Abstract
Folding intermediates play a key role in defining protein folding and assembly pathways as well as those of misfolding and aggregation. Yet, due to their transient nature, they are poorly accessible to high-resolution techniques. Here, we made use of the intrinsically slow folding reaction of an antibody domain to characterize its major folding intermediate in detail. Furthermore, by a single point mutation we were able to trap the intermediate in equilibrium and characterize it at atomic resolution. The intermediate exhibits the basic β-barrel topology, yet some strands are distorted. Surprisingly, two short strand-connecting helices conserved in constant antibody domains assume their completely native structure already in the intermediate, thus providing a scaffold for adjacent strands. By transplanting these helical elements into β2-microglobulin, a highly homologous member of the same superfamily, we drastically reduced its amyloidogenicity. Thus, minor structural differences in an intermediate can shape the folding landscape decisively to favor either folding or misfolding.
Keywords: amyloids, NMR, protein folding, antibodies, molecular dynamics
In the current view, almost all proteins are believed to populate partially folded species, so-called folding intermediates, along their pathways to the native state (1–6). The characteristics of folding intermediates are critical in determining whether a protein is able to fold robustly or has the tendency to misfold (6, 7). A detailed structural characterization of folding intermediates is thus key for the understanding of protein folding in general. Because they are transient, however, only very few folding intermediates have so far been described in atomic detail (8–13). In general, experimental data argue for near-native topology with incompletely folded or partially misfolded structural elements such as side-chain interactions (8–13). In this context it is of particular importance that partially folded states have recently been associated with a large variety of pathologies (14). In the case of amyloid diseases—e.g., transthyretin (TTR) familial amyloid polyneuropathy (15), light chain amyloidosis (16), or dialysis-related amyloidosis (17)—it is thought that folding intermediates are key precursors for the formation of amyloid fibrils (18). Amyloids are long-lived dissociation- and degradation-resistant structures. They are made up of β-strands that are arranged into sheets lying perpendicular to the long fiber axis and possess a core cross-β structure (19). Despite the large variety of native folds shown by amyloidogenic proteins, these structural features seem to be a recurring motif in amyloids suggesting a common assembly mechanism (20). A particularly well studied example in this respect is the MHCI component β2-microglobulin (β2m), a member of the widespread immunoglobulin (Ig) superfamily (21, 22). β2m has been shown to form amyloid fibrils if partially unfolded—e.g., at acidic pH (23), where the reaction is thought to be initiated by the population of a partially folded intermediate (23, 24). When such intermediates are populated for long periods, they are particularly susceptible to misfolding and misassembly reactions as has been shown for β2m. Its productive folding from a native-like intermediate to the native state is limited by an intrinsically slow trans-to-cis peptidyl-prolyl isomerization reaction (24, 25). Experiments in which the critical proline residue was held in a trans state confirmed that this intermediate is a major determinant in amyloid formation (24, 26). In this regard, several studies showed that the most probable amyloidogenic precursor already possesses a large part of the native β-sheet topology with only the outer strands and loop regions being distorted (24, 25, 27).
Bearing in mind that intermediates are a rather general aspect of a protein folding reaction and that most polypeptides are in principle susceptible to amyloid formation (28), the question arises of how proteins avoid aggregation in the majority of cases. To address this issue we set out to study the folding pathway of the constant domain of the antibody light chain (CL) with high structural resolution. The CL domain is a particularly instructive model system because it also belongs to the Ig superfamily and, like β2m, forms a β-sandwich composed of seven strands stabilized by a single disulfide bond between strands B and F (29, 30). The cis proline residue associated with the amyloidogenic potential of β2m is conserved in the CL domain (29). Furthermore, the overall folding mechanisms of the two proteins are highly similar (24, 30), each populating an intermediate state en route to the native state. Nevertheless, the CL domain has never been directly associated with amyloidogenic diseases even if present at much higher concentrations than β2m in the blood (31). By the structural characterization of its major folding intermediate, we show how the CL antibody domain might avoid such harmful misfolding reactions.
Results
The Major Kinetic Folding Intermediate of CL is Highly Structured.
The CL domain folds via an obligatory intermediate on two parallel pathways to its native state, the slower one being limited by the isomerization of the Y34–P35 bond to the native cis conformation (30, 32). This bond is predominantly trans in the unfolded state. As a consequence, only ≈10% of the molecules are able to fold to the native state within a few seconds (30, 32), and ≈90% of the molecules have to undergo the intrinsically slow isomerization reaction before complete folding to the native state (30, 32). At 2°C this reaction takes several hours to complete [see supporting information (SI) Fig. S1], allowing the major kinetic intermediate to be populated for a significant amount of time. CD spectra of the intermediate argue for a partially formed β-sheet framework and the absence of defined asymmetric environment around the aromatic amino acids (see Fig. S1).
