Abstract
We have engineered a mutant of Dictyostelium discoideum (Dicty) myosin II that contains the same fast-reacting “SH1” thiol as in muscle myosin, spin-labeled it, and performed electron paramagnetic resonance (EPR) to compare the structure of the force-generating region of the two myosins. Dicty myosin serves as a model system for muscle myosin because of greater ease of mutagenesis, expression, and crystallization. The catalytic domains of these myosins have nearly identical crystal structures in the apo state, but there are significant differences in ATPase kinetics, and there are no crystal structures of skeletal muscle myosin with bound nucleotides, so another structural technique is needed. Previous EPR studies, with a spin label attached to SH1 in muscle myosin, have resolved the key structural states of this region. Therefore, we have performed identical experiments on both myosins spin-labeled at equivalent sites. Spectra were identical for the two myosins in the apo and ADP-bound states. With bound ADP and phosphate analogs, (i) both proteins exhibit two resolved structural states (prepowerstroke, postpowerstroke) in a single biochemical state (defined by the bound nucleotide), and (ii) these structural states are essentially identical in the two myosins but (iii) are occupied to different extents as a function of the biochemical state. We conclude that (i) myosin structural and biochemical states do not have a one-to-one correspondence, and (ii) Dicty myosin can serve as a good analog for structural studies of muscle myosin only if differences in the coupling between biochemical and structural states are taken into account.
Keywords: Dictyostelium, electron paramagnetic resonance, EPR, site-directed spin labeling, SDSL
Myosin II is a motor enzyme involved in muscle contraction and cell motility through a cycle of actin-activated ATP hydrolysis. The coupling of ATP hydrolysis and force generation is determined by myosin's head domain, subfragment 1 (S1). The myosin ATPase cycle can be described to a first approximation by the following scheme:
where M, M*, and M** are myosin structural states, originally identified by intrinsic fluorescence (1). In this scheme, these structural states are assumed to be tightly coupled to biochemical states, which are defined by the nucleotide bound to the active site. Structural changes of muscle myosin have also been detected by electron paramagnetic resonance (EPR) of a nitroxide spin label (IASL) at SH1 (C707) (2–5), fluorescence of dyes attached to SH1 (6, 7), and fluorescence resonance energy transfer (FRET) between probes at SH1 and other sites (8, 9) or W510 (10). All these studies reflect structural changes of the force-generating region of myosin, which includes the converter domain, N-terminal domain, and the relay and SH1 helices. However, only EPR can resolve and quantify the distinct structural states depicted in the scheme shown above in solution (5).
The only crystal structure of myosin II from skeletal muscle was determined in the apo state (Rayment et al., 1993). Other crystal structures of myosin II, from Dictyostelium discoideum (Dicty), scallop and smooth muscle, have been obtained from myosin trapped in different states with nonhydrolyzable nucleotide analogs. Although there is considerable variation among these structures in the force-generating region, they appear to be divided into two classes, often denoted as postpowerstroke and prepowerstroke, which have been assigned to the M* and M** states. Thus in the scheme shown above, step 3 is the powerstroke (M** to M*) and step 2 is the recovery (repriming) stroke (M* to M**) (11). Crystal structures suggest that in the M*-to-M** transition, a kink is formed in the relay helix, and the converter domain, controlled by the relay helix, dramatically changes its orientation (12–14). These changes in the force-generating region are governed by ATP binding and hydrolysis, and it is accepted that they reflect allosteric effects from the nucleotide-binding site.
However, the correlation between bound nucleotide (biochemical state) and structural state is not entirely consistent. Dicty and smooth muscle myosin complexed with the ADP.BeFx nucleotide analog (12, 15) show a dramatic difference in crystal structure (M** in Dicty, M* in smooth); moreover, the same myosin isoform (Dicty), complexed with the same nucleotide analog (ADP.BeFx), has been observed in both structural states in crystals (12, 16). It is not clear whether these differences reflect intrinsic properties of different myosins or whether they arise from different experimental conditions used during protein preparation and crystal growth. These discrepancies must be resolved by using structural methods that do not require crystallization. This is particularly important because of the frequent usage of Dicty myosin as a model system to study the mechanism of muscle contraction. Compared with muscle myosin, it is easy to express Dicty myosin in large quantities, create truncated versions of the protein to facilitate crystallization, and mutate the protein for structure–function analysis and for site-directed labeling (12, 17–19). Nevertheless, the application of any model system requires validation. The high degree of homology between muscle and Dicty myosin II suggests that the two proteins have similar structures in solution; but direct experimental evidence is needed to test this assumption. The only crystal structure available for skeletal muscle myosin is the apo form (no nucleotide), which is nearly identical to the Dicty structure obtained under similar conditions (14, 20). However, some kinetic properties of Dicty and muscle myosin (based on intrinsic fluorescence, interaction with pyrene-labeled actin and MANT nucleotide) are different: Dicty myosin exhibits slower ATP hydrolysis (21), faster ADP release (21), and faster dissociation of phosphate analogs (22). It is not clear whether the observed differences are due to different kinetic constants or to different structural states in the two proteins. Unfortunately, most studies comparing Dicty and muscle myosins in solution are focused on kinetic rather than structural aspects. Our study provides the missing information, providing high-resolution spectroscopic data in solution that clarifies greatly the extent to which Dicty is (and is not) a valid muscle myosin analog.
