Abstract
The earliest kinetic folding events for (βα)8 barrels reflect the appearance of off-pathway intermediates. Continuous-flow microchannel mixing methods interfaced to small-angle x-ray scattering (SAXS), circular dichroism (CD), time-resolved Förster resonant energy transfer (trFRET), and time-resolved fluorescence anisotropy (trFLAN) have been used to directly monitor global and specific dimensional properties of the partially folded state in the microsecond time range for a representative (βα)8 barrel protein. Within 150 μs, the α-subunit of Trp synthase (αTS) experiences a global collapse and the partial formation of secondary structure. The time resolution of the folding reaction was enhanced with trFRET and trFLAN to show that, within 30 μs, a distinct and autonomous partially collapsed structure has already formed in the N-terminal and central regions but not in the C-terminal region. A distance distribution analysis of the trFRET data confirmed the presence of a heterogeneous ensemble that persists for several hundreds of microseconds. Ready access to locally folded, stable substructures may be a hallmark of repeat-module proteins and the source of early kinetic traps in these very common motifs. Their folding free-energy landscapes should be elaborated to capture this source of frustration.
Keywords: FRET, microsecond mixing, misfolding, small-angle x-ray scattering
The funnel shape of typical protein folding-energy landscapes suggests that sequences have evolved to minimize energetic and topological frustration (1, 2). This view is supported by the two-state folding of many small proteins (3) and the significant correlation of their folding rates with native topology (4, 5). More-complex mechanisms, typical of larger proteins, often place intermediates on a progressive path to the native conformation. Surprisingly, a few kinetic studies (6–9) and simulations (10) have revealed that off-pathway intermediates can be populated during the refolding of single-domain globular proteins (11). Delineating the relative contributions of sequence and topology to biases in the energy landscape that lead to these kinetic traps offers potential insights into factors responsible for protein misfolding and, potentially, for numerous devastating pathologies (12).
(β/α)8 TIM barrels (named after triosephosphate isomerase) are one of the most common motifs in biology (13) and are of particular interest in examining the factors leading to off-pathway folding. They constitute the most common structural class of substrates of the chaperonin GroEL, which captures and sequesters partially folded proteins to facilitate their folding to the native state (14). Although the specific partially folded forms of the Escherichia coli TIM barrels responsible for binding to GroEL are not known, in vitro studies suggest two potential candidates. The folding reactions of three TIM barrels of very low sequence identity, IOLI (a protein of unknown function corresponding to the ioll gene in Bacillus subtilis), IGPS (indole-3-glycerol phosphate synthase), and the α-subunit of tryptophan synthase (αTS), display early marginally-stable kinetic traps and subsequent on-pathway stable intermediates (6, 15, 16). These common features in the folding mechanisms are consistent with an important role for topology in dictating the essential features of the energy landscape. However, the structured regions of these intermediates for αTS and IGPS, as determined from HX protection experiments (16) and mutational analysis (17, 18), vary significantly. Clearly, the sequences of these proteins dictate the structures of these crucial partially folded states.
Detailed insights into the formation of the kinetically-trapped states in TIM barrels have thus far been limited by the time resolution of stopped-flow kinetic methods, ≈5 ms. In this report, the time resolution was extended to the microsecond time range with continuous-flow (CF) microchannel mixing devices and coupled with time-resolved Förster resonant energy transfer (trFRET) (19), time-resolved fluorescence anisotropy, small-angle x-ray scattering (SAXS) (20) and CD spectroscopy (21) to probe the dimensional, dynamic, and structural properties of the kinetically-trapped state of αTS. (Fig. 1). The complex responses reveal widespread and differential aspects of the initial collapse reaction that ultimately anneal into a global kinetic trap. The subsequent unfolding of this off-pathway species controls access to the productive folding channel and, ultimately, to the native TIM barrel structure.
Fig. 1.
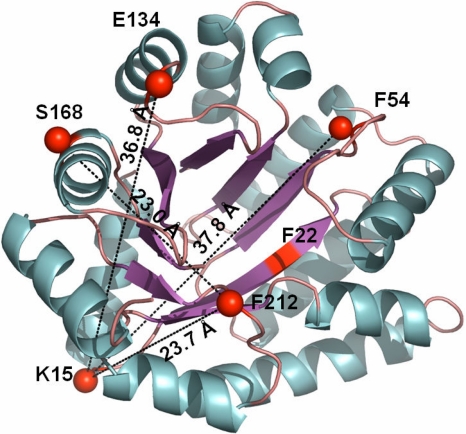
Structure of αTS and location of chromophores used as FRET probes. X-ray crystal structure of αTS [PDB ID code:1BKS (49)] with FRET pairs 15-54, 15-134, 168-212 and 15-212 highlighted. The figure was prepared by using PyMol (50).
Results
Global Structural Properties of αTS Probed by Microsecond Continuous-Flow SAXS and CD.
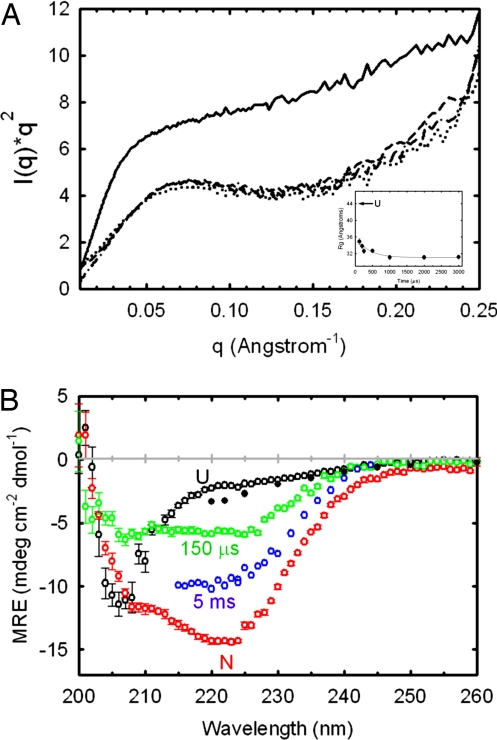
Global structural insights in the hundreds of microseconds time range were obtained by acquiring SAXS profiles upon refolding αTS after a 6.0–0.6 M urea jump (Fig. 2A). The data show that initial compaction of the protein takes place within the 150-μs time resolution of the CF-SAXS device. Compaction and collapse to a globular form are illustrated by two metrics: the radius of gyration (Rg) and the shape of the scattering profiles. Throughout the 150-μs to 3-ms time range, the Rg is significantly lower (Rg ≈ 31–34 Å) than in the urea-denatured state (Rg ≈ 43 Å) and exhibits a Kratky plot with a broad peak consistent with the presence of globular structure. The Kratky plot of unfolded αTS does not have a peak in this q-range, as expected for a statistical coil (22). The compaction is far from complete, however, with a Rg that is 50% larger than that of the native state (18.1 Å).