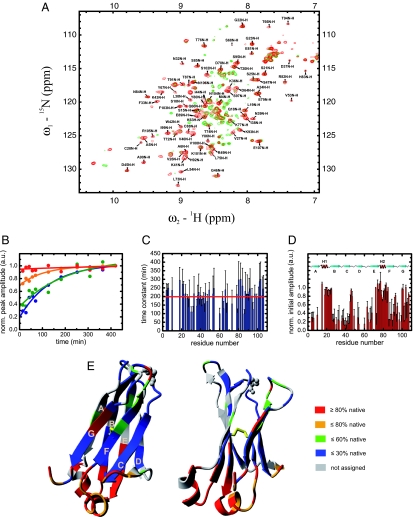
To structurally characterize the intermediate state as well as the folding process on a residue level, >70% of the CL domain backbone was assigned by standard NMR techniques (Fig. 1A), and real-time 15N-1H HSQC spectra were measured during refolding from the chemically denatured state. The first spectrum recorded after 14 min reflects almost exclusively the kinetic intermediate and had only to be corrected for 10% of the CL molecules possessing the correct Y34–P35 isomerization state (Fig. 1A; see Materials and Methods for details). Because the chemical shifts of the amide protons strongly depend on their molecular environment, overlaying the HSQC spectra of the intermediate and the native state reveals similarities and changes in their environment during the folding process (Fig. 1A). The HSQC spectra of the native CL domain and the folding intermediate are superimposable for some residues but non-superimposable for others where significant differences in the chemical shifts are observed (Fig. 1A). To obtain more insights into the structural properties of the intermediate, the change in the peak intensity at the native chemical shift position was followed over time for each assigned residue. In every case the change in peak intensity could be well described by a single exponential function, if not already showing a native-like intensity after the dead-time of the experiment (Fig. 1B). As can be seen in Fig. 1C, the time constants of the folding of the individual residues show stochastic behavior around a mean value of τ = 199 min at 2°C without any significant systematic deviations for any part of the protein. In contrast, initial HSQC amplitudes in the folding intermediate show interesting patterns: almost native initial amplitudes are found in correspondence of the two short helices connecting strand A and B as well as E and F and adjacent β-sheet termini suggesting that they are already in a native environment in the intermediate whereas low initial amplitudes are observed for some of the β-strands, in particular strands C and D, which suggests a lack of native structure (Fig. 1D). In Fig. 1E regions of high or low initial amplitudes are mapped on the crystal structure of CL revealing that the two helices and their local environment are highly structured in the intermediate.
Fig. 1.
Structural characterization of the major CL folding intermediate by NMR spectroscopy. (A) 15N-1H HSQC spectrum of the native CL domain including the backbone resonance assignment is shown in red and the one of the intermediate in green. The spectrum of the intermediate was derived from the first HSQC spectrum measured during refolding after 14 min and corrected for the 10% CL molecules that refold within the dead-time of the experiment. (B) The change in intensity over the time for each peak was fitted by a single exponential function and extrapolated to time 0 (blue, K36; green, E89; orange, E79; red, L19 selected as representative residues). The time constants for every assigned residue are shown in C. The red line denotes their mean value. Initial amplitudes are shown in D. (E) Initial HSQC amplitudes mapped on the crystal structure of the native CL domain (PDB entry 1FH5). The colors represent the initial HSQC amplitudes relative to the native HSQC amplitudes. All spectra were recorded at 2°C. Protein concentrations of 0.5–1.0 mM in PBS buffer with a GdmCl concentration of 0.2 M were used.
An Intermediate Structure at Equilibrium Trapped by a Single Point Mutation.
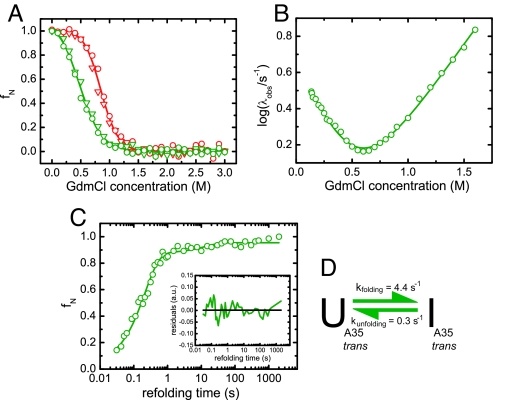
The initial HSQC amplitudes only provide hints on the structural properties of the intermediate. Equilibrium spectroscopic data could provide information more directly related to structure. Therefore, we tried to trap the intermediate at equilibrium by exploiting the isomerization reaction separating the intermediate from the native state. We hypothesized that mutating the P35 residue against another amino acid that preferentially adopts a trans peptide bond (33), such as Ala (CLP35A), might “trap” the kinetic intermediate making it populated at equilibrium. Indeed, far-UV and near-UV CD spectra of CLP35A were found to be very similar to the respective spectra of the kinetic intermediate (data not shown). To determine the stability of the mutant in comparison to the wild type (CLwt), denaturant-induced unfolding transitions were performed. The unfolding of both proteins, CLwt and CLP35A, was a two-state process because there was concurrent loss of secondary structure (monitored by far-UV CD-spectroscopy) and tertiary structure (monitored by the change in the intrinsic tryptophan fluorescence) (Fig. 2A). Consistent with a partially folded species being the major equilibrium state, the P35A mutation led to a stability reduction of the CL domain by >50% from ΔGunfolding = 13.4 ± 0.9 kJ·mol−1 for the wild-type protein to ΔGunfolding = 6.1 ± 0.5 kJ·mol−1 for the mutant (Fig. 2A). The cooperativity parameter of the transition decreased by 20% from meq = 15.9 ± 0.9 kJ·mol−1·M−1 for CLwt to meq = 12.8 ± 0.8 kJ·mol−1·M−1 for CLP35A. If the kinetic