The goal of the present study is to determine to what extent the structural dynamics of the force-generating region of Dicty myosin resembles that of muscle myosin in solution, and thus to assess the validity of extending structural information available for Dicty onto muscle myosin. Dicty myosin contains a Thr residue at position 688, which has been shown to be biochemically equivalent to the SH1 site (Cys 707) in skeletal myosin, because the mutation T688C permits Dicty myosin to undergo a similar nucleotide-dependent cross-linking reaction as observed for skeletal myosin (23). Therefore, starting from a Dicty construct lacking reactive Cys (19), we created a T688C mutant, attached a spin label (iodoacetamido-TEMPO, designated IASL) to both myosins, acquired EPR spectra in different biochemical states, and analyzed EPR spectra to determine the restriction of spin-label rotational motion. The resolved EPR spectral components were assigned to structural (conformational) states, allowing us to compare the two myosins directly with respect to the structural states of the force-generating region and the coupling of these structural states to biochemical states. The results not only answer important questions about the similarities and differences of the two myosins but also provide fundamental insights into the structural dynamics of force generation in myosin.
Results
Labeling Efficiency.
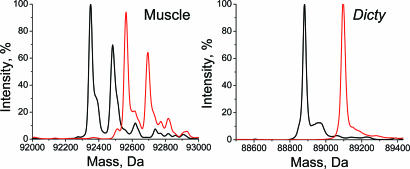
The extent of protein modification by the spin label was determined by mass spectrometry (Fig. 1). The two peaks of skeletal myosin and the single peak of Dicty myosin were each shifted by 213 Da, the mass of the attached spin label. Negligible peak intensity was observed at positions corresponding to unlabeled S1. Control experiments with mixtures showed that unlabeled and labeled proteins (both muscle and Dicty) were detected with similar sensitivity by MS. These results confirm complete labeling of myosin with only one label reacting per protein molecule.
Fig. 1.
MS spectra of unlabeled (black) and IASL labeled (red) S1.
Functional Properties of IASL-S1.
High-salt (Table 1) and actin-activated (Table 2) ATPase activities were measured to evaluate the effect of labeling on functional properties of myosin. The effects of labeling on high-salt ATPase activities are much less dramatic for Dicty than for muscle myosin—the decrease in the K/EDTA activity is less pronounced in Dicty, and the large increase of Ca activity is absent (Table 1).
Table 1.
High-salt ATPase activity
| Myosin | K/EDTA, s−1 |
Ca, s−1 |
||
|---|---|---|---|---|
| Unlabeled | Labeled | Unlabeled | Labeled | |
| Muscle | 8.5 ± 0.5 | 1.4 ± 0.3 | 1.0 ± 0.1 | 11.2 ± 1.3 |
| Dicty | 0.32 ± 0.12 | 0.11 ± 0.09 | 4.2 ± 0.9 | 4.7 ± 2.1 |
Table 2.
Actin-activated ATPase activity
| Myosin | Vbasal†, s−1 |
Vmax‡, s−1 |
Km§, μM |
|||
|---|---|---|---|---|---|---|
| Unlabeled | Labeled | Unlabeled | Labeled | Unlabeled | Labeled | |
| Muscle | 0.10 ± 0.02 | 0.32 ± 0.13 | 13.5 ± 0.5 | 2.4 ± 0.2 | 13.8 ± 2.3 | 7.2 ± 0.3 |
| Dicty | 0.15 ± 0.03 | 0.42 ± 0.05 | 4.9 ± 0.8 | 1.9 ± 0.4 | 12.8 ± 1.8 | 8.8 ± 1.8 |
†In the absence of actin.
‡At saturating actin.
§Actin concentration at which V = 0.5 Vmax.
Under physiological buffer conditions, the ATPase activities of the two myosins are quite similar, especially after spin labeling, which removes all significant differences between the two (Table 2). We confirm previous observations (4, 24) that modification of muscle myosin at the SH1 site alters its ATPase kinetics and show that qualitatively similar effects take place for Dicty myosin, including a substantial decrease in Vmax, the activity at saturating actin (Table 2). At the same time, these results demonstrate unambiguously that the completely labeled proteins (as shown by MS data in Fig. 1) are active and are activated by actin.
Binding of Nucleotide Analogs to IASL-S1.
To ensure that the concentrations of nucleotide analogs (ADP plus phosphate analog,) used in our experiments were saturating and the myosin population was biochemically homogeneous, EPR spectra were recorded at different ligand concentrations. Increasing the concentration >5 mM did not change EPR spectra, indicating that 5 mM is indeed a saturating concentration in all cases. The rate of ATP hydrolysis was measured for these same samples under EPR conditions. Binding of nucleotide analogs abolishes myosin ATPase activity (25). We found that incubation of myosin with 5 mM nucleotide analog decreased the ATPase rate to < 5% of the control value [supporting information (SI) Table S1 and SI Text], confirming that essentially every myosin formed a complex with nucleotide analog.
