Abstract
The anaphase-promoting complex/cyclosome (APC/C) controls the onset of anaphase by targeting securin for destruction. We report here the identification and characterization of a substrate of APC/C, RCS1, as a mitotic regulator that controls the metaphase-to-anaphase transition. We showed that the levels of RCS1 fluctuate in the cell cycle, peaking in mitosis and dropping drastically as cells exit into G1. Indeed, RCS1 is efficiently ubiquitinated by APC/C in vitro and degraded during mitotic exit in a Cdh1-dependent manner in vivo. APC/C recognizes a unique D-box at the N terminus of RCS1, as mutations of this D-box abolished ubiquitination in vitro and stabilized the mutant protein in vivo. RCS1 controls the timing of the anaphase onset, because the loss of RCS1 resulted in a faster progression from the metaphase to anaphase and accelerated degradation of securin and cyclin B. Biochemically, mitotic RCS1 associates with the NuRD chromatin-remodeling complex, and this RCS1 complex is likely involved in regulating gene expression or chromatin structure, which in turn may control anaphase onset. Our study uncovers a complex regulatory network for the metaphase-to-anaphase transition.
Keywords: HDAC1/2, mitosis, ubiquitin-dependent proteolysis, Cdh1
Ubiquitin-mediated proteolysis regulates key transitions in biology. For example, the anaphase-promoting complex/cyclosome (APC/C) is an essential ubiquitin ligase that controls the ordered degradation of over 20 cell-cycle regulators in mitosis and G1 (1). Degradation of securin by the APC/C at the onset of anaphase allows activation of separase and cleavage of the cohesin complex, leading to segregation of sister chromatids. Similarly, APC/C-dependent destruction of cyclin B at anaphase inactivates the cyclin B/Cdk1 kinase and results in mitotic exit. At telophase and cytokinesis, APC/C controls the degradation of a group of cell-cycle proteins, such as Aurora A/B, Plk1, TPX2, CKAP2 and anillin (2–5), to ensure the ordered mitotic exit and cytokinesis. Beyond mitosis, APC/C is also involved in the G1/S transition and DNA replication.
The ordered destruction of APC/C substrates is at least in part controlled by the association of APC/C with its regulators, Cdc20 and Cdh1 (1). Cdc20 binds to APC/C in mitosis and activates its ligase activity at the anaphase onset to trigger the degradation of securin, cyclin B, and Xkid, whereas Cdh1 is required for the activation of APC/C during mitotic exit and in G1. Cdc20 and Cdh1 recognize two motifs in the substrates: the destruction box (D-box; RxxL) and the KEN box.
The temporal control of APC/C activation, and hence the anaphase onset, is under complex regulation (1). Upon mitotic entry, APC/C is phosphorylated by mitotic kinases, such as Plk1, which is necessary but not sufficient for its activation. On the other hand, the spindle checkpoint, which delays the anaphase onset until all of the chromosomes are aligned at the metaphase plate, prevents the activation of APC/C through association of inhibitory checkpoint proteins Mad2 and BubR1 with APC/C-Cdc20 (6). This checkpoint mechanism ensures the fidelity of chromosome segregation and determines the timing of anaphase onset.
Given the central role of the APC/C in mitotic regulation, it is essential to identify its substrates to gain a mechanistic understanding of its function in the cell cycle. We report here the identification of a substrate of APC/C through a powerful functional genomic analysis (5) and the characterization of the physiological function of this substrate at the metaphase-to-anaphase transition.
Results
Functional Genomic Analyses Identified Substrates of APC/C.
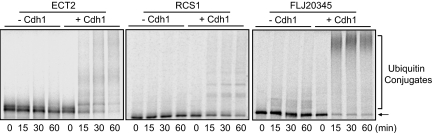
To develop a systematic approach for the identification of APC/C substrates (5), we analyzed 16 known substrates of the APC/C that have been characterized at the onset of this study. These substrates included cyclin A, cyclin B, Plk1, Aurora A, Aurora B, Nek2, Cdc20, UbcH10, securin, GTSE1, anillin, TPX2, Cdc6, geminin, ribonucleotide reductase M2, and SnoN (1–4, 7, 8). The first 15 substrates are cell-cycle regulators (1), whereas the last one, SnoN, acts in cell signaling (7). Genes involved in cell cycle regulation have been shown to covary transcriptionally during tumorigenesis, as these regulators are under selective pressures to act coordinately as one module for cell proliferation (9, 10). Indeed, the expression of the first 15 cell-cycle substrates of the APC/C covaries transcriptionally across hundreds of tumor tissues/cell lines (10), whereas the expression of SnoN does not (9, 10). Furthermore, of the 15 cell cycle substrates, the first 12 act in mitosis and cytokinesis (1–4, 7), and all 12 genes are transcriptionally induced in G2 or in mitosis (11). We concluded that a common theme for cell cycle substrates is their transcriptional covariation with each other across hundreds of tumor tissues. Substrates that only function in mitosis and cytokinesis are likely to be induced in G2 or in mitosis. Thus, we searched published microarray databases for genes that fulfill these two criteria (10, 11) and then tested the corresponding proteins for their ability to be ubiquitinated by the active APC/C-Cdh1 in a reconstituted assay (4, 12). From these experiments, we identified eight new substrates of the APC/C: ECT2, KIF2C/MCAK, KIFC1/HSET, MELK, MKLP2, Par-1a/MARK3, and two proteins, FLJ20354 and FLJ10156, all of which are efficiently ubiquitinated by the APC/C in a Cdh1-dependent manner [Fig. 1 and supporting information (SI) Fig. S1]. We named FLJ10156 as a regulator of chromosome segregation (RCS1) and reported here the characterization of FLJ10156 as a substrate of APC/C that controls the metaphase-to-anaphase transition.