intermediate is trapped by the Pro35Ala mutation, a simplified folding mechanism devoid of the kinetic phases leading to the native state is expected for CLP35A as compared to CLwt. To assess the folding mechanism of CLP35A, a chevron plot was determined. The mutant only showed one major folding/unfolding phase in the chevron plot (Fig. 2B) giving rise to one folding and one unfolding microscopic rate constant (kf = 4.4 ± 0.1 s−1/ku = 0.30 ± 0.02 s−1 at 20°C and 0 M GdmCl). In contrast to this, two separate folding phases had been reported for the major folding pathway of CLwt (30). From a global fit of the overall folding mechanism, the rate constants for each individual folding process could be determined for CLwt (30). It folds to its intermediate state with a very similar rate constant as CLP35A folds to its final state (30). From the derived rate constants, the stability of the CLwt folding intermediate could be calculated to 11 ± 2 kJ·mol−1, which is slightly higher than the stability of CLP35A (ΔGunfolding = 6.1 ± 0.5 kJ·mol−1). ΔGunfolding for the intermediate of CLwt, however, is only an indirect estimate. Additionally, CLP35A unfolds faster than the CLwt intermediate (30) pointing towards a certain kinetic destabilization of the intermediate structure by the P35A exchange itself.
Fig. 2.
Stability and folding mechanism of the CLP35A mutant. (A) Equilibrium unfolding transitions of CLP35A (green) and CLwt (red) determined by the intrinsic tryptophan fluorescence excited at 280 nm and detected at 360 nm (circles) as well as the far-UV CD-signal at 218 nm (inverted triangles). The data were fit to a two-state unfolding model. (B) Chevron plot for CLP35A determined by stopped-flow fluorescence spectroscopy. It could be described by a two-state folding reaction. (C) Formation of the final refolding species of CLP35A was followed by interrupted refolding experiments. The data were fit by a double exponential function (residuals shown as Inset) with 93% of the molecules folding via the fast pathway. (D) The folding mechanism of CLP35A can be described by a simple two-state model neglecting the 7% slow folding species. Transitions were measured at 10 μM protein concentration, and all kinetic experiments were performed at a final protein concentration of 2 μM. All measurements were carried out at 20°C in PBS.
Notably, for CLP35A ΔGkinetic = 6.6 ± 0.3 kJ·mol−1, the stability calculated from the rate constants obtained from the chevron plot, is in very good agreement with the value from the equilibrium unfolding experiment (ΔGunfolding = 6.1 ± 0.5 kJ·mol−1) confirming a two-state kinetic process. This was corroborated by interrupted refolding experiments (Fig. 2C), which show that 93% of the molecules fold to their final state with a rate constant of 4.8 ± 0.4 s−1 and only 7% with a rate constant of 0.08 ± 0.01 s−1, which can likely be attributed to the isomerization of one of the four remaining Proline residues from a nonnative cis to the native trans state. All data argue for the CLP35A mutant populating the kinetic intermediate in equilibrium, which is trapped by the trans state of the bond preceding A35. The data are summarized in the folding model shown in Fig. 2D.
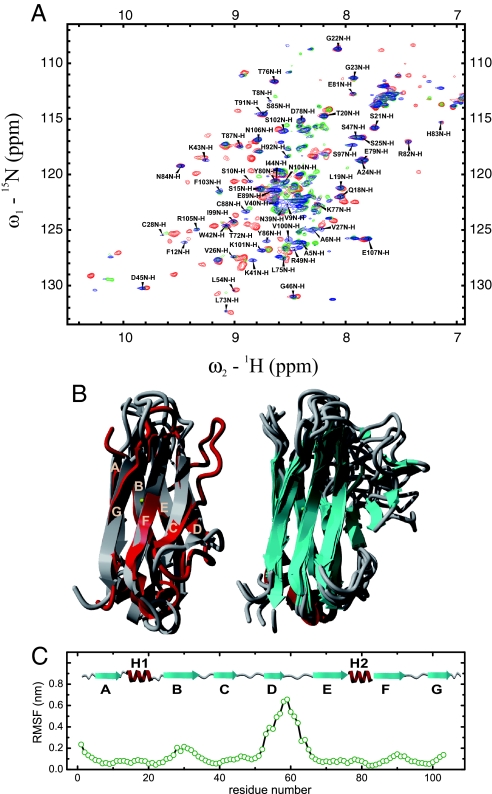
To confirm this, the structure of CLP35A was further characterized by NMR spectroscopy. 15N-1H HSQC spectra of the CLwt kinetic intermediate and CLP35A were almost completely superimposable (Fig. 3A), demonstrating equivalent secondary and tertiary structure in both species. Crucially, the trapping of the intermediate state allowed an NMR assignment to be carried out (Fig. 3A), which was not feasible for the transiently populated kinetic folding intermediate observed in the folding of CLwt. To further compare the native and the intermediate states, a set of NMR measurements was recorded for both proteins (CLwt and CLP35A). These included, besides the standard backbone experiments and triple resonance experiments for aliphatic side-chain assignment, 3D-NOESY spectra. These data revealed that large parts of the proteins possess almost identical carbon chemical shifts and, in addition, corresponding NOESY strips within the same range (Fig. S2 and data not shown), indicating that the intermediate already adopts a highly ordered structure. Notably, the two helices are fully formed, as judged from the NOESY pattern and stabilized by distinct helix capping motifs (34). Regions in CLP35A with significant differences in the carbon chemical shifts and the NOESY pattern from CLwt are all located around A35 and strands D and E. To gain further dynamic and structural information that cannot be deduced from the experiments alone, we performed molecular dynamics (MD) simulations with NMR-derived restraints (see Materials and Methods for details). Low-energy conformations in which restraints are collectively minimally violated can be sampled by this approach and provide an ensemble of structures that best represent the partially folded state. The simulations show that the overall topology of CLP35A is on average well retained (Fig. 3B). Additionally, both helices were fully