EPR of IASL-S1 Complexed with Nucleotide Analogs.
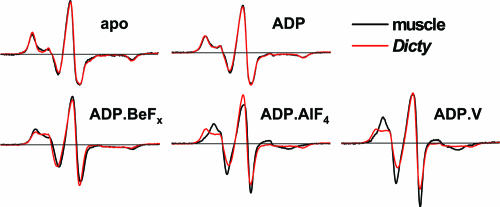
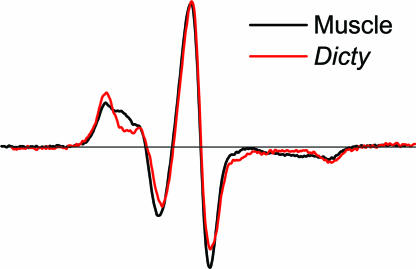
EPR spectra were recorded under conditions (defined above) intended to trap intermediate biochemical states in the ATPase cycle. In the absence of nucleotide (apo state), muscle and Dicty myosin have virtually identical EPR spectra Fig. 2(M), indicating no difference in spin-label dynamics and suggesting structural similarity of the force-generating regions. This agrees with crystallographic studies that show similar atomic structures for these proteins in the apo state (14, 20). The apo spectra are characteristic of strongly immobilized spin labels, having little or no nanosecond mobility relative to the protein, as shown previously for muscle myosin (3). The binding of ADP causes a slight but significant narrowing of the spectrum (Fig. S1), as shown previously for muscle myosin (3), indicating slight mobilization of the label, but there is still no significant difference between muscle and Dicty (Fig. 2, M.ADP). In contrast, there are clear differences between the two myosins in the presence of ADP and phosphate analogs (Fig. 2, bottom row). These differences are especially pronounced in the low-field portion of the spectrum. All of these spectra are significantly narrower than in the presence of ADP, indicating further mobilization of the spin label. The posthydrolysis analogs (ADP.AlF4 and ADP.V) induce greater probe mobility and show a larger difference between Dicty and muscle, compared with the prehydrolysis analog (ADP.BeFx).
Fig. 2.
EPR spectra of muscle and Dicty S1 in different biochemical states.
Quantitative Analysis of EPR Spectra: Resolved Structural States and Their Mole Fractions.
IASL-labeled muscle S1 exhibits single-component X-band EPR spectra in apo and ADP-bound states, and two-component spectra in the nucleotide analog-bound states (3–5). These spectra clearly resolve three structural states, presumably the same states detected by intrinsic fluorescence: M, M*, and M**. These three structural states have been resolved and characterized in terms of spin-label rotational dynamics, based on EPR spectra obtained at both X (9.4 GHz) and W (94 GHz) band (table 4 in ref. 5), reflecting the structure of the force-generating domain of muscle myosin. In the present study, experimental X-band EPR spectra, Vexp(H) of muscle and Dicty myosins were fitted as a linear combination of previously determined spectral components V, V*, and V**, corresponding to structural states M, M*, and M** (5):
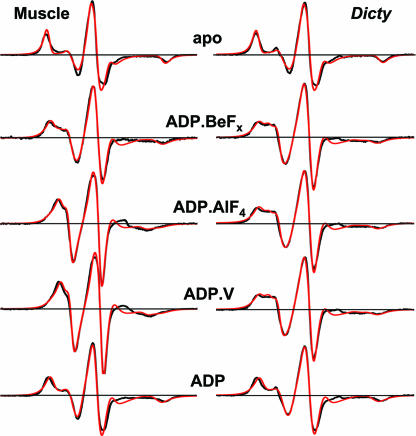
The only variable parameters in the fit were mole fractions (x, x*, and x**), and the quality of the fit was quantified with Pearson's χ2 test. Variation of parameters related to spin-label dynamics (rotational correlation times and order parameters) did not improve the fit. These fits confirmed that muscle and Dicty myosins have (i) a single structural state M (x = 1) in the apo biochemical state and (ii) a single structural state M* (x* = 1) in the ADP-bound biochemical state. In biochemical states containing bound ADP and phosphate analogs, spectra all exhibit two components, a combination of M* and M** structural states, with different mole fractions (x* and x**) for muscle and Dicty myosins (Table 3). Experimental spectra and fits are shown on Fig. 3, and the mole fractions obtained from these fits are given in Table 3.
Table 3.
Mole fractions of structural states derived from EPR spectra (Fig. 3)
| Myosin | apo | ADP | ADP.BeFx | ADP.AlF4 | ADP.V |
|---|---|---|---|---|---|
| muscle | 1, 0, 0 | 0, 1, 0 | 0, 0.84, 0.16 | 0, 0.15, 0.85 | 0, 0.15, 0.85 |
| Dicty | 1, 0, 0 | 0, 1, 0 | 0, 0.66, 0.34 | 0, 0.72, 0.28 | 0, 0.63, 0.37 |
Values are mole fractions (x, x*, x**) of structural states (M, M*, M**). Uncertainties are 0.02–0.04.