Fig. 1.
Substrates are specifically ubiquitinated by APC/C-Cdh1 in vitro. iAPC/C was immunopurified from Xenopus interphase extracts and activated by in vitro translated Cdh1. APC/C-dependent ubiquitination of 35S-labeled substrates was analyzed in the presence of E1, E2, ubiquitin, ubiquitin-aldehyde, and ATP at indicated times. The arrow points to the input substrates, and the bracket indicates the position of ubiquitin conjugates. The extent of ubiquitination can be judged by the disappearance of the input substrates and by the formation of ubiquitin conjugates migrating slower than the input substrates.
RCS1 Is a Substrate of the APC/C in Vitro.
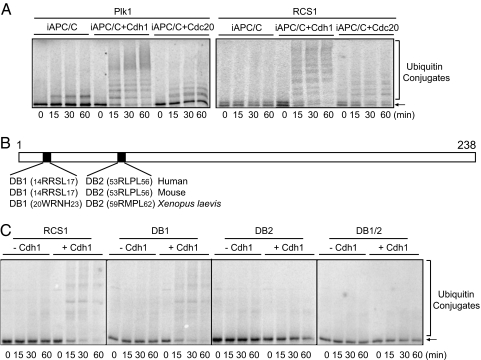
Two activators, Cdc20 and Cdh1, control the activity of APC/C in anaphase and from mitotic exit to G1, respectively (1, 7). In vitro ubiquitination assay indicated that RCS1 was ubiquitinated by both APC/C-Cdc20 and APC/C-Cdh1 (Fig. 2A), similar to securin and cyclin B (data not shown), whereas substrates degraded at the M-to-G1 transition, such as Plk1, were efficiently ubiquitinated by APC/C-Cdh1, but weakly by APC/C-Cdc20 (Fig. 2A).
Fig. 2.
RCS1 is specifically ubiquitinated by APC/C-Cdh1 and APC/C-Cdc20 in a D-box-dependent manner. (A) Ubiquitination assay for Plk1 and RCS1 by APC/C-Cdh1 and APC/C-Cdc20 as described in Fig. 1. (B) Schematic representation of RCS1 depicting two D-box sequences. (C) Ubiquitination assay for wild-type and mutant RCS1 by APC/C-Cdh1 as described in Fig. 1.
RCS1 contains two putative D-boxes at amino acids 14–17 and 53–56, but no KEN box (Fig. 2B). The second D-box is conserved among human, mouse, and Xenopus RCS1, whereas the first one is not. Mutation of the first D-box (DB1) (RxxL to AxxA) did not or only weakly affected the ubiquitination efficiency, whereas mutation of the second D-box (DB2) greatly reduced its ubiquitination by the APC/C-Cdh1 (Fig. 2C). Mutations of both D-boxes (DB1/2) abolished the ubiquitination of RCS1. Thus, APC/C-Cdh1 predominantly recognizes RCS1 through DB2, although DB1 also contributes to this recognition.
RCS1 Is a Substrate of the APC/C-Cdh1 in Vivo.
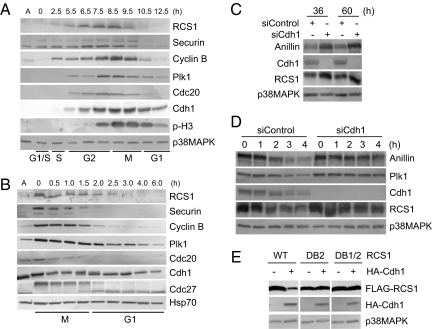
To determine whether RCS1 is a substrate of the APC/C-Cdh1 in vivo, we first analyzed the levels of the RCS1 protein in the cell cycle. HeLa S3 cells were arrested at the G1/S transition by a double-thymidine block and then released to allow synchronous progression through S, G2, and M. The cell cycle profile was determined by FACS, and the mitotic time points were identified by the presence of phospho-histone H3 (Fig. 3A and Fig. S2A). The levels of RCS1 accumulated in S and G2 peaked in mitosis and then dropped drastically as cells exited from mitosis into G1 (Fig. 3A). This profile of RCS1 in the cell cycle is similar to those of known substrates of the APC/C, such as cyclin B, securin, Plk1, and Cdc20 (Fig. 3A), suggesting a regulation of RCS1 protein stability during mitotic exit. The level of RCS1 was reduced before that of Cdh1 (Fig. 3A), which occurs in G1. To determine the exact cell-cycle stage for RCS1 destruction, HeLa S3 cells were synchronized at prometaphase by a thymidine-nocodazole block and then released into fresh media (Fig. 3B and Fig. S2B). As expected, RCS1 levels were down-regulated during mitotic exit, and the bulk of RCS1 was degraded after the destruction of securin at anaphase but before the degradation of Plk1 in G1.