Fig. 2.
SAXS and CD spectra of αTS as a function of refolding time. (A) Kratky plots at 150 μs (dotted line), 1 ms (dashed line), and 5 ms (dotted-dashed line). The unfolded Kratky plot recorded at 6 M urea (solid line), normalized to the concentration used for the kinetic data, is shown for reference. The Rg of the native state of αTS is 18.1 Å (51). Refolding was initiated by a urea concentration dilution from 6 to 0.6 M. The final protein concentration was 1 mg·ml−1. (Inset) Dependence of Rg on refolding time. Each scattering curve in A and time point in the Inset represents a total accumation time of ≈5 s. (B) CD spectra of αTS at various refolding times. Spectra at 150 μs (green open circles) and 5 ms (blue open circles), acquired after an 8–0.8 M urea concentration jump, are compared with the native spectrum (red open circles), the unfolded spectrum recorded at 8 M urea (black open circles) and the unfolded spectrum extrapolated to 0.8 M urea (black filled circles). The 150-μs spectrum, the native spectrum at 0.8 M urea, and the unfolded spectrum at 8 M urea were acquired with the same stock solutions in the continuous-flow mixer having a 127-μm optical path. The 5-ms time point spectrum was acquired by using the stopped-flow cuvette with a 1.5-mm optical path. Sample concentration was 60 μM for data recorded in the continuous-flow mixer and 10 μM for the stopped-flow data. The extrapolation of the unfolded baseline was performed by using published data and global fit parameters (52). Data were recorded at 21°C.
Far-UV CF-CD indicates that the compaction at 150 μs is accompanied by the formation of detectable secondary structure (Fig. 2B). The CF-CD spectrum at 150 μs shows significant spectral changes relative to the unfolded CD spectrum, exhibiting a diminished mean residue ellipticity (MRE) near 206 nm and an increase in MRE at ≈220 nm. Approximately 50% of the 222 nm MRE of the burst-phase intermediate at 5 ms and ≈25% of the signal from the native state is acquired within 150 μs. Although interpretation of CD spectra can be complicated by exciton interactions between spatially-adjacent aromatic residues, the K2D secondary structure prediction algorithm (23) predicts a larger fractional contribution of β-structure at 150 μs than in the native spectrum. Estimation of β-structure for the 5 ms intermediate is precluded by the limited wavelength range accessible in the stopped flow. The diminished helical content at 150 μs may reflect dynamic or disordered helices on the surface of the (βα)8 barrel at this early stage in folding.
In the time range from 150 μs to several milliseconds, a nearly twofold increase in the MRE is observed at 222 nm. By contrast, the Rg values and Kratky profiles from SAXS demonstrate that relatively minor rearrangements in global structure occur. The weak correlation of secondary-structure formation with compaction for αTS across the first few milliseconds of folding shows that the folding trajectory is intermediary between a Type II trajectory (concurrent compaction and secondary structure formation) and a Type III trajectory (collapse, followed by secondary-structure formation) (24–26).
Dimensional Analysis of Microsecond Folding Reactions in αTS by CF-trFRET.
The global structural insights from SAXS and CD during folding were complemented by intramolecular distance information from four Trp–5-({2-[(iodoacetyl)amino]ethyl}amino)naphthalene-1-sulfonic acid (IAEDANS) Förster resonant energy transfer (FRET) pairs. Results from hydrogen–deuterium exchange (HX) mass-spectrometry (27), HX-NMR (17), mutational analysis (17), fragment studies (28) and nativecentric Gō-modeling (29) led to the proposal that structure forms early in the N-terminal half of αTS. To test this hypothesis on the microsecond folding time scale, FRET pairs 15-54 and 15-134 were directed at probing structural changes in the N-terminal half of the protein. Structural changes within the C-terminal half of αTS and between the N- and C-terminal regions were probed by the 168-212 and 15-212 pairs, respectively [Fig. 1 and supporting information (SI) Fig. S1].
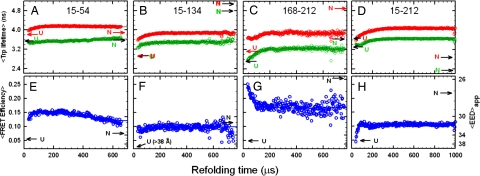
The trFRET data show that collapse of the protein is heterogeneous and very rapid, with substantial compaction occurring within 30 μs for the 15-54, 15-134, and 168-212 pairs. The 15-212 pair, in contrast, exhibits Trp lifetimes and average FRET efficiencies indistinguishable from the unfolded state (Fig. 3). The average apparent end-to-end distance (〈EED〉app) for the 15-54, 15-134, and 168-212 pairs are 5 Å smaller than the 〈EED〉app expected for the unfolded state under refolding conditions, assuming a linear dependence of the size of the unfolded state on the denaturant concentration. Single-molecule FRET studies on small two-state folding proteins have shown a nonlinear dependence of the unfolded baseline on denaturant concentration and may explain the contraction observed at 30 μs (30).