Fig. 3.
Computational simulations of EPR spectra, assuming a linear combination of spectra (Eq. 1), corresponding to M, M*, and M** states of myosin. Mole fractions are given in Table 3. Black, experiment; red, best-fit simulation.
These results indicate that when myosin is trapped by ADP and a phosphate analog in a single biochemical state, there remain significant populations of both M* and M** structural states in equilibrium. Thus there is clearly not a one-to-one correspondence between myosin's biochemical and structural states. This is much more clear for Dicty, in which the minor component makes up ≈1/3 of the population in these states, than for muscle, in which the minor component is ≈1/6 (Table 3). We conclude that the coupling between biochemical and structural states (as revealed by the coupling between the nucleotide-binding site and the force-generating region) is different for the two myosins. This difference in coupling is presumably related to the observed differences in ATPase kinetics between muscle and nonmuscle myosins (21, 26–28).
Temperature Dependence and Thermodynamic Analysis.
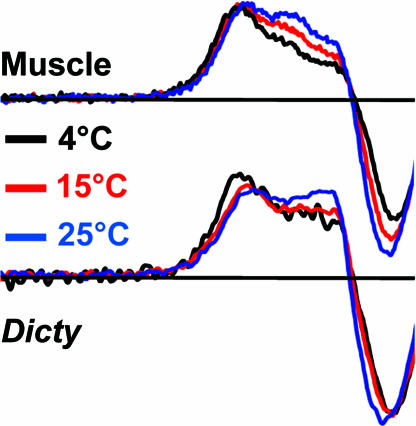
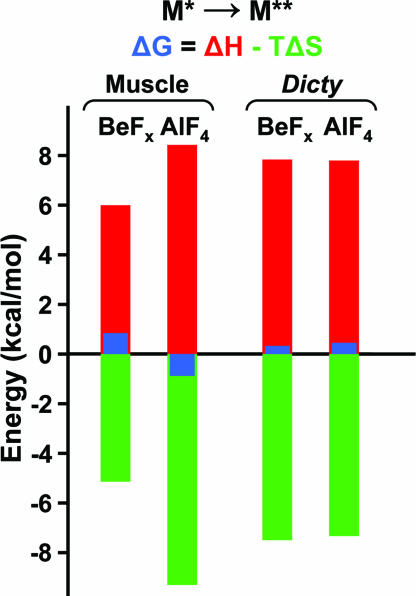
To test the hypothesis that the resolved spectral components observed in EPR experiments represent an equilibrium between structural states, we studied the effect of temperature on the distribution between the states (Fig. 4). It was demonstrated for muscle myosin (24) that an increase in temperature shifts the equilibrium from the prehydrolysis to the posthydrolysis biochemical state. Our EPR experiments revealed a similar temperature dependence on the distribution between structural states, without a change in the biochemical state—the mole fraction x** of the M** population increased with temperature at the expense of the M* population. Fig. 4 demonstrates this effect for S1ADP.BeFx; the effect was similar for S1.ADP.AlF4 (Fig. S2). Thus there is a temperature-dependent equilibrium between the M* and M** structural states within a single biochemical state.
Fig. 4.
Temperature dependence of EPR spectra (low-field region) of myosin with ADP.BeFx bound.
By deconvoluting EPR spectra (Eq. 1), we determined the mole fractions x* and x** of M* and M** states as a function of temperature and calculated the enthalpy and entropy of the M*-to-M** transition from the van't Hoff equation for both myosins (see SI Text, Fig. S2, and Fig. S3). Fig. 5 shows the changes in G, H, and S that occur at 20°C. Although the equilibrium between M* and M** is fairly well balanced (small ΔG in all cases), the values of ΔH and TΔS are large. This suggests that substantial structural changes occur in this transition, but in each case the unfavorable enthalpy change is balanced almost precisely by a favorable entropy change. Also, although the ∣ΔG∣ values are smaller for Dicty (more balanced equilibrium than in muscle), the ∣ΔH∣ and ∣TΔS∣ values are large for both myosins; i.e., even though the two biochemical states are more similar in Dicty than in muscle, the M*-to-M** structural transition is enthalpically and entropically large in both myosins.
Fig. 5.
Thermodynamics of the M*-to-M** transition at 20°C, based on van't Hoff plots (Fig. S3 and Table S2).
Myosin Conformation During Steady-State ATP Hydrolysis.