Fig. 3.
RCS1 is degraded in a Cdh1-dependent manner in vivo. (A and B) HeLa S3 cells were synchronized at the G1/S boundary by a double-thymidine arrest (A) or at prometaphase by a thymidine-nocodazole arrest (B). Cells were then released into fresh media and harvested at the indicated times. Levels of RCS1, securin, cyclin B, Plk1, Cdc20, Cdh1, Cdc27, phospho-histone H3 (Ser-10) (p-H3), p38MAPK (a loading control), and Hsp70 (a loading control) were analyzed by Western blotting. A, asynchronous cells. (C) HeLa cells were control-transfected or transfected with an siRNA against Cdh1 and harvested at the indicated times after transfection. Levels of anillin, Cdh1, RCS1, and p38MAPK were determined by Western blot analysis of total cell lysates. (D) HeLa cells were control-transfected or transfected with an siRNA against Cdh1 and arrested at prometaphase by incubating with nocodazole for 20 h. Cells were then released into fresh media in the presence of 100 μg/ml cycloheximide and harvested at the indicated times after release. Levels of anillin, Plk1, Cdh1, RCS1, and p38MAPK were determined by Western blot analysis. (E) FLAG-RCS1, FLAG-RCS1-DB2, or FLAG-RCS1-DB1/2 were transfected into HeLa cells with HA or HA-Cdh1. Levels of FLAG-RCS1, HA-Cdh1, and p38MAKP were determined by Western blot analysis.
To confirm that the APC/C-Cdh1 is required for the destruction of RCS1, we inhibited the activity of the APC/C-Cdh1 by RNAi. HeLa cells were transfected with an siRNA targeted against Cdh1 or control-transfected. Transfection of Cdh1-siRNA but not control-transfection reduced the Cdh1 protein levels and resulted in an increase in endogenous RCS1 and anillin, a known APC/C substrate (Fig. 3C) (4). To demonstrate that this increase is directly because of the stabilization of the RCS1 protein rather than indirectly through modulating RCS1 synthesis, we measured the half-life of RCS1 in knockdown cells. Transfected cells were arrested at prometaphase by nocodazole and then released into fresh media in the presence of cycloheximide, an inhibitor of protein synthesis. The kinetics of RCS1 degradation were much faster in mock-transfected cells than that in Cdh1-depleted cells (Fig. 3D). Similarly, the half-life of anillin was also increased in Cdh1-knockdown cells. Thus, Cdh1 controls the half-life of the RCS1 protein during mitotic exit. Even though RCS1 is ubiquitinated by APC/C-Cdc20 in vitro, this form of APC/C is not sufficient for complete degradation of RCS1 in vivo.
Because the second D-box is required for in vitro ubiquitination of RCS1 by the APC/C (Fig. 2C), we determined whether this D-box is recognized during the degradation of RCS1 in vivo. FLAG-tagged wild-type or DB2 RCS1 were cotransfected with a vector or HA-Cdh1 into HeLa cells. Overexpression of Cdh1, which hyperactivated the APC/C, reduced the levels of the wild-type RCS1 (Fig. 3E), confirming the degradation of RCS1 by the APC/C pathway in vivo. Interestingly, overexpression of Cdh1 did not affect the level of RCS1-DB2 or RCS1-DB1/2, indicating that the DB2 is important for RCS1 turnover in vivo, consistent with the in vitro ubiquitination results (Fig. 2C).
RCS1 Controls the Metaphase-to-Anaphase Transition.
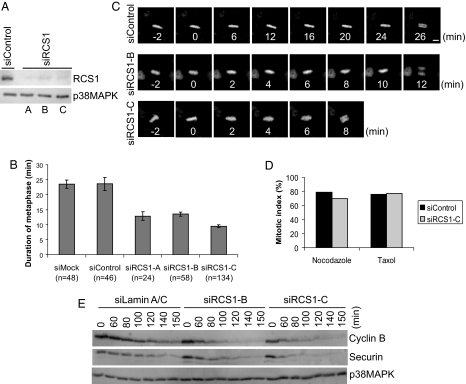
The cell cycle profile of the RCS1 protein suggests a function in mitosis. Thus, we tested whether the depletion of RCS1 leads to defects in mitosis. Three siRNAs targeting to different regions of the RCS1 gene all resulted in efficient depletion of the endogenous RCS1 protein (Fig. 4A), and yet no global defects in mitosis were observable: The mitotic index was not altered in knockdown cells, and spindle assembly, chromosome congression/segregation, and cytokinesis all proceeded normally as analyzed by immunofluorescence staining of various cellular markers as well as by time lapse imaging (data not shown).
