Abstract
MicroRNA 34a (miR-34a) is a tumor suppressor gene, but how it regulates cell proliferation is not completely understood. We now show that the microRNA miR-34a regulates silent information regulator 1 (SIRT1) expression. MiR-34a inhibits SIRT1 expression through a miR-34a-binding site within the 3′ UTR of SIRT1. MiR-34 inhibition of SIRT1 leads to an increase in acetylated p53 and expression of p21 and PUMA, transcriptional targets of p53 that regulate the cell cycle and apoptosis, respectively. Furthermore, miR-34 suppression of SIRT1 ultimately leads to apoptosis in WT human colon cancer cells but not in human colon cancer cells lacking p53. Finally, miR-34a itself is a transcriptional target of p53, suggesting a positive feedback loop between p53 and miR-34a. Thus, miR-34a functions as a tumor suppressor, in part, through a SIRT1-p53 pathway.
Keywords: microRNA, p53, cancer
Deletion of chromosome region 1p36 is associated with a variety of cancers including colon cancer (1–12). Transfection of a normal chromosome 1p36 region into human colon cancer cells decreases tumorigenicity, suggesting the presence of a tumor suppressor in this region (13). No conventional tumor suppressor genes have been identified within chromosome region 1p36. However, miR-34a is encoded within chromosome 1p36.23. Recent studies suggest that miRNA-34a may act as a tumor suppressor by regulating apoptosis in neuroblastoma cells and pancreatic cancer cells (14, 15). However, the mechanism by which miRNA-34a regulates cell proliferation is unclear.
MicroRNAs (miRNAs) are short 18–24 nt RNA that inhibit the transcription or translation of mRNA (16–20). MiRNA regulation of gene expression plays a role in development, differentiation, proliferation, and apoptosis (21–26). Expression of miRNAs are abnormal in a variety of cancers, and miRNA may play a role in tumorigenesis (27–34). Several miRNA have been identified that may act as tumor suppressor genes (35–40).
The tumor suppressor gene TP53 may serve as a link between miRNA and tumorigenesis. The p53 protein is a transcription factor that normally inhibits cell proliferation and stimulates cell death. However, disruption of the p53 pathway can promote tumorigenesis. One pathway through which p53 regulates cell growth is through miRNA. Cellular stress stabilizes p53 that in turn regulates the expression of a set of miRNA (15, 41–44). These miRNA in turn may control apoptosis and senescence. Recent reports show that one of the miRNA activated by p53 is miR-34a (15, 41–44).
We searched for downstream targets of miR-34a that might mediate p53 suppression of cell proliferation and found that miR-34a regulates silent information regulator 1 (SIRT1) expression. SIRT1 is an NAD-dependent deacetylase that regulates apoptosis in response to oxidative and genotoxic stress (45, 46). Recent data suggests that SIRT1 may function as an oncogene and play a role in tumorigenesis (47–50). Because SIRT1 is believed to regulate apoptotic thresholds by deacetylating molecular targets that include p53 (51–54), miR-34a might regulate cellular growth through a SIRT1-p53 pathway.
Results
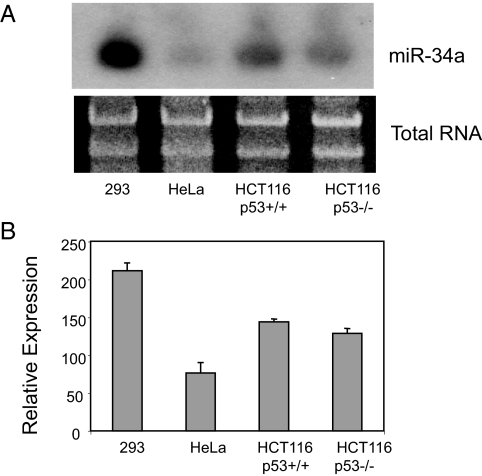
We hypothesized that miR-34a regulates the cell cycle by inhibiting SIRT1 expression based on an in silico analysis showing a potential response element for miR-34a in the 3′ UTR of SIRT1. We first measured the distribution of miR-34a in human tissue. Northern analysis shows that miR-34a is expressed in ovary, testis, bladder, lung, and other human tissues [supporting information (SI) Fig. S1]. MiR-34a is also expressed in gastrointestinal tissues and in the human colon cancer cell line HCT116 (Fig. 1). The HCT116 with WT p53 [HCT116(WT)] appear to express miR-34a, and HCT116(p53−/−) cells lacking p53 express similar levels (Fig. 1).
Fig. 1.
Expression of miR-34a. (A) Human cell lines, including human colon cancer (HCT116) cells by Northern blot (repeated three times with similar results). (B) Densitometric quantification of Northern analysis of miR-34a repeated three times. Both p53+/+ and p53−/− HCT116 cells have similar amounts of miR-34a, in the absence of genotoxic damage.