structured in the simulations and showed only minor fluctuations (Fig. 3 B and C). Strikingly, the edge strands A and G were found to be native in all simulations (Fig. 3 B and C). However, we observe considerable heterogeneity at the ends of some strands in the ensemble of structures (Fig. 3C). This is especially pronounced around A35, arguing for a partial distortion of one edge of the protein by the trans state of the peptide bond preceding residue 35. The only strand which was highly distorted was strand D (Fig. 3 B and C). Only residues V53 and L54 of this strand were relatively native-like in the ensemble of structures. This is in good agreement with NOE signals observed exclusively for these residues within strand D (data not shown). The N terminus of strand E and strand G were found to be flexible—i.e., a mixture of structures with full or partial ordering of the respective strands was observed in the MD simulations (Fig. 3 B and C). Interestingly, the partial disordering of strand E and one of its ends provides a rationale for the missing NMR assignment of some residues from its flanking strand B, which itself is found to be highly structured in the simulations (Fig. 3 B and C). The overall solvent-accessible surface area of CLP35A was found to be on average 10% larger than for the wild type, which is in good agreement with the observed 20% decrease in the cooperativity parameter, meq, for unfolding. Taken together, the NMR experiments in combination with the simulations provide a detailed picture of the major CL folding intermediate. The two small helices and their local environment are completely folded, and the intermediate exhibits a native-like core structure despite the presence of flexible regions that are able to adopt a variety of conformations.
Fig. 3.
NMR-spectroscopic characterization of the CLP35A mutant and structural comparison to the kinetic folding intermediate and CLwt. The 15N-1H HSQC spectrum of CLP35A including the backbone resonance assignment is shown in blue (A). For comparison, the HSQC spectrum of the kinetic folding intermediate is shown in green and the one of the wild type in red. All spectra were recorded at 2°C and protein concentrations of 0.5–1.0 mM in PBS buffer with a GdmCl concentration of 0.2 M. (B Left) Overlay of the energy-minimized native CL structure (gray) with the average of 30 structures from the NMR-restrained MD simulations of CLP35A (red). (B Right) Eight structures from the restrained CLP35A MD simulations are shown to highlight flexible parts. (C) Root mean square fluctuations (RMSF) for the last 30 CLP35A structures derived from the restrained simulations.
Dissection of the Amyloidogenic Properties of the CL Domain and β2-Microglobulin.
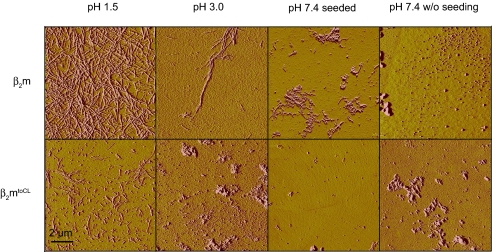
The constant domain of the antibody light chain has never been reported to be directly responsible for amyloidogenic processes even though it possesses the same topology as amyloidogenic variable antibody domains (VL) or β2-microglobulin (β2m). In both cases, amyloid formation is assumed to proceed from a partially folded intermediate state (27, 35). Interestingly, neither amyloidogenic protein possesses the short strand-connecting helices that we identified as highly structured elements in the CL folding intermediate. Accordingly, the sequence or structure of these helical elements might play a role in the inhibition of amyloid formation. To test this, we exchanged the unstructured loop regions connecting strands A and B as well as strands E and F in β2m against the corresponding helical elements of the CL domain (β2mtoCL) (see Fig. S3). The β2mtoCL exchange mutant folds to a well defined structure with similar far-UV CD-spectroscopic properties as wild-type β2m (see Fig. S3 for details). The helical elements destabilize β2mtoCL against thermal denaturation but have only a minor effect on its pH stability as compared to β2m (see Fig. S3). According to the TANGO algorithm (36), the aggregation propensity of its primary sequence is left unaffected by the mutations (data not shown). To assess the amyloidogenicity of the different proteins, we used established reaction conditions for β2m (37) and monitored fibril formation by atomic force microscopy (AFM). As expected, CLwt and the CLP35A mutant were not prone to fibril formation. In only one out of seven individual experiments were fibrils observed at pH 1.5, yet no fibrils were found at pH 3.0 or under physiological conditions, whether seeded or not (see Fig. S4). Importantly, a clear difference in amyloidogenicity is observed for wild-type β2m and the β2mtoCL mutant. Whereas β2m readily formed fibrils under all conditions tested, β2mtoCL only formed short fibrils at pH 1.5 (Fig. 4). At pH 3.0, fibrils were only detected in two out of seven individual experiments for β2mtoCL, and no fibrils were detected under physiological conditions for this protein, even when β2m fibrils were used for cross-seeding experiments (data not shown). The data clearly show that transplanting the sequences corresponding to the CL helices into the β2m framework significantly reduces its amyloidogenicity.