Fig. 4.
RCS1 controls the metaphase-to-anaphase transition. (A) Immunoblot analysis of RCS1-knockdown cells. HeLa cells were control-transfected or transfected with three siRNAs (A–C) against RCS1, synchronized by a thymidine-nocodazole arrest, and harvested as prometaphase cells at 36 h after transfection. Levels of RCS1 and p38MAPK were determined by Western blot analysis. (B) The duration (min) of metaphase in control and RCS1 knockdown cells was determined by analysis of timelapse imaging. Data represent the average duration from the time when all of the chromosomes were aligned at the metaphase plate to the onset of anaphase. Number of cells in each experiment is indicated. Error bars represent SEM. (C) Selected frames from timelapse movies of representative HeLa/GFP-Histone H2B cells control-transfected or transfected with siRCS1-B and siRCS1-C. GFP-H2B images are shown. (Scale bar, 5 μm.) (D) HeLa cells were either mock-transfected or transfected with siRCS1-C, followed by a thymidine-nocodazole or thymidine-taxol arrest. Mitotic index was determined by FACS analysis using a monoclonal antibody (MPM2) specific for mitotic phospho-proteins. (E) HeLa S3 cells were transfected with siControl, siRCS1-B, or siRCS1-C and synchronized at prometaphase by a thymidine-nocodazole arrest. Mitotic cells were collected by shake-off, released into fresh media, and then harvested at the indicated times after release. Levels of cyclin B, securin, and p38MAPK were determined by Western blot analysis.
Interestingly, time lapse analysis of mitotic progression in HeLa cells stably expressing GFP-histone H2B indicated that knockdown of RCS1 resulted in a faster metaphase-to-anaphase transition, a phenotype consistently generated by all three siRNAs, thereby confirming the specificity of phenotype (Fig. 4B). Whereas control cells stayed at metaphase for an average of 23.5 ± 2 min between the congression of the last chromosome and the onset of anaphase, knockdown of RCS1 by three siRNAs resulted in an average duration of metaphase for 12.9 ± 1.4 min, 13.5 ± 0.7 min, and 9.5 ± 0.4 min, respectively. This shortened mitosis in knockdown cells is specific to metaphase, as the average duration from the nuclear envelop breakdown to metaphase remained statistically unchanged between control- and RCS1-knockdown cells (10.1 ± 2.7 min for control cells and 10.1 ± 1.9 min, 9.9 ± 2.1 min, and 10.7 ± 2.7 min for three siRNAs, respectively). Although the metaphase is shortened in knockdown cells, anaphase initiated only after the successful congression of all chromosomes (Fig. 4C and data not shown). Furthermore, once chromosomes were segregated at anaphase in knockdown cells, two sets of chromosomes migrating synchronously toward opposite spindle poles and no lagging chromosome were detected in knockdown cells (Fig. 4C). This faster metaphase-to-anaphase transition is not because of the inactivation of the spindle checkpoint, as RCS1-depleted cells remained stably arrested at mitosis in the presence of either nocodazole or taxol, even after prolonged drug treatment (Fig. 4D and data not shown), indicating that both the attachment and the tension branches of the spindle checkpoint are functional in knockdown cells. Furthermore, control and RCS1-depleted cells responded similarly to monastrol, an inhibitor of Eg5, and to nocodazole at doses that partially activated the spindle checkpoint (Fig. S3 A–D), indicating that the shorter duration of metaphase in RCS1-depleted cells did not result from a weakened checkpoint response.
Biochemical experiments in knockdown cells independently confirmed the results from live cell analysis. Depletion of RCS1 resulted in a faster anaphase initiation as indicated by an accelerated kinetics for the degradation of APC/C substrates, such as securin and cyclin B, on release from a prometaphase arrest generated by nocodazole (Fig. 4E). Interestingly, the acceleration of anaphase onset in RCS1-depleted cells was greater on release from nocodazole (Fig. 4E) than that in unperturbed mitosis (Fig. 4B), suggesting that RCS1 plays a more important role in determining the kinetics of mitotic progression when microtubules are polymerized de novo in mitosis.
One possible explanation for the RCS1 knockdown phenotype is that the loss of RCS1, an APC/C-Cdc20 substrate, may accelerate the kinetics for the degradation of other substrates such as securin and cyclin B because of a lack of substrate competition, leading to an earlier anaphase onset. However, in an in vitro APC/C-Cdc20 ubiquitination assay, the presence of an equal molar amount of recombinant securin greatly reduced the levels of RCS1 ubiquitination, but not vice versa, indicating that APC/C-Cdc20 preferentially ubiquitinates securin (Fig. S3E). Thus, the accelerated anaphase onset in RCS1-knockdown cells does not simply result from RCS1 acting as a competitive inhibitor of APC/C in the wild-type cells. We conclude that RCS1 controls the metaphase-to-anaphase transition.
Expression of a Nondegradable RCS1 Does Not Affect M-Phase Progression.