To confirm that the structural states M* and M**, trapped by phosphate analogs, also exist during the ATPase cycle, as suggested by the scheme shown in the introductory materials, we recorded EPR spectra in the presence of ATP (with addition of 40 mM creatine phosphate and 1 mg/ml creatine phosphokinase). The resulting spectrum for muscle myosin (Fig. 6, black) clearly shows the presence of two structural states M* and M** (69% and 31%, respectively), in good agreement with published results (3). In the case of Dicty myosin, the EPR spectrum (Fig. 6, red) was remarkably similar to the one obtained for the ADP complex, showing (within experimental error, ± 6%) only the M* component. This result is not entirely surprising, because EPR of the ADP.AlF4 and ADP.V complexes (which predominantly generate the M** state in muscle myosin) showed a much larger fraction in the M* state for Dicty. It has been previously shown that the apparent forward rate constant of ATP hydrolysis (the step k2 that populates the M** state in the scheme shown above), is significantly slower in Dicty S1 compared with muscle S1 (21, 26, 29). In addition, the steady-state rate of ATP hydrolysis (k3 in the scheme shown above, because Pi release is rate-limiting) is greater for Dicty myosin (Table 2), again suggesting depopulation of M**, and modification of the SH1 site of muscle myosin shifts the equilibrium toward the M* state (4, 24). It appears that the combination of these factors leads to the domination of the M* conformation during steady-state ATP hydrolysis in Dicty S1.
Fig. 6.
EPR spectra during steady-state ATP hydrolysis.
Discussion
EPR Resolves Structural States.
We have used site-directed spin labeling and EPR to compare the structure of the force-generating region of Dicty and muscle myosins in different states of the ATPase cycle under physiological conditions. Compared with other structural techniques, EPR offers the resolution to distinguish several populations with different structures. This structural resolution has allowed us to determine that myosin can adopt two different conformations (structural states) in the presence of one nucleotide analog (biochemical state). Although this effect is present in both isoforms, it is particularly clear for Dicty myosin (Table 3). Unfortunately, other structural techniques (e.g., crystallography, electron microscopy) tend to trap a single structural state of a protein, or they are not capable of resolving distinct structural populations and report only average properties (e.g., steady-state fluorescence). Time-resolved fluorescence previously resolved three conformations of the lever arm in Dicty myosin (19); the present study resolves three conformations of the force-generating region.
The Force-Generating Region of Muscle and Nonmuscle Myosin: Similar Structures, Different Kinetic Coupling.
Quantitative analysis of EPR spectra reveals one structural state of the force-generating region in apo and ADP-bound biochemical states, and two structural states in the nucleotide analog-bound biochemical states of both muscle and Dicty myosin (Table 3). As in previous fluorescence and EPR studies (1, 3, 7), we assigned the structural state in apo S1 as M, in S1.ADP as M* and two structural states with ADP and phosphate analog bound as M* and M**. As detected by the probe at SH1, the states M, M*, and M** are essentially identical in muscle and Dicty myosins, indicating that the structural states of the force-generating region of muscle and Dicty myosins are quite similar. The two myosins differ significantly in the populations of structural states in identical biochemical states (Table 3), demonstrating differences in kinetic coupling between biochemical and structural states in these myosins.
Thermodynamics of the Power Stroke: Disorder-to-Order Transition.
Thermodynamic analysis shows that the M*-to-M** transition in the two myosins is remarkably similar: Large unfavorable enthalpy and favorable entropy changes balance to produce a small ΔG and, thus, a poised equilibrium that significantly populates both states (Fig. 5). Because the reverse of this transition (M** to M*) probably represents the power stroke, Fig. 5 is the most direct demonstration to date that the structural change occurring in the power stroke involves a small free-energy change, driven by a large negative ΔH but balanced by a large negative ΔS; i.e., the disorder-to-order transition that occurs in the power stroke (30) plays an important thermodynamic role.
Myosin's Biochemical States are Distinct from Structural States.
Our EPR results show that there is not a one-to-one correspondence between myosin's structural and biochemical states (Table 3), indicating that the scheme shown in the introductory materials requires more complexity, as previously suggested from kinetic studies. It has been proposed that myosin isomerization can precede the hydrolysis step, producing both M* and M** structural states with bound ATP (31, 32). Similarly, it has been suggested that M* and M** coexist in the posthydrolysis state (with ADP.P as a ligand) (1, 24):
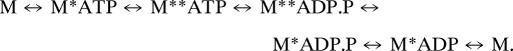 |
In this scheme, M* corresponds to the postpowerstroke (sometimes also designated “near-rigor”) state of myosin, and M** corresponds to the prepowerstroke state detected in crystallography (12, 13, 32). It is widely accepted that the binding of ADP and BeFx traps myosin in the M.ATP biochemical state. According to crystal structures, M.ADP.BeFx can be in either M* or M** structural states (PDB ID codes 1br4, 1kk8, 1mmd), suggesting the possibility of the M**.ATP structural state. This hypothesis is supported by the present study, in which EPR shows clearly that both M* and M** states coexist in the M.ADP.BeFx biochemical state in solution (Table 3). This suggests that ATP binding is sufficient to switch myosin conformation; hydrolysis is not required to energize myosin but is needed to continue the cycle. Similarly, our data demonstrate that both M* and M** structural states coexist in the M.ADP.V and M.ADP.AlF4 biochemical states, supporting the hypothesis that both structural states M* and M** are populated in myosin's posthydrolysis state (M.ADP.P) as well (1, 24), and P remains in the nucleotide-binding pocket during at least part of the powerstroke.
