. doi: 10.1111/j.1749-6632.1975.tb29211.x. [DOI] [PubMed] [Google Scholar]
- 30.Stoddard BL. Trapping reaction intermediates in macromolecular crystals for structural analyses. Methods. 2001;24:125–138. doi: 10.1006/meth.2001.1174. [DOI] [PubMed] [Google Scholar]
- 31.Moffat K, Henderson R. Freeze trapping of reaction intermediates. Curr Opin Chem Biol. 1995;5:656–663. doi: 10.1016/0959-440x(95)80059-x. [DOI] [PubMed] [Google Scholar]
- 32.Copeland RA. Enzymes: A Practical Introduction to Structure, Mechanism, and Data Analysis. 2nd ed. New York: Wiley-VCH; 2000. [Google Scholar]
- 33.Otwinowski Z, Minor W. In: Methods in Enzymology. Carter JCW, Sweet RM, editors. New York: Academic; 1997. pp. 307–326. [DOI] [PubMed] [Google Scholar]
- 34.Collaborative Computing Project #4. Acta Crystallogr D. 1994;50:760–763. [Google Scholar]
- 35.Murshudov G, Vagin AA, Dodson EJ . Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 36.Jones TA, Zou J-Y, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr A. 1991;47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 37.Brünger AT. Free R value: A novel statistical quantity for assessing the accuracy of crystal structures. Nature. 1992;355:472–474. doi: 10.1038/355472a0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.