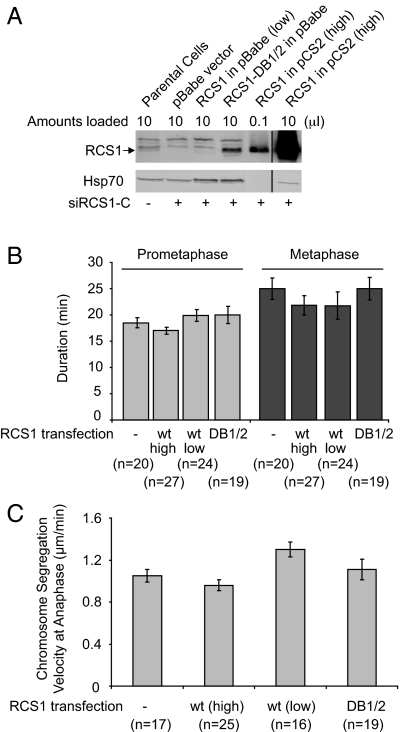
To analyze the functional requirement for degradation of RCS1 in mitosis, we expressed RCS1-DB1/2 mutant in HeLa cells at five times above the endogenous RCS1 level (Fig. 5). Expression of RCS1-DB1/2 did not affect the kinetics of prometaphase or metaphase progression, nor did it alter the rate of chromosome segregation at anaphase. Furthermore, the presence of RCS1-DB1/2 did not affect cytokinesis based on live cell imaging under DIC (data not shown). Similar, expression of wild-type RCS1 in a range from 0.3- (low) to over 100-fold (high) of the endogenous RCS1 did not change the kinetics of mitotic progression (Fig. 5). We conclude that degradation of RCS1 is not essential for normal mitotic progression within a single cell cycle.
Fig. 5.
Expression of a nondegradable RCS1 mutant does not affect M-phase progression. (A) Determination of the levels of ectopically expressed RCS1 and RCS1-DB1/2. HeLa cells were transfected with siRCS1-C to deplete endogenous RCS1 and then transfected with RCS1 or RCS1-DB1/2. Levels of RCS1 and Hsp70 were assayed by Western blotting. The pBabe vector drives the expression of RCS1 and RCS1-DB1/2 at relatively low levels with a weak LTR promoter, whereas pCS2 drives the expression of RCS1 at a high level with a strong CMV promoter. An anti-RCS1 antibody different from the one in Figs. 3 and 4 was used here, and bands above RCS1 correspond to cross-reacting bands. The vertical lines in the figure indicate where intervening (irrelevant) lanes had been removed from the gel images. (B and C) The pBabe vector, RCS1-DB1/2 in pBabe, RCS1 in pBabe, and RCS1 in pCS2 were cotransfected with a RFP vector into HeLa/GFP-H2B cells, and GFP-H2B was imaged at 44 h after transfection. As fusion of a GFP tag to either the N or C terminus of RCS1 interferes with its degradation (data not shown), cotransfection of RFP, which accounts for 5% of transfected DNA, was used here to identify transfected cells. The durations (min) of prometaphase and metaphase (B) as well as the rate of chromosome segregation at anaphase (C) in transfected RFP-positive cells were determined by analysis of timelapse images. The number of cells in each experiment is indicated. As depletion of RCS1 affects the kinetics of the metaphase-to-anaphase transition, experiments described here were done in the presence of endogenous wild-type RCS1. Error bars, SEM.
RCS1 Associates with the NuRD Complex During Mitosis.
To understand the biochemical mechanisms underlying the mitotic functions of RCS1, we identified cell-cycle regulators that function in the same pathway. A HeLa S3 cell line stably expressing tandem-tagged RCS1 was established. Proteins associated with RCS1 were tandem affinity-purified from cells arrested at prometaphase and analyzed by mass spectrometry (13) (Fig. S4A). RCS1 was found to associate with the subunits of the NuRD complex, an ATP-dependent chromatin-remodeling complex implicated in transcriptional regulation (14). The NuRD complex consists of Mi2, histone deacetylases 1 and 2 (HDAC1/2), RbAp46/48, MBP3, and MTA2. We identified by mass spectrometry MTA2, HDAC1/2, and RbAp46/48 as RCS1-interacting proteins, all with high percentages of sequence coverage (Fig. S4B).
We next investigated whether endogenous RCS1 interacts with the subunits of the NuRD complex in mitosis. HeLa S3 cells were synchronized by a thymidine-nocodazole block and then released from prometaphase into G1 (12). The levels of MTA2 and HDAC2 were constant from mitosis to early G1 (Fig. S4C). Next, RCS1 was immunoprecipitated and the immunocomplexes analyzed by Western blotting. Both MTA2 and HDAC2 coprecipitated with RCS1 in mitosis, even though they did not form a stoichiometric complex with RCS1 (Fig. S4C and data not shown). This association was strongest at prometaphase (0 h after release), decreased as the cell progressed through metaphase and anaphase (1–1.5 h after release), and was not detectable in G1 cells and in asynchronous cells. Thus, endogenous RCS1 forms a complex with MTA2 and HDAC2 in mitosis.