Fig. 3.
Average excited-state lifetimes of tryptophan as a function of refolding time following an 8 to 0.8 M urea concentration jump for four FRET pairs (green) and donor-only controls (red). (A) Pair 15-54. (B) Pair 15-134. (C) Pair 168-212. (D) Pair 15-212. (E–H) The corresponding FRET efficiencies are shown. The 〈EED〉app calculated from the Förster equation are indicated on the right vertical axis. The uncertainty in the average lifetimes, determined from controls in which no kinetics are present, is ≈0.015 ns. A consistent set of kinetics is observed in the average Trp lifetimes and the FRET efficiencies, suggesting that the observed kinetics are not an artifact of the mutations or acceptor chromophore labeling. Data were recorded at 21°C.
Over the course of the first few hundred microseconds, the distance between the N- and C-terminal regions, as monitored by the 15-212 pair, contracts (Fig. 3D). More modest contractions in the 〈EED〉app (≤2 Å) on the same timescale were observed for the 15-54 and 15-134 donor-acceptor pairs (Fig. 3A-C). The surprising increase of the 168-212 〈EED〉app suggests that the central region of the barrel rearranges in the same ≈100-μs time frame that the 15-212 pair distance contracts. These coincident temporal changes reflect the coalescence of semiautonomously folded regions into a more globular structure, as shown by the SAXS profiles at 150–500 μs. The 〈EED〉app for all FRET pairs achieve their native values on a considerably longer time scale, 1–100 s (Y.W., C.K., C.R.M., and O.B., unpublished work), consistent with previous stopped-flow and manual-mixing results (6).
Distance Distributions from trFRET.
Although analysis of the trFRET data by average lifetimes is a robust method of calculating average FRET efficiencies, the ensemble-average efficiency and distance can potentially mask the presence of subpopulations. Additionally, the average distance can be significantly biased toward shorter distances reflecting the insensitivity of the FRET probes to distances ≥1.5 Ro. Potential ambiguities in 〈EED〉app can be resolved because the excited state decay kinetics of the donor and acceptor in trFRET data also contain information about the distribution of donor–acceptor distances (31, 32).
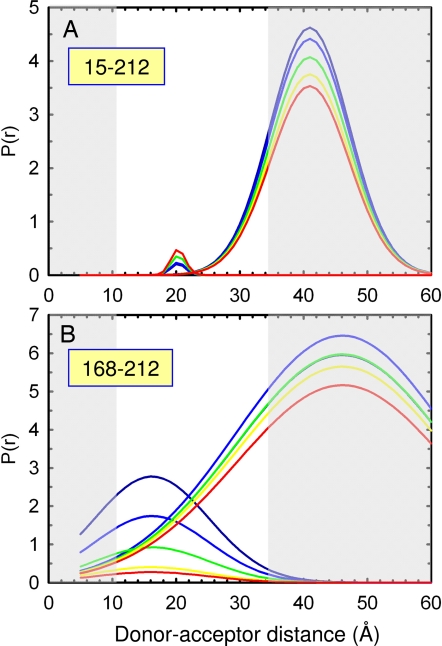
The time dependence of the donor–acceptor distance distribution for the 15-212 and 168-212 FRET pairs obtained from an analysis using a double-Gaussian function are shown in Fig. 4. Use of a single-Gaussian function gave a significantly poorer fit as judged by the randomness of residuals and reduced-χ2 ratios (χ12/χ22>2). The presence of two peaks in the distance distribution suggests that two populations are present in the burst-phase of the continuous-flow mixing device. For the 15-212 FRET pair (Fig. 4A), the analysis clearly shows that extended conformations, with a peak beyond 35 Å, dominate at the earliest time point (53 μs). Surprisingly, a minor high-FRET efficiency subpopulation with a peak near 20 Å, close to the 21 Å observed for the native conformation, appears on the same 75- to 100-μs time scale. The interconversion time of these distinct populations must exceed that of the lifetime of the Trp donor, ≈3–7 ns.
Fig. 4.
Gaussian distance distribution analysis of the time-resolved FRET data shown in Fig. 3. Results of global nonlinear least-squares analysis of tryptophan excited-state decays at various refolding time points using a double-Gaussian function are shown for the 15-212 (A) and 168-212 (B) samples. Distance distributions are shown at 53 (dark blue), 82 (light blue), 112 (green), 141 (yellow), and 170 (red) μs. Five pairs of donor-only and donor-acceptor-labeled decay traces were fit. The gray areas correspond to regions where the Trp–AEDEANS FRET pair is insensitive to distance changes. Based on the measured anisotropy of W212 (Fig. 5) and estimated anisotropy of AEDANS, values of κ2 can range between 0.4 and 1.5, however, leading to an uncertainty in the distances of approximately ±10%. The Gaussians in A have peaks at 20 and 41 Å and corresponding full-width at half-maximum of 2.7 and 14.3 Å. The Gaussians in B have peaks at 16 and 46 Å and corresponding full-width at half-maximum of 21 and 39 Å, respectively.
Examination of the distance distribution for the 168-212 pair shows that a heterogeneous population in the central region of αTS is present at the earliest time point, 53 μs (Figs. 4B). Populations having less-than-native distances (≈16 Å) coexist with those having more extended (≈35–50 Å) and broader distributions. The compact high-FRET efficiency subpopulation preferentially “melts” over the same 75- to 100-μs time scale observed for the appearance of the minor compact population for the 15-212 FRET pair (Fig. 4A). The coincidence of these processes links the unfolding of the overfolded subpopulation seen with the 168-212 FRET pair with the folding of the minor compact conformer for the 15-212 pair. The distance distribution analysis reveals that unfolding of an overly-compact subpopulation is responsible for the increase in the 〈EED〉app observed for the 168-212 pair in the average lifetime analysis (Fig. 3G). A complementary model-independent analysis using a maximum entropy method along both the donor decay rate and energy transfer rate dimensions gave consistent results (Figs. S2 and S3 and SI Text).