The present study resolves ambiguities raised by crystallographic (12, 15, 33) and fluorescence experiments (34), which suggested the absence of a direct correlation between biochemical and structural states of myosin. The presence of a certain ligand at the active site does not necessarily imply a specific protein structure. This principle was demonstrated most clearly in muscle fibers, where mechanical strain produced direct evidence for kinetically nonequivalent states having the same biochemical ligands (35). The present study shows this to be the case in solution, especially in Dicty myosin. This result emphasizes that caution should be exercised in assigning existing crystal structures to myosin conformations at certain steps of the ATPase cycle and suggests that many of the structural differences observed for different isoforms are likely to be due to different experimental conditions, or to differences in kinetic coupling of structural states, rather than to intrinsic differences in structural states.
Conclusions
Through protein engineering and the high resolution of EPR spectroscopy, we compared directly the structures of muscle and nonmuscle myosins in the force-generating region. We found that these myosins share the same three structural states during the ATPase cycle. However, the structural states are populated differently in the two myosins, suggesting differences in the kinetic coupling of biochemical and structural states. We observed two-component EPR spectra, showing that the biochemical state, defined by the bound nucleotide, does not correspond to a single structural state and reflects, rather, a balanced equilibrium between structural states. We conclude that Dicty myosin can be considered as structurally similar to muscle myosin, when differences in the coupling between biochemical and structural states are taken into account. In both myosins, thermodynamic analysis reveals that the power stroke involves a large favorable enthalpy change that is balanced by a large entropy decrease, supporting the hypothesis that a disorder-to-order transition plays an important role in force generation.
Materials and Methods
Protein Preparation and Labeling.
Actin and myosin were prepared from rabbit skeletal muscle (36), and myosin S1 was prepared and selectively labeled at SH1 (Cys 707) with IASL (5). The T688C mutant of Dictyostelium S1 was constructed and purified as described in (36), except that a mutation was introduced at the T688 residue. The base construct for our studies was a Cys-lite S1 mutant that contains no reactive Cys residues (gift from James Spudich, Stanford University, Stanford, CA). Protein concentrations were determined spectrophotometrically by assuming the following extinction coefficients A280 nm = 0.74 (mg/ml)−1cm−1 for skeletal S1, A280 nm = 0.69 (mg/ml)−1cm−1 for Dicty S1, and A290 nm = 0.63 (mg/ml)−1cm−1 for actin. Dicty S1 was labeled at Cys-688 with IASL by incubating 100 μM S1 with a 4-fold excess of IASL for 12 h on ice. Labeling buffer contained 20 mM EPPS (pH 8.0), 50 mM KCl, 5 mM MgCl2, 1 mM EDTA.
Formation of Stable S1.ADP, S1.ADP.V, S1.ADP.AlF4, and S1.ADP.BeFx Complexes.
Complexes of S1 with nucleotide analogs were obtained by incubation of S1 with 5 mM MgADP (or 5 mM MgADP plus 20 mM NaF in case of MgADP.AlF4, and MgADP.BeFx) for 5 min at 25°C. After that, 5 mM Na3VO4 (or 5 mM AlCl3 or 5 mM BeCl2) were added, and the incubation was continued at 25°C for an additional 20 min.
EPR Spectroscopy.
X-band EPR spectra were acquired with an E-500 spectrometer (Bruker BioSpin) using a SHQ EPR cavity (ER4122 ST). Instrument settings were as follows: incident power 2 mW, modulation amplitude 1 G, modulation frequency 100 kHz, scan width 120 G, time constant 20.5 ms, temperature 20°C. Buffer contained 20 mM EPPS (pH 8.0), 6 mM MgCl2, 1 mM EGTA; during steady-state ATP hydrolysis, 40 mM creatine phosphate and 1 mg/ml (≥150 units/mg) creatine phosphokinase were also present. Before plotting, each spectrum was normalized by dividing by its double integral, and are all displayed at the same aspect ratio (vertical scale relative to horizontal scale).
Mass Spectrometry.
Mass spectrometry used a QSTAR quadrupole-TOF mass spectrometer with an electrospray ionization source. The protein sample (2 mg/ml S1 in 10 mM NH4HCO3 buffer at pH 7.9) was injected into the solvent stream by using a 10-μl injection loop installed in the integrated loop injector. Three to five injections were performed for every sample, with 2-min intervals between them. Data were acquired continuously during load buffer infusion and protein infusions over the range 500–2,000 m/z. Spectra were analyzed with AnalystQS (Applied Biosystems) software.
Supplementary Material
Acknowledgments.
We thank Sarah Blakely, Erin M. Hoffman, Eunice Song, Igor V. Negrashov, and Octavian Cornea for excellent technical assistance. Software was kindly provided by Dr. Z. Liang (Cornell University, Ithaca, NY). This work was supported by National Institutes of Health Grants AR32961 (to D.D.T.) and AR53562 (to Y.E.N.), a grant from the Minnesota Medical Foundation (to Y.E.N.), and the University of Minnesota Supercomputing Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801342105/DCSupplemental.