It has been reported that the histones H3 and H4 are globally hypoacetylated during mitosis and that their acetylation becomes increased as cells exit from mitosis (15). Furthermore, treatments with HDAC inhibitors lead to mitotic defects such as chromosome bridges, breaks, and mis-segregation (16). Our finding on the role of RCS1 in the metaphase-to-anaphase transition and on its association with the HDAC1/2 complex during mitosis prompted us to investigate a potential role for RCS1 in mitotic chromatin structure by determining the levels of histone acetylation during mitosis in RCS1-depleted cells. Consistent with previous findings, histones H3 and H4 were only hypoacetylated in nocodazole-treated cells when analyzed with antibodies against histone H3 acetylated on Lys 9 and 14 and against histone H4 acetylated on Lys 5, 8, 12, and 16 (Fig. S4D). However, we did not observe an obvious change in the levels of global histone deacetylation in RCS1-depleted mitotic cells vs. control mitotic cells (Fig. S4D). Thus, RCS1 does not seem to regulate the global acetylation levels for Histone H3 and H4 during mitosis, although we cannot exclude the possibility that RCS1 only controls the histone deacetylation on a particular lysine residue in the histone octomer complex associated with local chromatin structures, such as centromeres.
The RCS1-NuRD complex may be involved in regulating the expression of cell cycle genes, which in turn control the kinetics of mitotic progression. Western blot analysis in synchronized cells indicated that depletion of RCS1 did not alter the expression of spindle checkpoint proteins, such as Bub1, Mps1, BubR1, Bub3, Mad1, and Mad2 or G2/M regulators, such as Plk1, Aurora A, cyclin B, and Wee1 (Fig. S5). Similarly, overexpression of RCS1, either the wild-type protein or the DB1/2 mutant, did not altered the levels of cell-cycle or checkpoint proteins, such as Mps1, Mad1, Bub3, and Wee1 (unpublished results).
Discussion
Through an analysis of the gene expression profile common for substrates of APC/C, we identified a transcriptional covariation pattern for known cell cycle substrates during tumorigenesis (5). Furthermore, we found that M-phase substrates are usually induced in G2 or early mitosis before its function and that subsequent degradation of these proteins by APC/C at the M-to-G1 transition restricts their function to M-phase. Thus, we searched for potential mitotic substrates of APC/C that fulfill these two criteria and identified eight previously unrecognized substrates. These substrates have diverse function in M-phase, ranging from mitotic entry (MELK), spindle dynamics and function (MCAK and HSET), and cytokinesis (ECT2 and MKLP2) to cell polarity and neurodegeneration (Par-1a/MARK3). We also identified two functionally uncharacterized proteins, FLJ20354 and RCS1. FLJ20354, a protein of unknown function with a DEP-domain and a RhoGAP-like domain, is a spindle-pole-associated protein (unpublished data). We reported here that RCS1 is a substrate of APC/C involved in regulating the metaphase-to-anaphase transition. RCS1 is degraded during mitotic exit and destruction of RCS1 depends on its D-box 2 (DB2), both in vitro and in vivo.
RCS1 controls the kinetics of the metaphase-to-anaphase transition, as depletion of RCS1 shortened the duration of metaphase by 40–50%. Cell biological studies with specific anti-RCS1 antibodies as well as with RCS1-GFP-stable cell lines indicated that RCS1 is diffusively distributed throughout mitotic cells without specific association with chromosomes, kinetochores, spindle, or centrosomes (unpublished results), indicating that RCS1 acts in cytosol to control the anaphase onset. Biochemically, the timing of the metaphase-to-anaphase transition is controlled by a series of reactions. At prometaphase, the spindle checkpoint proteins BubR1 and Mad2 specifically bind to and inhibit APC/C-Cdc20 to prevent premature separation of sister chromatids. Upon the attachment of microtubules to every kinetochore and the establishment of tension across sister kinetochores, the checkpoint signal is off and active APC/C degrades securin, leading to the activation of separase, which then cleaves the cohesin complex in inner centrosomeres and initiates anaphase (1). In addition, complete chromosome separation also requires active topoisomerase II to resolve DNA catenation between sister chromatin, a process regulated by an unknown mechanism (17). RCS1 may control one of these biochemical steps. We noted that the RCS1 regulatory circuit is unique to somatic cells, as Xenopus RCS1 is not expressed in egg extracts (H. Du and G.F., unpublished results).