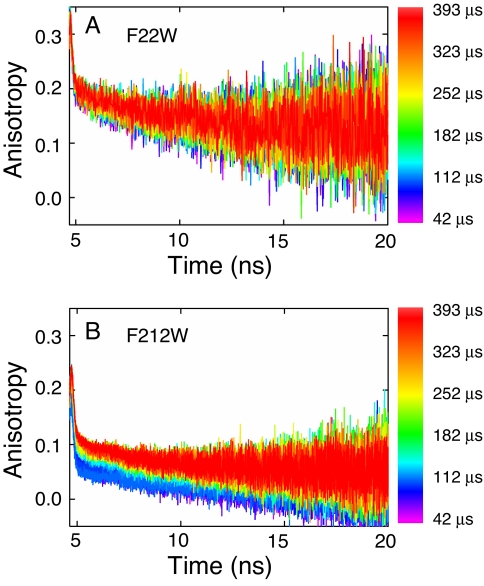
Microsecond Collapse of αTS Probed by Continuous-Flow Time-Resolved Fluorescence Anisotropy.
As a complement to the trFRET dimensional studies, information on local packing and regional hydrodynamics were obtained from time-resolved fluorescence anisotropy analysis of the single tryptophan variants, F22W and F212W, in the microsecond refolding time frame (Fig. 5). It has been shown for W22, located at the beginning of β1 and on the interior of the barrel (Fig. 1 and Fig. S1), that the anisotropy decay at the earliest time point (≈42 μs) is reminiscent of the behavior expected for a hindered side chain. A fast subnanosecond anisotropy decay component and a larger-amplitude slower component with a rotational correlation time ≥15 ns are observed (33). The anisotropy decay is similar, within error, for the duration of the continuous-flow experiment (≈300 μs) (33) and stopped-flow experiments (5 ms to 12 s, data not shown). In contrast, W212, which resides in a loop between β7 and α7 and protrudes into the solvent (Fig. 1 and Fig. S1), exhibits a slow anisotropy decay component that is faster (rotational correlation time <10 ns) and comprises a significantly smaller fraction of the anisotropy decay (Fig. 5). The faster correlation time and smaller amplitude imply a smaller folded structure at the C-terminal region and a more flexible environment with looser tertiary packing than its N-terminal counterpart.
Fig. 5.
Time-resolved anisotropy as a function of refolding time for F22W (A) and F212W (B) after an 8–0.8 M urea concentration jump. TCSPC curves were acquired every 10.93 μs in refolding time, and refolding times are indicated by the color-coding shown on the right. The total flow rate was 17.5 ml·min−1 (1.75 ml·min−1 protein and 15.75 ml·min−1 buffer), which corresponds to a 32.65 μs·mm−1 linear flow velocity in the channel. Data were recorded at 21°C in 10 mM phosphate buffer containing 0.2 mM K2EDTA and 1 mM βME. Final protein concentration was 10 μM.
Organization of structure in the vicinity of W212 occurs with kinetics comparable with those for the contraction of the 15-212 pair distance (≈75–100 μs) (Fig. 3). The amplitude of the slow anisotropy decay component for W212 systematically increases over ≈100 μs. A concomitant increase is observed in the average tryptophan lifetime. Based on the greater relative amplitude of the fast anisotropy decay in W212, however, the packing in the vicinity of W212 is more fluid than that near W22. Consistent with these results, tight packing in the N-terminal region of αTS by 5 ms was suggested by a mutational analysis study (17). The mutational analysis also indicated a molten-globule-like packing in the C-terminal region of αTS, including β7 where W212 resides (17).
Discussion
The various geometric and hydrodynamic probes presented here suggest that a specific structure, possibly guided by the repeat topology of TIM barrels and formation of hydrophobic clusters, forms very rapidly (<30 μs) but is localized preferentially to the N-terminal half of the protein. A subpopulation of the early intermediates is nonproductive for folding and melts away in the first observable kinetic step. The role of analogous misfolded species in aggregation of repeat-sequence motifs is discussed below.
Structural Picture of Early Misfolded Forms of αTS.
The compact dimensions and tight packing in the N-terminal region but not the C-terminal region at ≈30 μs show that the earliest folded forms of αTS are structurally heterogeneous. This conclusion is consistent with a previous mutational analysis (17) and with the protection of main-chain amide hydrogens against exchange with solvent via NMR analysis (17). The further reduction of the 168-212 trFRET data by a distance distribution analysis revealed that the C-terminal region is itself heterogeneous. A minor conformer, whose donor–acceptor distance is substantially smaller than the distance for the native conformation, is present after 30 μs and melts away over the first few hundred microseconds as the N and C termini coalesce. Curiously, a minor, overly-compact conformer in the N terminus appears during the coalescence reaction. A cartoon schematic of the reaction is shown in Fig. S4.
Specificity of the Initial Collapse Reaction.
Distance distributions obtained from time-resolved FRET and SAXS can in principle distinguish between a generalized nonspecific contraction and distinct intermediate formation (30, 34). The presence of distinct intermediates in the earliest observable intermediates is supported by the observation of two subpopulations in the donor–acceptor distance distribution and the preferential “melting” of the more compact population (Fig. 4A). The bimodal distance distribution (Fig. 4A) is difficult to reconcile with merely a generalized contraction of the polypeptide. However, although the more compact population may represent a distinct intermediate, the more extended subpopulation could still represent a contracted form of the unfolded ensemble in equilibrium with the marginally-stable intermediate.