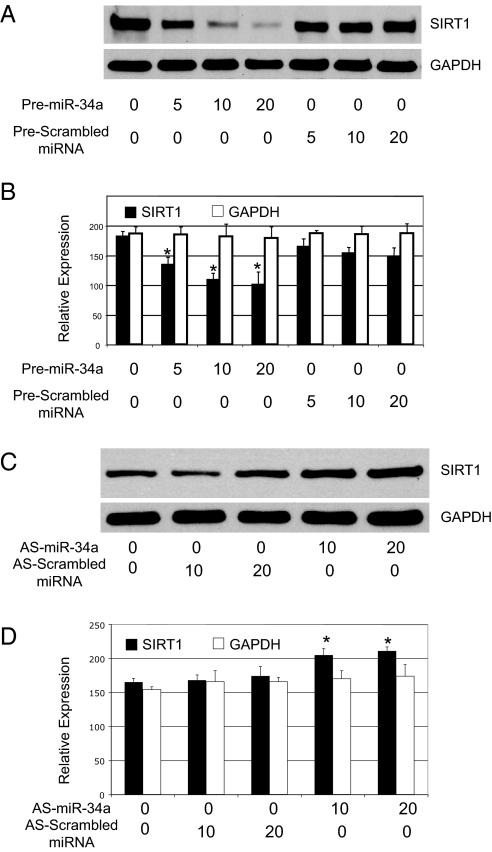
To test the hypothesis that miR-34a decreases SIRT1 expression, we transfected HCT116 cells with the precursor to miR-34a (designated premiR-34a). As a control, some cells were transfected with precursor to other miR or with scrambled miRNA. The precursor to miR-34a decreases SIRT1 protein levels in a dose-dependent manner (Fig. 2 A and B). Conversely, antisense miR-34a increases SIRT1 protein levels (Fig. 2 C and D). Thus, miR-34a regulates SIRT1 expression.
Fig. 2.
SIRT1 is a target of microRNA miR-34. (A) Dose-dependent suppression of SIRT1 by miR-34a in HeLa cells. HeLa cells were transfected with 5, 10, or 20 nM miRNA precursors and immunoblotted as above. (B) Quantification by densitometry of effects of premiR-34a on SIRT1 expression (n = 3 ± SD; *, P < 0.05 compared with scrambled mRNA). (C) Dose-dependent increase of SIRT1 by miR-34a in HEK293 cells. HEK293 cells were transfected with 15 or 30 nM antisense miRNA and immunoblotted as above. (D) Quantification by densitometry of effects of knocking down endogenous miR-34a on SIRT1 expression (n = 3 ± SD; *, P < 0.05 compared with scrambled mRNA).
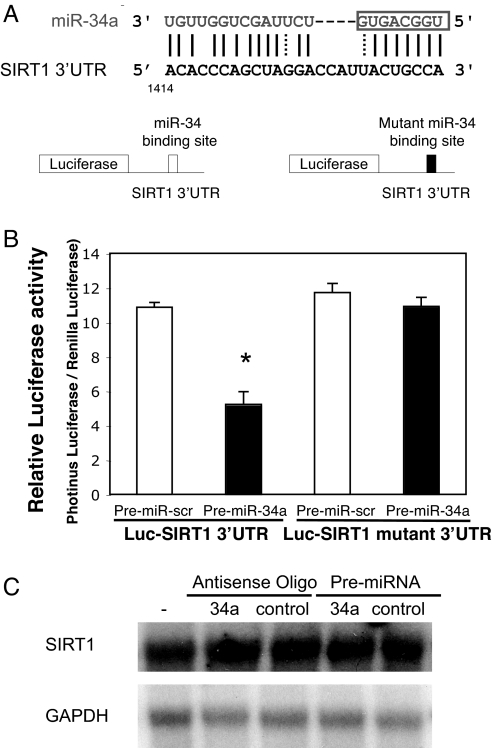
We hypothesized that miR-34a inhibits SIRT1 expression by repressing its translation. By computer analysis, we found a potential binding site for miR-34a within the 3′ UTR of SIRT1, extending from bp 1414–1439 (Fig. 3A Upper). To test the idea that miR-34a represses SIRT1 through this site, we constructed a reporter vector consisting of a luciferase cDNA followed by the 3′ UTR of SIRT1 (Fig. 3A Lower). We also constructed a luciferase reporter vector fused to the SIRT1 3′ UTR but with a mutant miR-34a response element. We then transfected into HEK293 cells this luciferase reporter vector with a WT or mutant miR-34a response element. We cotransfected these cells with control or premiR-34a and measured luciferase activity. PremiR-34a decreases luciferase activity of the reporter vector containing the miR-34a response element (Fig. 3B). In contrast, miR-34a has a minimal effect on a reporter vector with a mutated miR-34a response element. Furthermore, miR-34a does not affect SIRT1 RNA (Fig. 3C). Taken together, these data suggest that miR-34a decreases SIRT1 mRNA translation by acting on a response element in the SIRT1 3′ UTR.