In contrast to the checkpoint proteins BubR1 and Mad2, RCS1 neither functions as a stoichiometric inhibitor of APC/C nor is required for the inhibition of APC/C by checkpoint proteins (unpublished results). Furthermore, RCS1 does not affect the kinetochore localization of the spindle-checkpoint proteins, such as BubR1 and Mad2 (unpublished results). Indeed, the spindle checkpoint is fully established and maintained in the absence of RCS1, and depletion of RCS1 does not cause a slippage of the checkpoint. As RCS1-depleted cells degraded securin faster than control cells on release from nocodazole, RCS1, either directly or indirectly, regulates the kinetics of APC/C activation after the disappearance of spindle-checkpoint signals. Although RCS1 is a substrate of APC/C, endogenous RCS1 does not act as a competitive inhibitor against degradation of securin, as securin competitively inhibits the ubiquitination of RCS1, but not vice versa, in the in vitro ubiquitination assay. Consistent with this, RCS1 is degraded during mitotic exit, whereas securin is degraded at the onset of anaphase in vivo. The exact molecular mechanism of RCS1 function in regulating the kinetics of APC/C activation remains to be determined.
Because RCS1 specifically interacts with the NuRD complex during mitosis, RCS1 may control the anaphase onset through regulating gene expression, although the targets of this regulation remain to be characterized. As the kinetics of anaphase onset are determined by the spindle checkpoint, checkpoint proteins are logical targets. However, we did not observe any detectable change in the steady-state levels of Mad1, Mad2, BubR1, Bub1, Bub3, and Mps1 in RCS1-depleted cells. Alternatively, RCS1 may control the chromatin structure through the NuRD complex, but the exact nature of this regulation remains to be determined. The identification of RCS1 as a regulator of sister-chromatid segregation and the demonstration of its association with the HDAC1/2 complex uncover a complex regulatory network that controls the anaphase onset, a process essential for the mitotic fidelity and genomic stability.
Materials and Methods
Recombinant Proteins and Antibodies.
Recombinant RCS1-His6 was expressed in Sf9 cells and recombinant His6-securin was expressed in Escherichia coli. Recombinant RCS1 protein fragments were expressed in E. coli and used for the production of anti-RCS1 sera in rabbits. Anti-anillin and anti-Mad2 antibodies were described (4, 18). The following antibodies were from commercial sources: anti-FLAG antibody (Sigma), anti-HA antibody (Covance), anti-securin (Zymed), anti-Cdh1 (Lab Vision), anti-phospho-Ser/Thr-Pro (MPM-2), anti-Histone H3, anti-phospho-Histone H3 (Ser-10), anti-acetyl-Histone H3, anti-Histone H4 and anti-acetyl-Histone H4 (Upstate), anti-Bub1, BubR1, and Mps1 (Chemicon), anti-Cdc20, anti-Cdc27, anti-cyclin B1, anti-Aurora A, anti-HDAC2, anti-MTA2, anti-Plk1, anti-Hsp70, and anti-p38MAPK antibodies (Santa Cruz Biotechnology).
In Vitro Ubiquitination Assays.
Interphase extracts were made from Xenopus eggs as described in ref. 12. Extracts were immunoprecipitated with anti-Cdc27 antibody/protein A beads for 2 h at 4°C to purify interphase APC/C (iAPC/C). The iAPC/C beads were collected by centrifugation and washed five times in the XB buffer [10 mM Hepes-KOH (pH 7.8) 100 mM KCl, 1 mM MgCl2, 0.1 mM CaCl2, and 50 mM sucrose] containing 500 mM KCl and 0.5% Nonidet P-40 and two times in XB buffer. Purified iAPC/C beads were then incubated with in vitro translated Cdh1 or Cdc20 for 1 h at 25°C and washed twice with XB buffer. Ubiquitination reactions were initiated by mixing 35S-labeled substrate with E1 (50 μg/ml), E2 (50 μg/ml), ubiquitin (1.25 mg/ml), ubiquitin aldehyde (1 μM), and an energy regeneration mix (4, 12). Reactions were performed at 25°C and stopped at various times by the addition of the SDS sample buffer. Samples from each time point were then analyzed by SDS/PAGE and a PhosphorImager (Molecular Dynamics).
Cell Culture, Transfection, and Time Lapse Microscopy.
To determine the levels of the RCS1 protein across the cell cycle, cells were synchronized at the G1/S boundary by a double-thymidine treatment (an 18-h thymidine arrest and a 9-h release, followed by an 18-h thymidine arrest) and then released (12). Alternatively, cells were synchronized at prometaphase by a thymidine-nocodazole arrest (an 18-h thymidine arrest and a 5-h release, followed a 14-h nocodazole arrest) (12).
DNA transfection in Figs. 3E and 5 was carried out with Lipofectamine 2000 (Invitrogen) and Effectene (Qiagen), respectively. siRNAs against RCS1 and Cdh1 were synthesized by Dharmacon, Inc. Three RNAs targeting RCS1 all gave efficient knockdown and similar phenotypes. The target sequences against RCS1 are (in the sense orientation): 5′-CATGTGACTTTCCTAGATA-3′ (sequence A), 5′-CTTAGAAACTCATCGTACA-3′ (sequence B), and 5′-ACACTGCTTTCTCATAATA-3′ (sequence C), and the target sequence against Cdh1 is 5′-GCAACGATGTGTCTCCCTA-3′. Control RNA is the nontargeting siRNA #1 (cat no.: D-001210-01, Dharmacon). RNAs were transfected into HeLa cells by using DharmaFECT 1 reagent (Dharmacon).