The formation of distinct intermediates is also consistent with the time-resolved anisotropy data (Fig. 5) and extensive mutational analysis that suggests that the N-terminal half of αTS adopts a structure with specific tertiary packing very early in the folding reaction. Very small changes in FRET efficiency are observed for the two N-terminal-half FRET pairs (15-54 and 15-134) between ≈30 μs and ≈1 ms, and the mobility of Trp-22 (Fig. 5) is also similar over the ≈45-μs to 5-ms time range. It is likely that the tertiary packing, particularly in the N-terminal (βα)1–3 region of the protein, demonstrated from extensive mutational analysis on the intermediate populated at 5 ms (17, 18) is mostly present in the continuous-flow burst-phase intermediate at 30–40 μs.
Factors Leading to Off-Pathway Misfolding.
An increasing body of evidence points to the propensity of the βα-repeat fold, both the TIM barrel (6) and the flavodoxin motifs (7, 8), to sample off-pathway misfolded forms early in their refolding pathway. Mutational analysis and hydrogen exchange experiments have highlighted a role for large clusters of branched aliphatic side chains located on consecutive elements of secondary structure in αTS (17, 35). Isoleucine, leucine, and valine (ILV) residues are especially favorable participants in stable hydrophobic clusters in globular proteins because they are the only residues with side chains with unfavorable transfer free energies from the gas phase to water (36). This enables these side chains to effectively exclude water from the interiors of ILV clusters and from their underlying peptide bonds. As a consequence, they are preferentially sequestered in the interior of proteins (37), and their amide hydrogens are invariably among those most strongly protected against exchange with solvent (17, 38–40). When a group of ILV side chains are also clustered in sequence, rapid kinetic access to stable (on- or off-pathway) substructures is feasible. This view is supported by the elimination of the off-pathway kinetic folding reaction for αTS by alanine replacements for all 10 of the isoleucine and leucine side chains examined in an extensive hydrophobic cluster spanning β1α1β2α2β3 (17).
Protein amino acid sequences selected by evolution for specificity and stability of the native state are expected to have a funneled, typically smooth, energy landscape. However, our results indicate that energetic frustration can remain. The deviation from a smooth funnel landscape is reconciled by inhomogeneities in the magnitude of native contacts (2). The pronounced sensitivity of the folding pathway to selective mutation of ILV residues (17) may indicate a disproportionate energetic contribution of branched aliphatics to the stability of early intermediates of αTS, resulting in energetic frustration.
An additional factor predisposing αTS to misfold may be persistent nonrandom structure in the unfolded ensemble. The experimental methods used in this report are not overly sensitive to local structure. However, the near-UV CD spectra of αTS and its 188-residue N-terminal fragment comprising the first 188 residues suggest that nonrandom tertiary structure may persist under high denaturant concentrations (28), where the overall chain dimensions remain within expectations of a statistical coil (41). These structures could serve as nucleation sites that further bias αTS, and possibly TIM barrels in general, toward an off-pathway trajectory at the earliest stages of folding.
Implications for Protein-Misfolding Diseases.
It is now well recognized that protein-misfolding reactions can give rise to a plethora of heritable diseases (12). However, the identification of the specific conformers responsible for the soluble oligomers and larger-scale aggregates remains an open issue for most proteins. The complex folding behavior of the TIM barrel family of proteins, involving partitioning between off-pathway and on-pathway intermediates, raises the intriguing possibility that the early nonproductive species under examination in the present communication may be a key to aggregation in repeat-sequence motifs. Hartl and colleagues (14) have previously observed that TIM barrels are the most common motif bound to the GroEL chaperonin in E. coli. Scoring of the αTS sequence by TANGO (42) shows that β2, α2, and β3 are the most likely candidates for aggregation and, presumably, recognition by a chaperone. That these same sequences support the early collapse of the N terminus of αTS shows that, for the isolated wild-type protein, the intramolecular association of these marginally soluble segments effectively competes with intermolecular association reactions and aggregation. Mutations that increase the population and/or lifetime of the off-pathway species or that decrease its solubility would be candidates for aggregation.
Materials and Methods
Mutagenesis, Protein Preparation, and Labeling.
Four variants (K15C/F54W, K15C/E134W, S168C/F212W, and K15C/F212W) introducing an additional exposed cysteine and single tryptophan were constructed in two rounds of site-directed mutagenesis as previously described. Protein expression and purification followed standard protocols described in ref. 18. The three intrinsic cysteines present in wild-type αTS were not mutated, leaving the variants with four cysteine residues. Extensive efforts to remove the intrinsic cysteines, which are highly buried and inaccessible to solvent, were unsuccessful. The significant difference in solvent accessibility, however, allowed selective labeling with IAEDANS of the exposed artificially introduced cysteine, which has a reactivity ≈80-fold higher than the three highly buried native cysteines (43). Standard labeling protocols were followed and are detailed in SI Text.
The thermodynamic properties of all labeled and unlabeled αTS variants were checked by absorption, CD, fluorescence/anisotropy spectra, and time-correlated single-photon counting (TCSPC) by urea titrations on a JASCO V-560 UV/VIS spectrophotometer, JASCO J-810 spectropolarimeter, Fluorolog3 spectrofluorimeter (Jobin Yvon) and custom TCSPC instrument, respectively. The CD spectra of the unlabeled and labeled variants were consistent with those of wild-type αTS (Fig. S5).
All experiments were conducted in 10 mM phosphate buffer at pH 7.8 containing 0.2 mM K2EDTA and 1 mM βME. The temperature for all experiments was 25°C unless indicated otherwise.
TCSPC.
Details of the TCSPC apparatus equipped with a microsecond continuous-flow mixer have been described (Fig. S6) (33, 44). The mixer consisted of 75 μm channels laser-machined in 127-μm-thick polyetheretherketone (PEEK) and held between two ¼-inch fused silica windows by compression. Flow to the microchannel mixer was provided by two syringe pumps (Isco) operating at a combined flow rate of 15 or 20 ml·min−1.