Fig. 4.
Amyloidogenic properties of β2m and the β2mtoCL exchange mutant. Both proteins were incubated at pH 1.5 or 3.0 at 37°C for 7 days at a concentration of 50 μM. Additionally, the proteins were incubated under physiological conditions (PBS, 37°C) either seeded with β2m or β2mtoCL fibrils or not. Formation of amyloid fibrils was assessed by AFM measurements. Representative pictures of each sample are shown.
Discussion
From a combination of NMR experiments and MD simulations we determined the ensemble of structures making up the major kinetic folding intermediate on the CL folding pathway at atomic resolution. The structures provide hints on what sets an aggregation-prone folding intermediate apart from a structurally similar yet productive intermediate. On average, the overall β-sheet topology is well established for a major part of the protein, but most aromatic residues are still solvent-exposed or adjacent to dynamic structural elements. The only strand that seems to be highly disordered is strand D, despite native interactions of V53 and L54 with strand E. All other strands and in particular strands B, C, E, and F, which constitute the folding nucleus of Ig proteins (38, 39), exhibit some dynamics but are already well structured.
The most striking structural features of the intermediate are the two completely folded small helices. Although they are strongly conserved in constant antibody domains, their role in the folding process has not yet been recognized. These helices seem to fulfill a spacer and orienting function between strand pairs A–B and E–F and provide hydrogen bond donors and acceptors for adjacent strands and loops. In addition, the helices appear to position hydrophobic residues (e.g., Y80 in helix 2) so that they can participate in the formation of the hydrophobic core. Our data suggest that the two helices are able to fold efficiently and autonomously to their native structures in the context of the intermediate. Hence, these two helices can be regarded as a scaffold within the CL intermediate favoring the formation of a native-like topology by correctly positioning important parts of the molecule.
An important protective role against amyloid formation has been attributed to the edge strands of β-sheet proteins (40, 41). Our finding that the edge strands A and G on one side of the CL intermediate are highly structured provides one possible explanation for the marked difference in amyloidogenicity between CL and β2m. Grafting the sequences corresponding to the CL helices onto the corresponding positions in β2m significantly reduces the propensity of β2m to form amyloids. This reduction is most pronounced at physiological pH, and the helices seem to be robust folding elements, which suggests a structural explanation for this effect but cannot completely rule out sequence effects of the transplanted elements themselves. Similar approaches to study the determinants of amyloid formation have been used previously but were targeted at increasing the amyloidogenicity of the “host” proteins rather than suppressing it as done here (42, 43). The edge strands A, D, and G have been reported to be disturbed in the amyloidogenic intermediate of β2m (24–27, 44). This leaves edge strands on both sides of the protein molecule unprotected, making a linear arrangement of monomers into fibrils more likely than when just one side of the protein is partially distorted, as observed for CL. For the amyloidogenic TTR, one complete sheet of the native β-sandwich structure is destabilized in the amyloidogenic precursor (45), and for VL, the major folding intermediate is less structured than the CL intermediate (38). In this context, we also note that β2m forms fibrils more readily than CL even though it is significantly more stable than CL (24, 30). Accordingly, it is not the stability of the native state per se that is the most important factor determining the amyloid forming propensity of a protein, but rather the sequence of the protein (46) and structural characteristics of partially folded species that may be populated along the folding pathway (37).
In conclusion, our data show how a high degree of local structuring in a protein folding intermediate can significantly influence the folding landscape and favor robust folding over harmful misfolding. The different characteristics of CL and β2m can be understood in evolutionary terms. Selection of antibodies took place under harsh extracellular conditions with high concentrations of the multimeric protein present (47), whereas β2m is found at much lower concentrations and usually associated with the MHCI complex (21). Thus, small differences, acquired over the course of evolution, between members of the same protein superfamily can lead to the avoidance of pathogenic misfolding reactions while preserving an identical protein topology.
Materials and Methods
Protein Production and Purification.
Proteins were expressed and purified as described in SI Methods.
Optical Spectroscopy.