HeLa cells stably expressing GFP-histone H2B were cultured in Leibovitz's L-15 medium (Invitrogen) supplemented with 10% FBS (Invitrogen) and 2 mM l-glutamine (Invitrogen). Cells were placed in a 37°C heated microscope chamber and observed under green fluorescence channel on a Zeiss Axiovert 200M microscope with a X20 lens. Images were acquired every 2 minutes. The duration of prometaphase and metaphase was determined by morphology and positioning of chromosomes in mitotic cells.
For additional methods, please see SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Haining Du, Bradley Tangonan, and Bill Wang for technical assistance and members of the G.F. lab for discussions. This work was supported by a Burroughs–Wellcome Career Award in Biomedical Research (to G.F.) and National Institutes of Health Grants GM062852 (to G.F.) and HL079442 and RR11823–10 (to J.Y.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0709227105/DCSupplemental.
References
- 1.Peters JM. The anaphase-promoting complex: Proteolysis in mitosis and beyond. Mol Cell. 2002;9:931–943. doi: 10.1016/s1097-2765(02)00540-3. [DOI] [PubMed] [Google Scholar]
- 2.Stewart S, Fang G. Destruction box-dependent degradation of aurora B is mediated by the anaphase-promoting complex/cyclosome and Cdh1. Cancer Res. 2005;65:8730–8735. doi: 10.1158/0008-5472.CAN-05-1500. [DOI] [PubMed] [Google Scholar]
- 3.Stewart S, Fang G. Anaphase-promoting complex/cyclosome controls the stability of TPX2 during mitotic exit. Mol Cell Biol. 2005;25:10516–10527. doi: 10.1128/MCB.25.23.10516-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Zhao WM, Fang G. Anillin is a substrate of anaphase-promoting complex/cyclosome (APC/C) that controls spatial contractility of myosin during late cytokinesis. J Biol Chem. 2005;280:33516–33524. doi: 10.1074/jbc.M504657200. [DOI] [PubMed] [Google Scholar]
- 5.Seki A, Fang G. CKAP2 is a spindle-associated protein degraded by APC/C-Cdh1 during mitotic exit. J Biol Chem. 2007;282:15103–15113. doi: 10.1074/jbc.M701688200. [DOI] [PubMed] [Google Scholar]
- 6.Peters JM. The anaphase promoting complex/cyclosome: A machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- 7.Harper JW, Burton JL, Solomon MJ. The anaphase-promoting complex: It's not just for mitosis any more. Genes Dev. 2002;16:2179–2206. doi: 10.1101/gad.1013102. [DOI] [PubMed] [Google Scholar]
- 8.Chabes AL, Pfleger CM, Kirschner MW, Thelander L. Mouse ribonucleotide reductase R2 protein: A new target for anaphase-promoting complex-Cdh1-mediated proteolysis. Proc Natl Acad Sci USA. 2003;100:3925–3929. doi: 10.1073/pnas.0330774100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rhodes DR, et al. ONCOMINE: A cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Segal E, Friedman N, Koller D, Regev A. A module map showing conditional activity of expression modules in cancer. Nat Genet. 2004;36:1090–1098. doi: 10.1038/ng1434. [DOI] [PubMed] [Google Scholar]
- 11.Whitfield ML, et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol Biol Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fang G, Yu H, Kirschner MW. Direct binding of CDC20 protein family members activates the anaphase-promoting complex in mitosis and G1. Mol Cell. 1998;2:163–171. doi: 10.1016/s1097-2765(00)80126-4. [DOI] [PubMed] [Google Scholar]
- 13.Tabb DL, McDonald WH, Yates JR., III DTASelect and Contrast: Tools for assembling and comparing protein identifications from shotgun proteomics. J Proteome Res. 2002;1:21–26. doi: 10.1021/pr015504q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Denslow SA, Wade PA. The human Mi-2/NuRD complex and gene regulation. Oncogene. 2007;26:5433–5438. doi: 10.1038/sj.onc.1210611. [DOI] [PubMed] [Google Scholar]
- 15.Kruhlak MJ, et al. Regulation of global acetylation in mitosis through loss of histone acetyltransferases and deacetylases from chromatin. J Biol Chem. 2001;276:38307–38319. doi: 10.1074/jbc.M100290200. [DOI] [PubMed] [Google Scholar]
- 16.Taddei A, Maison C, Roche D, Almouzni G. Reversible disruption of pericentric heterochromatin and centromere function by inhibiting deacetylases. Nat Cell Biol. 2001;3:114–120. doi: 10.1038/35055010. [DOI] [PubMed] [Google Scholar]
- 17.Shamu CE, Murray AW. Sister chromatid separation in frog egg extracts requires DNA topoisomerase II activity during anaphase. J Cell Biol. 1992;117:921–934. doi: 10.1083/jcb.117.5.921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fang G. Checkpoint protein BubR1 acts synergistically with Mad2 to inhibit anaphase-promoting complex. Mol Biol Cell. 2002;13:755–766. doi: 10.1091/mbc.01-09-0437. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.