Fig. 3.
MiR-34a binding site within SIRT1 3′ UTR mediates miR-34a translational repression. (A Top) miR-34a and the miR-34a-binding site in the 3′ UTR of SIRT1. (Bottom) Design of a miR-34a reporter vector containing a CMV promoter driving expression of a luciferase cDNA fused to the SIRT1 3′ UTR or to a mutated SIRT1 3′ UTR. (B) The 3′ UTR of SIRT1 mediates miR-34 control of SIRT1 expression. WT HCT116 cells were transfected with a reporter vector consisting of a luciferase cDNA fused to the 3′ UTR of SIRT1 which contains a binding site of miR-34. Another vector contained the luciferase cDNA fused to a SIRT1 3′ UTR with a mutant miR-34a-binding site. The cells were also transfected with a CMV-Renilla luciferase vector as an internal standard. PremiR-34a decreases expression of luciferase containing a WT miR-34a binding site (Left) but not a mutant binding site (Right). (C) miR-34 does not change SIRT1 mRNA. HCT116 cells were transfected with antisense-miR-34a or PremiR-34a, and total RNA was harvested and analyzed for SIRT1 by Northern blotting.
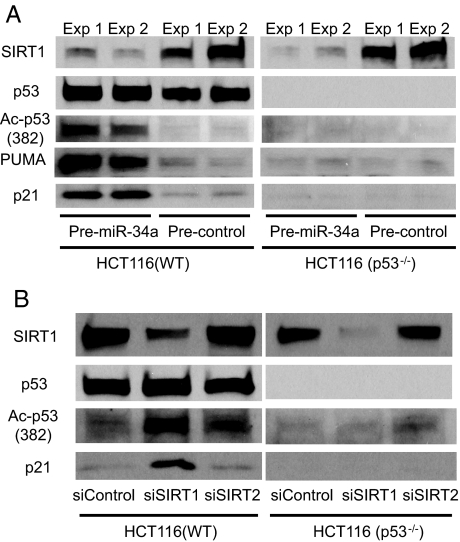
We next searched for SIRT1 signaling pathways affected by miR-34a. P53 is a transcription factor that inhibits cell growth in response to DNA damage, and inactivation of p53 pathways is a common feature of many cancers (55, 56). Because SIRT1 can inactivate p53 by deacetylating it, we examined the effect of miR-34a on acetylation of p53. Transfecting premiR-34a into WT HCT116 cells decreases SIRT1 and increases acetylated p53 (Fig. 4A). Furthermore, premiR-34a increases expression of p21 and PUMA, both transcriptional targets of p53 (Fig. 4A). In contrast, premiR-34a has no effect on p21 in HCT116 cells lacking p53. These data suggest that miR-34a increases activation of the p53 signaling pathway. Although miR-34a might have additional targets, these experiments taken together suggest that miR-34a indirectly regulates p53 through SIRT1.
Fig. 4.
miR-34a increases acetylation of p53 by decreasing SIRT1 expression. (A) miR-34 decreases SIRT1, increases acetylated p53, and increases two transcriptional targets of p53. In duplicate experiments, HCT(WT) and HCT(p53−/−) were transfected with precursors to microRNA and incubated for 36 h. SIRT1, acetylated p53, PUMA, and p21 were measured by immunoblotting. These experiments were performed in duplicate. (B) SIRT1 regulates acetylation of p53. HCT(WT) and HCT(p53−/−) were transfected with siRNA for SIRT1 or SIRT2, incubated for 36 h, and lysates were immunoblotted. Knockdown of SIRT1 permits an increase in p53 acetylation and an increase in p21, a transcriptional target of p53.
To confirm reports that SIRT1 regulates acetylation of p53, we transfected HCT cells with siRNA to knockdown SIRT1 (or knockdown SIRT2 as a control) and then measured acetylated p53. Knockdown of SIRT1 increases acetylation of p53 and activates p21 expression (Fig. 4B).