CD kinetics and spectra were measured as described in SI Methods. Equilibrium unfolding transitions, stopped-flow, and interrupted refolding experiments were performed as described in ref. 30. For the interrupted refolding experiments, CLP35A was unfolded in 1.5 M GdmCl, refolded in 136 mM GdmCl for different times, and finally unfolded again in 1.5 M GdmCl.
NMR Spectroscopy.
If not stated otherwise, all spectra were recorded at 25°C on Bruker DMX600, DMX750, and AVANCE900 spectrometers as described in SI Methods. For folding studies, 15N-labeled unfolded CL in PBS containing 2 M GdmCl was diluted 10-fold by adding ice-cold PBS without GdmCl. Real-time 15N-1H HSQC spectra were recorded at 2°C every 14 min by using selective proton flip-back pulses (48). Identical processing of all of the spectra was performed by using the program TOPSPIN 1.3 (Bruker BioSpin). To obtain the 15N-1H HSQC of the intermediate, 10% of the final spectrum was subtracted from the first recorded HSQC spectrum. Peak intensities were analyzed by using the program SPARKY (www.cgl.ucsf.edu/home/sparky). For kinetic studies, the intensities of every amino acid during the folding process were corrected for 10% native molecules and normalized to the corresponding intensity in the final spectrum after 7 h. Backbone resonance assignments were transferred from 25°C to 2°C recording a temperature series of spectra referenced to the internal standard TSP.
MD Simulations.
The CLP35A mutant was created from the crystal structure of the CL domain (PDB entry 1FH5). The bond preceding A35 was set to trans and the protein was energy-minimized with the steepest-descent and the adopted Newton–Rhapson methods (49). Distance and dihedral restraints derived from a comparison of the NMR chemical shifts for CLwt and the CLP35A mutant were used in conjunction with a simulated annealing (SA)-like protocol to derive a set of structures best representing the CLP35A mutant. One hundred cycles of SA were performed, and the final structures from the last 30 cycles were used for further analysis. Details about the restraints and the SA protocol are presented in SI Methods.
All simulations were performed with CHARMM (49), using the CHARMM19 force field, the EEF1 implicit solvent model (50), a Langevin dynamics scheme with a friction coefficient of 1 ps−1, a time step of 2 fs, and holonomic constraints (SHAKE) on all bonds involving hydrogen atoms.
AFM Measurements.
Fibrillization experiments and AFM measurements were performed as described in SI Methods.
Supplementary Material
Acknowledgments.
We thank Helmut Krause for performing the MS analysis and Emma R. Simpson for helpful comments on the manuscript. We are grateful for financial support of the Max-Buchner Forschungsstiftung, Deutsche Forschungsgemeinschaft Sonderforschungsbereich 749, and the Fonds der Chemischen Industrie. M.J.F. and M.M. acknowledge a Ph.D. scholarship from the Studienstiftung des deutschen Volkes. Z.T.Y. acknowledges a Ph.D. scholarship from the Wellcome Trust and the University of Leeds.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0802809105/DCSupplemental.
References
- 1.Dill KA, Chan HS. From Levinthal to pathways to funnels. Nat Struct Biol. 1997;4:10–19. doi: 10.1038/nsb0197-10. [DOI] [PubMed] [Google Scholar]
- 2.Sanchez IE, Kiefhaber T. Evidence for sequential barriers and obligatory intermediates in apparent two-state protein folding. J Mol Biol. 2003;325:367–376. doi: 10.1016/s0022-2836(02)01230-5. [DOI] [PubMed] [Google Scholar]
- 3.Korzhnev DM, et al. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2004;430:586–590. [Google Scholar]
- 4.Neuweiler H, Doose S, Sauer M. A microscopic view of miniprotein folding: Enhanced folding efficiency through formation of an intermediate. Proc Natl Acad Sci USA. 2005;102:16650–16655. doi: 10.1073/pnas.0507351102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bai Y. Energy barriers, cooperativity, and hidden intermediates in the folding of small proteins. Biochem Biophys Res Commun. 2006;340:976–983. doi: 10.1016/j.bbrc.2005.12.093. [DOI] [PubMed] [Google Scholar]
- 6.Brockwell DJ, Radford SE. Intermediates: Ubiquitous species on folding energy landscapes? Curr Opin Struct Biol. 2007;17:30–37. doi: 10.1016/j.sbi.2007.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jahn TR, Radford SE. Folding versus aggregation: Polypeptide conformations on competing pathways. Arch Biochem Biophys. 2008;469:100–117. doi: 10.1016/j.abb.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Balbach J, et al. Following protein-folding in real-time using NMR-spectroscopy. Nat Struct Biol. 1995;2:865–870. doi: 10.1038/nsb1095-865. [DOI] [PubMed] [Google Scholar]