References
- 1.Bagshaw CR, Trentham DR. The characterization of myosin-product complexes and of product-release steps during the magnesium ion-dependent adenosine triphosphatase reaction. Biochem J. 1974;141:331–349. doi: 10.1042/bj1410331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seidel J, Chopek M, Gergely J. Effect of nucleotides and pyrophosphate on spin labels bound to S1 thiol groups of myosin. Biochemistry. 1970;9:3265–3272. doi: 10.1021/bi00818a021. [DOI] [PubMed] [Google Scholar]
- 3.Barnett VA, Thomas DD. Resolution of conformational states of spin-labeled myosin during steady-state ATP hydrolysis. Biochemistry. 1987;26:314–323. doi: 10.1021/bi00375a044. [DOI] [PubMed] [Google Scholar]
- 4.Ostap EM, White HD, Thomas DD. Transient detection of spin-labeled myosin subfragment 1 conformational states during ATP hydrolysis. Biochemistry. 1993;32:6712–6720. doi: 10.1021/bi00077a026. [DOI] [PubMed] [Google Scholar]
- 5.Nesmelov YE, Agafonov RV, Burr A, Weber RT, Thomas DD. Structure and dynamics of the force generating domain of myosin probed by multifrequency electron paramagnetic resonance. Biophys J. 2008;95:247–256. doi: 10.1529/biophysj.107.124305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Phan BC, Cheung P, Stafford WF, Reisler E. Complexes of myosin subfragment-1 with adenosine diphosphate and phosphate analogs: probes of active site and protein conformation. Biophys Chem. 1996;59:341–349. doi: 10.1016/0301-4622(95)00127-1. [DOI] [PubMed] [Google Scholar]
- 7.Maruta S, et al. Solution structure of myosin-ADP-MgFn ternary complex by fluorescent probes and small-angle synchrotron x-ray scattering. J Biochem. 2000;128:687–694. doi: 10.1093/oxfordjournals.jbchem.a022802. [DOI] [PubMed] [Google Scholar]
- 8.Cheung HC, Gonsoulin F, Garland F. An investigation of the SH1-SH2 and SH1-ATPase distances in myosin subfragment-1 by resonance energy transfer using nanosecond fluorimetry. Biochim Biophys Acta. 1985;832:52–62. doi: 10.1016/0167-4838(85)90173-6. [DOI] [PubMed] [Google Scholar]
- 9.Mizukura Y, Maruta S. Analysis of the conformational change of myosin during ATP hydrolysis using fluorescence resonance energy transfer. J Biochem. 2002;132:471–482. doi: 10.1093/oxfordjournals.jbchem.a003245. [DOI] [PubMed] [Google Scholar]
- 10.Park S, Ajtai K, Burghardt TP. Optical activity of a nucleotide-sensitive tryptophan in myosin subfragment 1 during ATP hydrolysis. Biophys Chem. 1996;63:67–80. doi: 10.1016/s0301-4622(96)02203-x. [DOI] [PubMed] [Google Scholar]
- 11.Koppole S, Smith JC, Fischer S. The structural coupling between ATPase activation and recovery stroke in the myosin II motor. Structure (London) 2007;15:825–837. doi: 10.1016/j.str.2007.06.008. [DOI] [PubMed] [Google Scholar]
- 12.Fisher AJ, et al. X-ray structures of the myosin motor domain of Dictyostelium discoideum complexed with MgADP. BeFx and MgADP.AlF4. Biochemistry. 1995;34:8960–8972. doi: 10.1021/bi00028a004. [DOI] [PubMed] [Google Scholar]
- 13.Smith CA, Rayment I. X-ray structure of the magnesium(II). ADP.vanadate complex of the Dictyostelium discoideum myosin motor domain to 1.9 Å resolution. Biochemistry. 1996;35:5404–5417. doi: 10.1021/bi952633+. [DOI] [PubMed] [Google Scholar]
- 14.Bauer CB, Holden HM, Thoden JB, Smith R, Rayment I. X-ray structures of the apo and MgATP-bound states of Dictyostelium discoideum myosin motor domain. J Biol Chem. 2000;275:38494–38499. doi: 10.1074/jbc.M005585200. [DOI] [PubMed] [Google Scholar]
- 15.Dominguez R, Freyzon Y, Trybus KM, Cohen C. Crystal structure of a vertebrate smooth muscle myosin motor domain and its complex with the essential light chain: Visualization of the pre-power stroke state. Cell. 1998;94:559–571. doi: 10.1016/s0092-8674(00)81598-6. [DOI] [PubMed] [Google Scholar]
- 16.Holmes KC. Muscle contraction. Novartis Found Symp. 1998;213:76–89. discussion 89–92. [PubMed] [Google Scholar]
- 17.Manstein DJ, Titus MA, De Lozanne A, Spudich JA. Gene replacement in Dictyostelium: Generation of myosin null mutants. EMBO J. 1989;8:923–932. doi: 10.1002/j.1460-2075.1989.tb03453.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schroder RR, et al. Three-dimensional atomic model of F-actin decorated with Dictyostelium myosin S1. Nature. 1993;364:171–174. doi: 10.1038/364171a0. [DOI] [PubMed] [Google Scholar]