Excitation at 293 nm with a repetition rate of 3.8 MHz was provided by the vertically polarized third harmonic of a Ti/sapphire laser. Typical excitation power was ≈0.5 to 1 mW. The mixer flow channel was aligned to yield excitation power that was uniform along the flow channel within 5%. This variation in excitation intensity was corrected by using a standard N-Acetyl-l-tryptophanamide (NATA) as described (44). Typical total count rates at ≈5 μM protein concentration were between 5 × 104 and 1 × 105 counts per second, well below count rates at which pile-up effects begin to affect the measured lifetimes (45, 46). Photon counts in the peak channel were typically ≈5,000 counts. Separate instrument responses were recorded for each channel by recording a scattered light signal or by numerical deconvolution from the NATA decay curve.
TCSPC Data Analysis.
Moment analysis of the Trp lifetime decays were used to obtain a model-independent estimate of the approximate average FRET efficiency as a function of time. FRET efficiencies were transformed to 〈EED〉app by using the Förster equation (47), using an experimentally determined Ro of 22 Å and an orientation factor of 2/3. The donor–acceptor pair distance distribution analysis was carried out as described (47, 48). The donor-only and donor–acceptor time-resolved fluorescence decays were globally fit to an analytic function described by a double-Gaussian function. The quality of the fits was significantly better for the double-Gaussian function compared with just a single-Gaussian function. To obtain sufficient photon counts for this analysis (>30,000 photons in the peak channel), 7 TCSPC decays along the folding dimension (≈3.7 μs per decay) were averaged, limiting the folding-time resolution to ≈26 μs. Because this time window is less than the time for complete mixing (≈35 μs), the averaging does not adversely interfere with the detection of kinetic folding events. Additional details of the distance distribution analysis are provided in SI Text.
SAXS Measurements.
SAXS measurements were performed at the BioCAT undulator beamline at the Advanced Photon Source (APS), Argonne National Laboratory (Argonne, IL) as described (26). The total flow rate was either 10 ml·min−1 for longer time points or 20 ml·min−1 for the early time points. The refolding reaction was initiated by a 10-fold dilution of the urea concentration from 6 M to 0.6 M urea. The temperature was 24°C for both equilibrium and kinetic measurements.
The SAXS data were analyzed by using IGOR Pro (WaveMetrics) macros written by the BioCAT staff at APS. The Rg and I (0) were obtained based on the Guinier approximation within the Guinier region (RgQ < 1.3) (22). Because of beam-focusing constraints and larger channels in the SAXS mixer, the time resolution of the SAXS experiment (≈150 μs) is lower than the 30-μs resolution achieved in the CF-TCSPC experiments.
Continuous-Flow CD.
CD spectra were acquired by interfacing the same microsecond mixer used for the fluorescence studies to an Aviv model 202 CD instrument (Aviv Biomedical). CD measurements were performed by positioning the mixer at a suitable time point, determined by the flow rate and distance from the mixing region, and acquiring scans in the standard way. Buffer blanks were acquired for each measurement and subtracted from the data acquired with protein. Typically, three overlaying spectra were acquired and averaged. Data for native and unfolded protein (at 0.8 M and 8 M urea, respectively) were acquired in the continuous-flow mixer by using the same buffer and protein stock solutions. Comparison of the native spectrum and that of (1S)-(+)-10-camphorsulfonic acid (Sigma–Aldrich) with cuvette-based measurements showed excellent agreement, showing that birefringence offsets from the fused silica windows and PEEK mixer could be subtracted out with confidence. Scans were collected in step-scan mode with sufficient time (2 s) for stabilization of the photomultiplier tube voltage at each wavelength, particularly in the blue end of the spectrum. The short optical path (127 μm) permitted the collection of spectra down to 200 nm, even in the presence of 8 M urea. A 20 ml·min−1 total flow rate was used, yielding a mixing time of ≈27 μs and a total instrument dead time (accounting for the width of the beam) of ≈70 μs. Spectra were acquired with a final sample concentration of 2 mg·ml−1.
Supplementary Material
Acknowledgments.
We thank J. Zitzewitz, R. Vadrevu, and T. Irving (BioCAT) for many helpful discussions; R. Heurich, M. Vukonich, and D. Gore of BioCAT for technical assistance with SAXS; S. Kathuria for technical assistance with continuous-flow CD experiments; T. Partington and A. Allard (University of Massachusetts Medical School Machine Shop) and D. Sachs (TEAM Specialty Products, Albequerque, NM) for machining many mixer components and numerous helpful design suggestions. This work was supported by National Institutes of Health (NIH) Grant GM23303 and National Science Foundation Grants MCB0327504 and MCB0721312. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under contract No. W-31-109-ENG-38. BioCAT is a NIH-supported Research Center (RR-08630).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802788105/DCSupplemental.