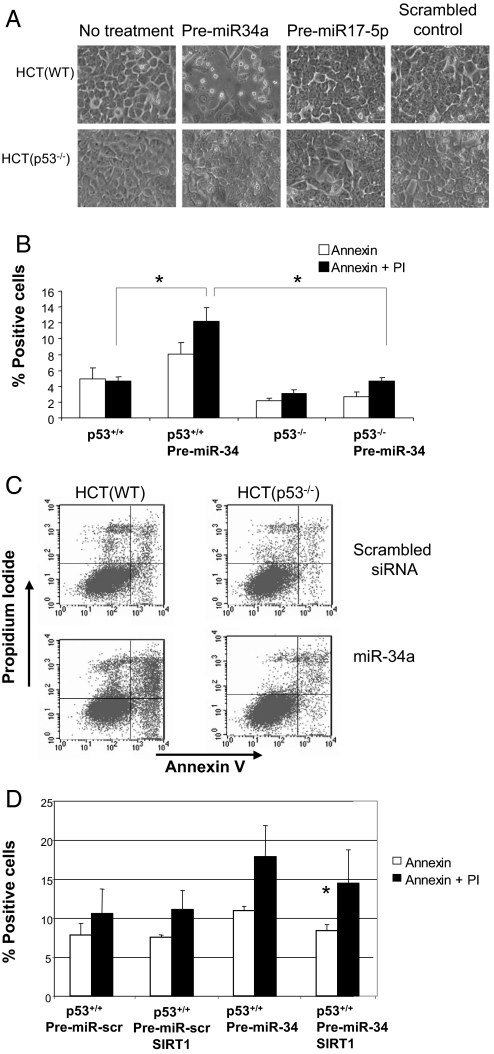
To explore the functional effect of miR-34a on cell survival, we measured apoptosis in cells transfected with premiR-34a. PremiR-34a increases cell death, as assessed by cell morphology (Fig. 5A). Premir-34a induces cell death in a p53-dependent manner (Fig. 5A). Overexpression of miR-34a increases apoptosis, as measured by FACS analysis of cells for annexin V and propidium iodide staining (Fig. 5 B and C). However, miR-34a does not activate apoptosis in cells lacking p53. These data suggest that miR-34a activates apoptosis in a p53-dependent pathway.
Fig. 5.
miR-34 activates apoptosis. (A) p53 mediates miR-34 induced apoptosis. HCT(WT) and HCT(p53−/−) cells were transfected with premiR-34 or control and then photographed. Overexpression of miR-34a causes cells to detach and to round up but only in cells expressing p53. (B) HCT(WT) and HCT(p53−/−) cells were treated as above, and apoptosis was measured by Annexin V and propidium iodine staining. (C) HCT(WT) and HCT(p53−/−) cells were treated as above, and apoptosis was measured by FACS (n = 3 ± SD; *, P = 0.003). Overexpression of miR-34a induces apoptosis but only in the presence of p53. (D) SIRT1 rescues cells from apoptosis after overexpression of miR-34. HCT(WT) cells were transfected with premiR-34 or control and with a vector expressing SIRT1 or control, and apoptosis was measured by FACS (n = 3 ± SD; *, P = 0.01 vs. PremiR-34 without SIRT1). Overexpression of SIRT1 blocks apoptosis induced by miR-34a.
To show that SIRT1 mediates miR-34a activation of p53 and apoptosis, we rescued cells with a vector expressing SIRT1. We transfected HCT116 cells with premiR-34a, a vector overexpressing SIRT1, and then measured apoptosis. As before, expression of miR-34a increases apoptosis (Fig. 5D). However, expression of SIRT1 partially blocks apoptosis induced by miR-34a. These data suggest that SIRT1 mediates miR-34a activation of cell death.
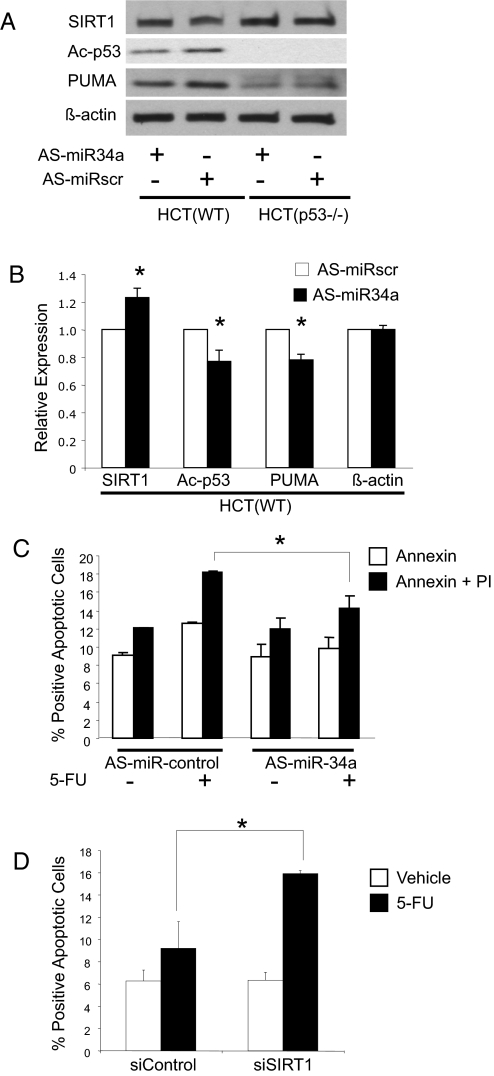
We next explored the effect of endogenous miR-34a on SIRT1. We transfected HCT116 cells with antisense oligonucleotides to miR-34a, treated them with 5-FU to activate p53, and measured apoptosis. Knockdown of endogenous miR-34 increases SIRT1 expression by ≈20% (Fig. 6 A and B). Knockdown of endogenous miR-34a also diminishes acetylated p53 levels by ≈55% and decreases steady-state levels of PUMA, a transcriptional target of p53 (Fig. 6 A and B). Furthermore, knockdown of endogenous miR-34a also decreases apoptosis (Fig. 6C). Finally, knockdown of SIRT1 permits an increase in 5-FU induced apoptosis, suggesting that SIRT1 blocks apoptosis in this pathway (Fig. 6D). The overexpression data and knockdown data together suggest that miR-34a decreases SIRT1 expression, which in turn modulates the p53 pathway.