- 19.Shih WM, Gryczynski Z, Lakowicz JR, Spudich JA. A FRET-based sensor reveals large ATP hydrolysis-induced conformational changes and three distinct states of the molecular motor myosin. Cell. 2000;102:683–694. doi: 10.1016/s0092-8674(00)00090-8. [DOI] [PubMed] [Google Scholar]
- 20.Rayment I, et al. Three-dimensional structure of myosin subfragment-1: A molecular motor. Science. 1993;261:50–58. doi: 10.1126/science.8316857. [DOI] [PubMed] [Google Scholar]
- 21.Ritchie MD, Geeves MA, Woodward SK, Manstein DJ. Kinetic characterization of a cytoplasmic myosin motor domain expressed in Dictyostelium discoideum. Proc Natl Acad Sci USA. 1993;90:8619–8623. doi: 10.1073/pnas.90.18.8619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bobkov AA, Sutoh K, Reisler E. Nucleotide and actin binding properties of the isolated motor domain from Dictyostelium discoideum myosin. J Muscle Res Cell Motil. 1997;18:563–571. doi: 10.1023/a:1018667319386. [DOI] [PubMed] [Google Scholar]
- 23.Liang W, Spudich JA. Nucleotide-dependent conformational change near the fulcrum region in Dictyostelium myosin II. Proc Natl Acad Sci USA. 1998;95:12844–12847. doi: 10.1073/pnas.95.22.12844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sleep JA, Trybus KM, Johnson KA, Taylor EW. Kinetic studies of normal and modified heavy meromyosin and sto even subfragment-1. J Muscle Res Cell Motil. 1981;2:373–399. [Google Scholar]
- 25.Werber MM, Peyser YM, Muhlrad A. Characterization of stable beryllium fluoride, aluminum fluoride, and vanadate containing myosin subfragment 1-nucleotide complexes. Biochemistry. 1992;31:7190–7197. doi: 10.1021/bi00146a023. [DOI] [PubMed] [Google Scholar]
- 26.Woodward SK, Geeves MA, Manstein DJ. Kinetic characterization of the catalytic domain of Dictyostelium discoideum myosin. Biochemistry. 1995;34:16056–16064. doi: 10.1021/bi00049a020. [DOI] [PubMed] [Google Scholar]
- 27.Reynoso JR, Jr, Bobkov A, Muhlrad A, Reisler E. Solution properties of full length and truncated forms of myosin subfragment 1 from Dictyostelium discoideum. J Muscle Res Cell Motil. 2001;22:657–664. doi: 10.1023/a:1016306409345. [DOI] [PubMed] [Google Scholar]
- 28.Van Dijk J, et al. Differences in the ionic interaction of actin with the motor domains of nonmuscle and muscle myosin II. Eur J Biochem. 1999;260:672–683. doi: 10.1046/j.1432-1327.1999.00172.x. [DOI] [PubMed] [Google Scholar]
- 29.Kuhlman PA, Bagshaw CR. ATPase kinetics of the Dictyostelium discoideum myosin II motor domain. J Muscle Res Cell Motil. 1998;19:491–504. doi: 10.1023/a:1005304408812. [DOI] [PubMed] [Google Scholar]
- 30.Thomas DD, Ramachandran S, Roopnarine O, Hayden DW, Ostap EM. The mechanism of force generation in myosin: a disorder-to-order transition, coupled to internal structural changes. Biophys J. 1995;68:135S–141S. [PMC free article] [PubMed] [Google Scholar]
- 31.Urbanke C, Wray J. A fluorescence temperature-jump study of conformational transitions in myosin subfragment 1. Biochem J. 2001;358:165–173. doi: 10.1042/0264-6021:3580165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malnasi-Csizmadia A, Woolley RJ, Bagshaw CR. Resolution of conformational states of Dictyostelium myosin II motor domain using tryptophan (W501) mutants: Implications for the open-closed transition identified by crystallography. Biochemistry. 2000;39:16135–16146. doi: 10.1021/bi001125j. [DOI] [PubMed] [Google Scholar]
- 33.Himmel DM, et al. Crystallographic findings on the internally uncoupled and near-rigor states of myosin: Further insights into the mechanics of the motor. Proc Natl Acad Sci USA. 2002;99:12645–12650. doi: 10.1073/pnas.202476799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Suzuki Y, Yasunaga T, Ohkura R, Wakabayashi T, Sutoh K. Swing of the lever arm of a myosin motor at the isomerization and phosphate-release steps. Nature. 1998;396:380–383. doi: 10.1038/24640. [DOI] [PubMed] [Google Scholar]
- 35.Takagi Y, Shuman H, Goldman YE. Coupling between phosphate release and force generation in muscle actomyosin. Philos Trans R Soc London Ser B. 2004;359:1913–1920. doi: 10.1098/rstb.2004.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Korman VL, Anderson SE, Prochniewicz E, Titus MA, Thomas DD. Structural dynamics of the actin-myosin interface by site-directed spectroscopy. J Mol Biol. 2006;356:1107–1117. doi: 10.1016/j.jmb.2005.10.024. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.