References
- 1.Brooks CL, Gruebele M, Onuchic JN, Wolynes PG. Chemical physics of protein folding. Proc Natl Acad Sci USA. 1998;95:11037–11038. doi: 10.1073/pnas.95.19.11037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Onuchic JN, Wolynes PG. Theory of protein folding. Curr Opin Struct Biol. 2004;14:70–75. doi: 10.1016/j.sbi.2004.01.009. [DOI] [PubMed] [Google Scholar]
- 3.Jackson SE. How do small single-domain proteins fold? Fold Des. 1998;3:R81–R91. doi: 10.1016/S1359-0278(98)00033-9. [DOI] [PubMed] [Google Scholar]
- 4.Gillespie B, Plaxco KW. Using protein folding rates to test protein folding theories. Annu Rev Biochem. 2004;73:837–859. doi: 10.1146/annurev.biochem.73.011303.073904. [DOI] [PubMed] [Google Scholar]
- 5.Istomin AY, Jacobs DJ, Livesay DR. On the role of structural class of a protein with two-state folding kinetics in determining correlations between its size, topology, and folding rate. Protein Sci. 2007;16:2564–2569. doi: 10.1110/ps.073124507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bilsel O, Zitzewitz JA, Bowers KE, Matthews CR. Folding mechanism of the alpha-subunit of tryptophan synthase, an alpha/beta barrel protein: Global analysis highlights the interconversion of multiple native, intermediate, and unfolded forms through parallel channels. Biochemistry. 1999;38:1018–1029. doi: 10.1021/bi982365q. [DOI] [PubMed] [Google Scholar]
- 7.Fernandez-Recio J, Genzor CG, Sancho J. Apoflavodoxin folding mechanism: An alpha/beta protein with an essentially off-pathway lntermediate. Biochemistry. 2001;40:15234–15245. doi: 10.1021/bi010216t. [DOI] [PubMed] [Google Scholar]
- 8.Bollen YJM, van Mierlo CPM. Protein topology affects the appearance of intermediates during the folding of proteins with a flavodoxin-like fold. Biophys Chem. 2005;114:181–189. doi: 10.1016/j.bpc.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 9.Chekmarev SF, Krivov SV, Karplus M. Folding of ubiquitin: A simple model describes the strange kinetics. J Phys Chem B. 2006;110:8865–8869. doi: 10.1021/jp056799o. [DOI] [PubMed] [Google Scholar]
- 10.Clementi C, Nymeyer H, Onuchic JN. Topological and energetic factors: What determines the structural details of the transition state ensemble and “en-route” intermediates for protein folding? An investigation for small globular proteins. J Mol Biol. 2000;298:937–953. doi: 10.1006/jmbi.2000.3693. [DOI] [PubMed] [Google Scholar]
- 11.Krishna MM, Englander SW. A unified mechanism for protein folding: predetermined pathways with optional errors. Protein Sci. 2007;16:449–464. doi: 10.1110/ps.062655907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 13.Thornton JM, Orengo CA, Todd AE, Pearl FM. Protein folds, functions and evolution. J Mol Biol. 1999;293:333–342. doi: 10.1006/jmbi.1999.3054. [DOI] [PubMed] [Google Scholar]
- 14.Kerner MJ, et al. Proteome-wide analysis of chaperonin-dependent protein folding in Escherichia coli. Cell. 2005;122:209–220. doi: 10.1016/j.cell.2005.05.028. [DOI] [PubMed] [Google Scholar]
- 15.Forsyth WR, Bilsel O, Gu Z, Matthews CR. Topology and sequence in the folding of a TIM barrel protein: Global analysis highlights partitioning between transient off-pathway and stable on-pathway folding intermediates in the complex folding mechanism of a (beta alpha)(8) barrel of unknown function from B-subtilis. J Mol Biol. 2007;372:236–253. doi: 10.1016/j.jmb.2007.06.018. [DOI] [PubMed] [Google Scholar]
- 16.Gu Z, Rao MK, Forsyth WR, Finke JM, Matthews CR. Structural analysis of kinetic folding intermediates for a TIM barrel protein, indole-3-glycerol phosphate synthase, by hydrogen exchange mass spectrometry and Gō model simulation. J Mol Biol. 2007;374:528–546. doi: 10.1016/j.jmb.2007.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu Y, Vadrevu R, Kathuria S, Yang XY, Matthews CR. A tightly packed hydrophobic cluster directs the formation of an off-pathway sub-millisecond folding intermediate in the alpha subunit of tryptophan synthase, a TIM barrel protein. J Mol Biol. 2007;366:1624–1638. doi: 10.1016/j.jmb.2006.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu Y, Vadrevu R, Yang X, Matthews CR. Specific structure appears at the N terminus in the sub-millisecond folding intermediate of the alpha subunit of tryptophan synthase, a TIM barrel protein. J Mol Biol. 2005;351:445–452. doi: 10.1016/j.jmb.2005.06.006. [DOI] [PubMed] [Google Scholar]
- 19.Kimura T, Lee JC, Gray HB, Winkler JR. Site-specific collapse dynamics guide the formation of the cytochrome c′ four-helix bundle. Proc Natl Acad Sci USA. 2007;104:117–122. doi: 10.1073/pnas.0609413103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Akiyama S, et al. Conformational landscape of cytochrome c folding studied by microsecond-resolved small-angle x-ray scattering. Proc Natl Acad Sci USA. 2002;99:1329–1334. doi: 10.1073/pnas.012458999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Uzawa T, et al. Collapse and search dynamics of apomyoglobin folding revealed by submillisecond observations of α-helical content and compactness. Proc Natl Acad Sci USA. 2004;101:1171–1176. doi: 10.1073/pnas.0305376101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Svergun DI, Koch MHJ. Small-angle scattering studies of biological macromolecules in solution. Rep Prog Phys. 2003;66:1735–1782. [Google Scholar]
- 23.Andrade MA, Chacon P, Merelo JJ, Moran F. Evaluation of secondary structure of proteins from uv circular-dichroism spectra using an unsupervised learning neural-network. Protein Eng. 1993;6:383–390. doi: 10.1093/protein/6.4.383. [DOI] [PubMed] [Google Scholar]
- 24.Brooks CL. Protein and peptide folding explored with molecular simulations. Acc Chem Res. 2002;35:447–454. doi: 10.1021/ar0100172. [DOI] [PubMed] [Google Scholar]