- 9.Mayor U, et al. The complete folding pathway of a protein from nanoseconds to microseconds. Nature. 2003;421:863–867. doi: 10.1038/nature01428. [DOI] [PubMed] [Google Scholar]
- 10.Feng HQ, Zhou Z, Bai YW. A protein folding pathway with multiple folding intermediates at atomic resolution. Proc Natl Acad Sci USA. 2005;102:5026–5031. doi: 10.1073/pnas.0501372102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Religa TL, Markson JS, Mayor U, Freund SMV, Fersht AR. Solution structure of a protein denatured state and folding intermediate. Nature. 2005;437:1053–1056. doi: 10.1038/nature04054. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura C, Dyson HJ, Wright PE. Identification of native and non-native structure in kinetic folding intermediates of apomyoglobin. J Mol Biol. 2006;355:139–156. doi: 10.1016/j.jmb.2005.10.047. [DOI] [PubMed] [Google Scholar]
- 13.Mizuguchi M, Kroon GJ, Wright PE, Dyson HJ. Folding of a β-sheet protein monitored by real-time NMR spectroscopy. J Mol Biol. 2003;328:1161–1171. doi: 10.1016/s0022-2836(03)00349-8. [DOI] [PubMed] [Google Scholar]
- 14.Chiti F, Dobson CM. Protein misfolding, functional amyloid, and human disease. Annu Rev Biochem. 2006;75:333–366. doi: 10.1146/annurev.biochem.75.101304.123901. [DOI] [PubMed] [Google Scholar]
- 15.Hou X, Aguilar MI, Small DH. Transthyretin and familial amyloidotic polyneuropathy—Recent progress in understanding the molecular mechanism of neurodegeneration. FEBS J. 2007;274:1637–1650. doi: 10.1111/j.1742-4658.2007.05712.x. [DOI] [PubMed] [Google Scholar]
- 16.Bellotti V, Mangione P, Merlini G. Review: Immunoglobulin light chain amyloidosis—The archetype of structural and pathogenic variability. J Struct Biol. 2000;130:280–289. doi: 10.1006/jsbi.2000.4248. [DOI] [PubMed] [Google Scholar]
- 17.Eakin CM, Miranker AD. From chance to frequent encounters: Origins of β2-microglobulin fibrillogenesis. Biochim Biophys Acta Proteins Proteomics. 2005;1753:92–99. doi: 10.1016/j.bbapap.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 18.Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 19.Fandrich M. On the structural definition of amyloid fibrils and other polypeptide aggregates. Cell Mol Life Sci. 2007;64:2066–2078. doi: 10.1007/s00018-007-7110-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Dobson CM. Protein folding and misfolding. Nature. 2003;426:884–890. doi: 10.1038/nature02261. [DOI] [PubMed] [Google Scholar]
- 21.Guo HC, et al. Different length peptides bind to Hla-Aw68 similarly at their ends but bulge out in the middle. Nature. 1992;360:364–366. doi: 10.1038/360364a0. [DOI] [PubMed] [Google Scholar]
- 22.Bork P, Holm L, Sander C. The immunoglobulin fold—Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 23.McParland VJ, et al. Partially unfolded states of β2-microglobulin and amyloid formation in vitro. Biochemistry. 2000;39:8735–8746. doi: 10.1021/bi000276j. [DOI] [PubMed] [Google Scholar]
- 24.Jahn TR, Parker MJ, Homans SW, Radford SE. Amyloid formation under physiological conditions proceeds via a native-like folding intermediate. Nat Struct Mol Biol. 2006;13:195–201. doi: 10.1038/nsmb1058. [DOI] [PubMed] [Google Scholar]
- 25.Kameda A, et al. Nuclear magnetic resonance characterization of the refolding intermediate of β2-microglobulin trapped by non-native prolyl peptide bond. J Mol Biol. 2005;348:383–397. doi: 10.1016/j.jmb.2005.02.050. [DOI] [PubMed] [Google Scholar]
- 26.Eakin CM, Berman AJ, Miranker AD. A native to amyloidogenic transition regulated by a backbone trigger. Nat Struct Mol Biol. 2006;13:202–208. doi: 10.1038/nsmb1068. [DOI] [PubMed] [Google Scholar]
- 27.McParland VJ, Kalverda AP, Homans SW, Radford SE. Structural properties of an amyloid precursor of β2-microglobulin. Nat Struct Biol. 2002;9:326–331. doi: 10.1038/nsb791. [DOI] [PubMed] [Google Scholar]
- 28.Fandrich M, Fletcher MA, Dobson CM. Amyloid fibrils from muscle myoglobin—Even an ordinary globular protein can assume a rogue guise if conditions are right. Nature. 2001;410:165–166. doi: 10.1038/35065514. [DOI] [PubMed] [Google Scholar]
- 29.Augustine JG, de la Calle A, Knarr G, Buchner J, Frederick CA. The crystal structure of the Fab fragment of the monoclonal antibody MAK33—Implications for folding and interaction with the chaperone BiP. J Biol Chem. 2001;276:3287–3294. doi: 10.1074/jbc.M005221200. [DOI] [PubMed] [Google Scholar]