Fig. 6.
Endogenous miR-34a regulates SIRT1. (A) HCT cells were transfected with antisense oligonucleotides to miR-34 (antisense-miR-34a) or scrambled oligonucleotides. Cells were treated with 5-FU for 16 h to activate p53, and cell lysates were immunoblotted for SIRT1, acetylated p53, and the p53 regulated gene product PUMA. Knockdown of endogenous miR-34a increases SIRT1, decreases acetylation of p53, and decreases protein levels of PUMA. (B) Quantitative effects of knockdown of endogenous miR-34a on Ac-p53. The above immunoblots were repeated three times and quantified by densitometry (n = 3 ± SD; *, P < 0.05 vs. scrambled miRNA). (C) Genotoxic stress increases apoptosis in HCT(WT) cells, and miR-34a mediates part of the genotoxic effect. HCT(WT) cells were transfected with antisense-miR-34a or control oligonucleotides, exposed to 5-FU, and apoptosis was measured as above. Knockdown of miR-34a decreases apoptosis (n = 3 ± SD; *, P < 0.05). (D) Knockdown of SIRT1 increases apoptosis. HCT(WT) cells were transfected with siControl or siSIRT1 then treated with vehicle or control. Annexin V staining was measured by FACS (n = 3 ± SD; *, P < 0.005).
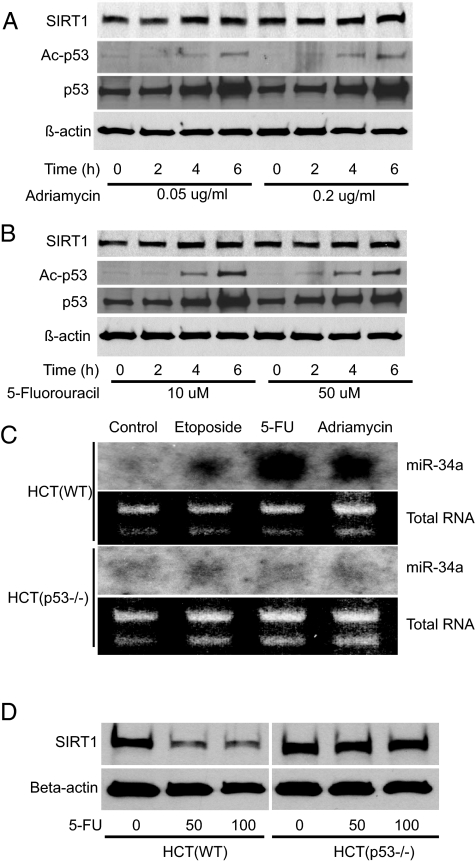
We and others have found that p53 regulates expression of a set of microRNA, including miR-34a (15, 41–44). If p53 directly regulates miR-34a and miR-34a indirectly regulates p53, then in theory, a positive feedback loop exists (Fig. S1). Activation of p53 increases miR-34a transcription; miR-34a suppresses SIRT1, permitting an increase in p53 activation which induces still more miR-34a. To search for p53 regulation of miR-34a, we activated p53 in HCT cells by using various agents that damage DNA (Fig. 7 A and B). We found that genotoxic agents increase miR-34a levels but only in HCT WT cells (Fig. 7C). Genotoxic agents fail to alter miR-34a levels in HCT cells lacking p53 (Fig. 7C). Furthermore, genotoxic agents that can induce miR-34a (Fig. 7C) could also decrease SIRT1 (Fig. 7D). However, in cells lacking p53, genotoxic agents did not induce miR-34a (Fig. 7C) and did not suppress SIRT1 (Fig. 7D).
Fig. 7.
p53 regulates miR-24a expression. HCT cells were treated with genotoxic agents, and cell lysates were assayed for p53, SIRT1, and miR-34a. Acetylation of p53 is increased after treatment with (A) adriamycin and (B) 5-FU. (C) Genotoxic agents increase miR-34a only in HCT cells expressing p53. (D) Genotoxic agents lead to a decrease in SIRT1 levels but only in cells expressing p53.