- 25.Kimura T, et al. Specific collapse followed by slow hydrogen-bond formation of β-sheet in the folding of single-chain monellin. Proc Natl Acad Sci USA. 2005;102:2748–2753. doi: 10.1073/pnas.0407982102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arai M, et al. Microsecond hydrophobic collapse in the folding of Escherichia coli dihydrofolate reductase, an alpha/beta-type protein. J Mol Biol. 2007;368:219–229. doi: 10.1016/j.jmb.2007.01.085. [DOI] [PubMed] [Google Scholar]
- 27.Wintrode PL, Rojsajjakul T, Vadrevu R, Matthews CR, Smith DL. An obligatory intermediate controls the folding of the alpha-subunit of tryptophan synthase, a TIM barrel protein. J Mol Biol. 2005;347:911–919. doi: 10.1016/j.jmb.2005.01.064. [DOI] [PubMed] [Google Scholar]
- 28.Zitzewitz JA, Matthews CR. Molecular dissection of the folding mechanism of the alpha subunit of tryptophan synthase: An amino-terminal autonomous folding unit controls several rate-limiting steps in the folding of a single domain protein. Biochemistry. 1999;38:10205–10214. doi: 10.1021/bi9909041. [DOI] [PubMed] [Google Scholar]
- 29.Finke JM, Onuchic JN. Equilibrium and kinetic folding pathways of a TIM barrel with a funneled energy landscape. Biophys J. 2005;89:488–505. doi: 10.1529/biophysj.105.059147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schuler B, Eaton WA. Protein folding studied by single-molecule FRET. Curr Opin Struct Biol. 2008;18:16–26. doi: 10.1016/j.sbi.2007.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Haas E. The study of protein folding and dynamics by determination of intramolecular distance distributions and their fluctuations using ensemble and single-molecule FRET measurements. Chem Phys Chem. 2005;6:858–870. doi: 10.1002/cphc.200400617. [DOI] [PubMed] [Google Scholar]
- 32.Lyubovitsky JG, Gray HB, Winkler JR. Mapping the cytochrome c folding landscape. J Am Chem Soc. 2002;124:5481–5485. doi: 10.1021/ja017399r. [DOI] [PubMed] [Google Scholar]
- 33.Bilsel O, Wu Y, Kayatekin C, Matthews CR. Becker W, editor. Proceedings of SPIE, Advanced Photon Counting Techniques II. 2007;6771:677103-1–677103-10. [Google Scholar]
- 34.Bilsel O, Matthews CR. Molecular dimensions and their distributions in early folding intermediates. Curr Opin Struct Biol. 2006;16:86–93. doi: 10.1016/j.sbi.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 35.Vadrevu R, Wu Y, Matthews CR. NMR analysis of partially folded states and persistent structure in the alpha subunit of tryptophan synthase: implications for the equilibrium folding mechanism of a 29-kDa TIM barrel protein. J Mol Biol. 2008;377:294–306. doi: 10.1016/j.jmb.2007.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wolfenden R, Andersson L, Cullis PM, Southgate CC. Affinities of amino acid side chains for solvent water. Biochemistry. 1981;20:849–855. doi: 10.1021/bi00507a030. [DOI] [PubMed] [Google Scholar]
- 37.Miller S, Janin J, Lesk AM, Chothia C. Interior and surface of monomeric proteins. J Mol Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 38.Bai Y, Sosnick TR, Mayne L, Englander SW. Protein folding intermediates: Native-state hydrogen exchange. Science. 1995;269:192–197. doi: 10.1126/science.7618079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chamberlain AK, Handel TM, Marqusee S. Detection of rare partially folded molecules in equilibrium with the native conformation of RNaseH. Nat Struct Biol. 1996;3:782–787. doi: 10.1038/nsb0996-782. [DOI] [PubMed] [Google Scholar]
- 40.Bedard S, Mayne LC, Peterson RW, Wand AJ, Englander SW. The foldon substructure of staphylococcal nuclease. J Mol Biol. 2008;376:1142–1154. doi: 10.1016/j.jmb.2007.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kohn JE, et al. Random-coil behavior and the dimensions of chemically unfolded proteins. Proc Natl Acad Sci USA. 2004;101:12491–12496. doi: 10.1073/pnas.0403643101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 43.Ratner V, Kahana E, Eichler M, Haas E. A general strategy for site-specific double labeling of globular proteins for kinetic FRET studies. Bioconjug Chem. 2002;13:1163–1170. doi: 10.1021/bc025537b. [DOI] [PubMed] [Google Scholar]
- 44.Bilsel O, Kayatekin C, Wallace LA, Matthews CR. A microchannel solution mixer for studying microsecond protein folding reactions. Rev Sci Instrum. 2005;76 014302-1–014302-8. [Google Scholar]
- 45.Becker W. Advanced Time-Correlated Single Photon Counting Techniques. Berlin: Springer; 2005. [Google Scholar]
- 46.Suhling K, McLoskey D, Birch DJS. Multiplexed single-photon counting. 2. The statistical theory of time-correlated measurements. Rev Sci Instrum. 1996;67:2238–2246. [Google Scholar]
- 47.Lakowicz J. Principles of Fluorescence Spectroscopy. New York: Plenum; 1999. [Google Scholar]
- 48.Haran G, Haas E, Szpikowska BK, Mas MT. Domain motions in phosphoglycerate kinase: Determination of interdomain distance distributions by site-specific labeling and time-resolved fluorescence energy transfer. Proc Natl Acad Sci USA. 1992;89:11764–11768. doi: 10.1073/pnas.89.24.11764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rhee S, et al. Crystal structures of a mutant (betaK87T) tryptophan synthase alpha2beta2 complex with ligands bound to the active sites of the alpha- and beta-subunits reveal ligand-induced conformational changes. Biochemistry. 1997;36:7664–7680. doi: 10.1021/bi9700429. [DOI] [PubMed] [Google Scholar]
- 50.DeLano W. The PyMOL Molecular Graphics System. Palo Alto, CA: DeLano Scientific; 2002. http://www.pymol.org. [Google Scholar]
- 51.Gualfetti PJ, et al. Apparent radii of the native, stable intermediates and unfolded conformers of the alpha-subunit of tryptophan synthase from E. coli, a TIM barrel protein. Biochemistry. 1999;38:13367–13378. doi: 10.1021/bi991296s. [DOI] [PubMed] [Google Scholar]
- 52.Gualfetti PJ, Bilsel O, Matthews CR. The progressive development of structure and stability during the equilibrium folding of the alpha subunit of tryptophan synthase from Escherichia coli. Protein Sci. 1999;8:1623–1635. doi: 10.1110/ps.8.8.1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.