- 30.Feige MJ, Hagn F, Esser J, Kessler H, Buchner J. Influence of the internal disulfide bridge on the folding pathway of the CL antibody domain. J Mol Biol. 2007;365:1232–1244. doi: 10.1016/j.jmb.2006.10.049. [DOI] [PubMed] [Google Scholar]
- 31.van Rhee F, et al. High serum-free light chain levels and their rapid reduction in response to therapy define an aggressive multiple myeloma subtype with poor prognosis. Blood. 2007;110:827–832. doi: 10.1182/blood-2007-01-067728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goto Y, Hamaguchi K. Unfolding and refolding of the constant fragment of the immunoglobulin light chain. J Mol Biol. 1982;156:891–910. doi: 10.1016/0022-2836(82)90146-2. [DOI] [PubMed] [Google Scholar]
- 33.Pappenberger G, et al. Nonprolyl cis peptide bonds in unfolded proteins cause complex folding kinetics. Nat Struct Biol. 2001;8:452–458. doi: 10.1038/87624. [DOI] [PubMed] [Google Scholar]
- 34.Aurora R, Rose GD. Helix capping. Protein Sci. 1998;7:21–38. doi: 10.1002/pro.5560070103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Qin ZJ, Hu DM, Zhu M, Fink AL. Structural characterization of the partially folded intermediates of an immunoglobulin light chain leading to amyloid fibrillation and amorphous aggregation. Biochemistry. 2007;46:3521–3531. doi: 10.1021/bi061716v. [DOI] [PubMed] [Google Scholar]
- 36.Fernandez-Escamilla AM, Rousseau F, Schymkowitz J, Serrano L. Prediction of sequence-dependent and mutational effects on the aggregation of peptides and proteins. Nat Biotechnol. 2004;22:1302–1306. doi: 10.1038/nbt1012. [DOI] [PubMed] [Google Scholar]
- 37.Smith DP, Jones S, Serpell LC, Sunde M, Radford SE. A systematic investigation into the effect of protein destabilisation on β2-microglobulin amyloid formation. J Mol Biol. 2003;330:943–954. doi: 10.1016/s0022-2836(03)00687-9. [DOI] [PubMed] [Google Scholar]
- 38.Freund C, Honegger A, Hunziker P, Holak TA, Pluckthun A. Folding nuclei of the scFv fragment of an antibody. Biochemistry. 1996;35:8457–8464. doi: 10.1021/bi952764a. [DOI] [PubMed] [Google Scholar]
- 39.Hamill SJ, Steward A, Clarke J. The folding of an immunoglobulin-like Greek key protein is defined by a common-core nucleus and regions constrained by topology. J Mol Biol. 2000;297:165–178. doi: 10.1006/jmbi.2000.3517. [DOI] [PubMed] [Google Scholar]
- 40.Richardson JS, Richardson DC. Natural β-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc Natl Acad Sci USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Monsellier E, Chiti F. Prevention of amyloid-like aggregation as a driving force of protein evolution. EMBO Rep. 2007;8:737–742. doi: 10.1038/sj.embor.7401034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Otzen DE, Kristensen O, Oliveberg M. Designed protein tetramer zipped together with a hydrophobic Alzheimer homology: A structural clue to amyloid assembly. Proc Natl Acad Sci USA. 2000;97:9907–9912. doi: 10.1073/pnas.160086297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ventura S, et al. Short amino acid stretches can mediate amyloid formation in globular proteins: The Src homology 3 (SH3) case. Proc Natl Acad Sci USA. 2004;101:7258–7263. doi: 10.1073/pnas.0308249101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoshino M, et al. Mapping the core of the β2-microglobulin amyloid fibril by H/D exchange. Nat Struct Biol. 2002;9:332–336. doi: 10.1038/nsb792. [DOI] [PubMed] [Google Scholar]
- 45.Liu K, Cho HS, Lashuel HA, Kelly JW, Wemmer DE. A glimpse of a possible amyloidogenic intermediate of transthyretin. Nat Struct Biol. 2000;7:754–757. doi: 10.1038/78980. [DOI] [PubMed] [Google Scholar]
- 46.Chiti F, Stefani M, Taddei N, Ramponi G, Dobson CM. Rationalization of the effects of mutations on peptide and protein aggregation rates. Nature. 2003;424:805–808. doi: 10.1038/nature01891. [DOI] [PubMed] [Google Scholar]
- 47.Han JH, Batey S, Nickson AA, Teichmann SA, Clarke J. The folding and evolution of multidomain proteins. Nat Rev Mol Cell Biol. 2007;8:319–330. doi: 10.1038/nrm2144. [DOI] [PubMed] [Google Scholar]
- 48.Diercks T, Daniels M, Kaptein R. Extended flip-back schemes for sensitivity enhancement in multidimensional HSQC-type out-and-back experiments. J Biomol NMR. 2005;33:243–259. doi: 10.1007/s10858-005-3868-4. [DOI] [PubMed] [Google Scholar]
- 49.Brooks BR, et al. CHARMM—A program for macromolecular energy, minimization, and dynamics calculations. J Comput Chem. 1983;4:187–217. [Google Scholar]
- 50.Lazaridis T, Karplus M. Effective energy function for proteins in solution. Proteins Struct Funct Genet. 1999;35:133–152. doi: 10.1002/(sici)1097-0134(19990501)35:2<133::aid-prot1>3.0.co;2-n. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.