Discussion
The major finding of this study is that miR-34a inhibits SIRT1 expression, thereby triggering pathways downstream of SIRT1. Our study extends the results of others showing that SIRT1 deacetylates p53, limiting the ability of p53 to transactivate its target genes (51, 52, 57). We and others have shown that p53 regulates miR-34a (15, 41–44), and our study extends these results by suggesting the existence of a positive feedback loop in which p53 induces miR-34a, which through SIRT1, increases p53 activity. These results suggest that miR-34a can function as a tumor suppressor gene (Fig. S1). These data also provide one mechanism by which miR-34a can regulate cell proliferation.
MiR-34a may have other targets besides SIRT1 that can regulate cell survival (15). For example, miR-34a can decrease expression of the transcription factor E2F3, which regulates cell-cycle progression (14). Thus, SIRT1 may be one of several distinct targets of miR-34a that contribute to its ability to promote apoptosis.
Although HCT cells contain miR-34a, they still express SIRT1, a target of miR-34a. The miR-34a does not suppress all SIRT1 expression in resting cells (Fig. 2). Overexpression of miR-34a does not completely suppress SIRT1 translation (Fig. 2A). Furthermore, overexpression of miR-34a decreases expression of a luciferase reporter gene by ≈50% but does not completely eliminate luciferase activity (Fig. 3B). Perhaps intracellular levels of miR-34a do not saturate its binding site in the SIRT1 mRNA, so that some SIRT1 mRNA are still translated. Another possibility is that even though miR-34a concentrations exceed concentrations of SIRT1 mRNA, because miR-34a does not exactly match its SIRT1 binding site (Fig. 3A), perhaps every SIRT1 binding site does not interact with miR-34a.
Our data suggest that SIRT1 mediates, in part, the tumor suppressor activity of miR-34a. Several lines of evidence implicate SIRT1 in tumorigenesis. First, SIRT1 is overexpressed in human colon cancer, breast cancer, prostate cancer, squamous cell carcinoma, and human non-small-cell lung cancer cell lines (47–50). Next, by deacetylating p53, SIRT1 decreases the ability of p53 to promote cell cycle arrest (51, 52). Increased expression of SIRT1 in normal or cancer cells inactivates p53, decreasing cellular apoptosis (58). Finally, small molecules that inhibit SIRT1 increase acetylated p53 and activate apoptosis of breast cancer and lymphoma cells (59, 60). Accumulating data thus suggest that SIRT1 activity may increase the risk of cancer. Our findings suggest that miR-34a may act as a tumor suppressor by blocking SIRT1, thereby permitting increased p53 activity (Fig. S2).
Materials and Methods
Cell Culture, Northern Blot analysis, and Western Blot analysis.
(See SI Text).
Transfection.
Precursor miRNAs were obtained from Applied Biosystems). Small interfering RNA, siSIRT1, and scrambled si-oligonucleotide were obtained from Dharmacon. For precursor miRNA, cells were transfected with siPORT NeoFX (Applied Biosystems) with precursor miRNA 0–20 nM or with antisense miRNA 0–20 nM and harvested 48 or 72 h later. To silence SIRT1 expression, cells were transfected with Lipofectamine 2000 (Invitrogen) and SIRT1 antisense oligonucleotides 0–10 nM.
Plasmid Construction.
The 3′ UTR of SIRT1 (1420 bp) containing the SIRT1-miR-34a response element was cloned into the PmeI/SacI site of pMIR-REPORT Luciferase vector (Applied Biosystems). A mutant 3′ UTR of SIRT1 was synthesized by PCR, whose sequence contained 5′-ACACCCACCAAGGACCATTAGTCCGAGA-3′ (the five italic nucleotides are mutated).
Luciferase Assays.
HCT cells were plated at 5 × 104 cells per well in 24-well plates. The next day, 200 ng pMIR-REPORT Luciferase vector, including 3′ UTR of SIRT1 (with the WT or mutant miR-34a response element) and precursor miR-34a or scrambled oligonucleotide, were transfected by using Lipofectamine 2000 (Invitrogen). Luciferase assays were performed by using the dual luciferase reporter assay system (Promega) 48 h after transfection.
Apoptosis Analysis.
HCT116 cells were transfected with precursor miR-34a or scrambled oligonucleotides as a control. After an additional incubation of 72 h, the cells were harvested, stained with propidium iodide and anti-annexin-V antibody, and analyzed by FACS.
Supplementary Material
Acknowledgments.
Supported by National Institutes of Health Grants P01HL56091, R01HL074061, R01HL78635, and P01HL65608, American Health Association Scientist Development Grant 0835446N (to M.Y), the Ciccarone Center, the John and Cora H. Davis Foundation, and the Clarence P. Doodeman Professorship in Cardiology (C.J.L.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. F.S. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0801613105/DCSupplemental.
References
- 1.Fong CT, et al. Loss of heterozygosity for the short arm of chromosome 1 in human neuroblastomas: Correlation with N-myc amplification. Proc Natl Acad Sci USA. 1989;86:3753–3757. doi: 10.1073/pnas.86.10.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rigaud G, et al. Allelotype of pancreatic acinar cell carcinoma. Int J Cancer. 2000;88:772–777. doi: 10.1002/1097-0215(20001201)88:5<772::aid-ijc14>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 3.Bello MJ, et al. Allelic status of chromosome 1 in neoplasms of the nervous system. Cancer Genet Cytogenet. 1995;83:160–164. doi: 10.1016/0165-4608(95)00064-v. [DOI] [PubMed] [Google Scholar]
- 4.Moley JF, et al. Consistent association of 1p loss of heterozygosity with pheochromocytomas from patients with multiple endocrine neoplasia type 2 syndromes. Cancer Res. 1992;52:770–774. [PubMed] [Google Scholar]
- 5.Genuardi M, Tsihira H, Anderson DE, Saunders GF. Distal deletion of chromosome Ip in ductal carcinoma of the breast. Am J Hum Genet. 1989;45:73–82. [PMC free article] [PubMed] [Google Scholar]
- 6.Fang W, et al. Mapping of a minimal deleted region in human hepatocellular carcinoma to 1p36.13-p36.23 and mutational analysis of the RIZ (PRDM2) gene localized to the region. Genes Chromosomes Cancer. 2000;28:269–275. [PubMed] [Google Scholar]
- 7.Wozniak A, et al. Array CGH analysis in primary gastrointestinal stromal tumors: Cytogenetic profile correlates with anatomic site and tumor aggressiveness, irrespective of mutational status. Genes Chromosomes Cancer. 2007;46:261–276. doi: 10.1002/gcc.20408. [DOI] [PubMed] [Google Scholar]
- 8.Leister I, et al. Human colorectal cancer: High frequency of deletions at chromosome 1p35. Cancer Res. 1990;50:7232–7235. [PubMed] [Google Scholar]
- 9.Di Vinci A, et al. Deletions at chromosome 1p by fluorescence in situ hybridization are an early event in human colorectal tumorigenesis. Gastroenterology. 1996;111:102–107. doi: 10.1053/gast.1996.v111.pm8698188. [DOI] [PubMed] [Google Scholar]
- 10.Bomme L, et al. Allelic imbalance and cytogenetic deletion of 1p in colorectal adenomas: A target region identified between DIS199 and DIS234. Genes Chromosomes Cancer. 1998;21:185–194. doi: 10.1002/(sici)1098-2264(199803)21:3<185::aid-gcc2>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 11.Thorstensen L, et al. Evaluation of 1p losses in primary carcinomas, local recurrences and peripheral metastases from colorectal cancer patients. Neoplasia. 2000;2:514–522. doi: 10.1038/sj.neo.7900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Knosel T, Schluns K, Dietel M, Petersen I. Chromosomal alterations in lung metastases of colorectal carcinomas: Associations with tissue specific tumor dissemination. Clin Exp Metastasis. 2005;22:533–538. doi: 10.1007/s10585-005-5239-7. [DOI] [PubMed] [Google Scholar]
- 13.Tanaka K, et al. Suppression of tumourigenicity in human colon carcinoma cells by introduction of normal chromosome 1p36 region. Oncogene. 1993;8:2253–2258. [PubMed] [Google Scholar]
- 14.Welch C, Chen Y, Stallings RL. MicroRNA-34a functions as a potential tumor suppressor by inducing apoptosis in neuroblastoma cells. Oncogene. 2007;26:5017–5022. doi: 10.1038/sj.onc.1210293. [DOI] [PubMed] [Google Scholar]
- 15.Chang TC, et al. Transactivation of miR-34a by p53 broadly influences gene expression and promotes apoptosis. Mol Cell. 2007;26:745–752. doi: 10.1016/j.molcel.2007.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bartel DP. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 17.Meister G, Tuschl T. Mechanisms of gene silencing by double-stranded RNA. Nature. 2004;431:343–349. doi: 10.1038/nature02873. [DOI] [PubMed] [Google Scholar]
- 18.Zamore PD, Haley B. Ribo-gnome: The big world of small RNAs. Science. 2005;309:1519–1524. doi: 10.1126/science.1111444. [DOI] [PubMed] [Google Scholar]
- 19.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–355. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 20.Cullen BR. Transcription and processing of human microRNA precursors. Mol Cell. 2004;16:861–865. doi: 10.1016/j.molcel.2004.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 22.Xu P, Vernooy SY, Guo M, Hay BA. The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol. 2003;13:790–795. doi: 10.1016/s0960-9822(03)00250-1. [DOI] [PubMed] [Google Scholar]
- 23.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 24.Reinhart BJ, et al. The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature. 2000;403:901–906. doi: 10.1038/35002607. [DOI] [PubMed] [Google Scholar]
- 25.Brennecke J, et al. Bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell. 2003;113:25–36. doi: 10.1016/s0092-8674(03)00231-9. [DOI] [PubMed] [Google Scholar]
- 26.Hatfield SD, et al. Stem cell division is regulated by the microRNA pathway. Nature. 2005;435:974–978. doi: 10.1038/nature03816. [DOI] [PubMed] [Google Scholar]
- 27.Michael MZ, et al. Reduced accumulation of specific microRNAs in colorectal neoplasia. Mol Cancer Res. 2003;1:882–891. [PubMed] [Google Scholar]
- 28.Calin GA, et al. Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci USA. 2004;101:2999–3004. doi: 10.1073/pnas.0307323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iorio MV, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 30.Lu J, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 31.He L, et al. A microRNA polycistron as a potential human oncogene. Nature. 2005;435:828–833. doi: 10.1038/nature03552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Volinia S, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc Natl Acad Sci USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pekarsky Y, et al. Tcl1 expression in chronic lymphocytic leukemia is regulated by miR-29 and miR-181. Cancer Res. 2006;66:11590–11593. doi: 10.1158/0008-5472.CAN-06-3613. [DOI] [PubMed] [Google Scholar]
- 34.Slack FJ, Weidhaas JB. MicroRNAs as a potential magic bullet in cancer. Future Oncol. 2006;2:73–82. doi: 10.2217/14796694.2.1.73. [DOI] [PubMed] [Google Scholar]
- 35.Cimmino A, et al. miR-15 and miR-16 induce apoptosis by targeting BCL2. Proc Natl Acad Sci USA. 2005;102:13944–13949. doi: 10.1073/pnas.0506654102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Johnson SM, et al. RAS is regulated by the let-7 microRNA family. Cell. 2005;120:635–647. doi: 10.1016/j.cell.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 37.Takamizawa J, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–3756. doi: 10.1158/0008-5472.CAN-04-0637. [DOI] [PubMed] [Google Scholar]
- 38.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Morris JP, McManus MT. Slowing down the Ras lane: miRNAs as tumor suppressors? Sci STKE. 2005;41 doi: 10.1126/stke.2972005pe41. [DOI] [PubMed] [Google Scholar]
- 40.He L, He X, Lowe SW, Hannon GJ. MicroRNAs join the p53 network–another piece in the tumour-suppression puzzle. Nat Rev Cancer. 2007;7:819–822. doi: 10.1038/nrc2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tazawa H, Tsuchiya N, Izumiya M, Nakagama H. Tumor-suppressive miR-34a induces senescence-like growth arrest through modulation of the E2F pathway in human colon cancer cells. Proc Natl Acad Sci USA. 2007;104:15472–15477. doi: 10.1073/pnas.0707351104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.He L, et al. A microRNA component of the p53 tumour suppressor network. Nature. 2007;447:1130–1134. doi: 10.1038/nature05939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tarasov V, et al. Differential regulation of microRNAs by p53 revealed by massively parallel sequencing: miR-34a is a p53 target that induces apoptosis and G1-arrest. Cell Cycle. 2007;6:1586–1593. doi: 10.4161/cc.6.13.4436. [DOI] [PubMed] [Google Scholar]
- 44.Raver-Shapira N, et al. Transcriptional activation of miR-34a contributes to p53-mediated apoptosis. Mol Cell. 2007;26:731–743. doi: 10.1016/j.molcel.2007.05.017. [DOI] [PubMed] [Google Scholar]
- 45.Haigis MC, Guarente LP. Mammalian sirtuins–emerging roles in physiology, aging, and calorie restriction. Genes Dev. 2006;20:2913–2921. doi: 10.1101/gad.1467506. [DOI] [PubMed] [Google Scholar]
- 46.Longo VD, Kennedy BK. Sirtuins in aging and age-related disease. Cell. 2006;126:257–268. doi: 10.1016/j.cell.2006.07.002. [DOI] [PubMed] [Google Scholar]
- 47.Kuzmichev A, et al. Composition and histone substrates of polycomb repressive group complexes change during cellular differentiation. Proc Natl Acad Sci USA. 2005;102:1859–1864. doi: 10.1073/pnas.0409875102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huffman DM, et al. SIRT1 is significantly elevated in mouse and human prostate cancer. Cancer Res. 2007;67:6612–6618. doi: 10.1158/0008-5472.CAN-07-0085. [DOI] [PubMed] [Google Scholar]
- 49.Hida Y, Kubo Y, Murao K, Arase S. Strong expression of a longevity-related protein, SIRT1, in Bowen's disease. Arch Dermatol Res. 2007;299:103–106. doi: 10.1007/s00403-006-0725-6. [DOI] [PubMed] [Google Scholar]
- 50.Yeung F, et al. Modulation of NF-kappaB-dependent transcription and cell survival by the SIRT1 deacetylase. EMBO J. 2004;23:2369–2380. doi: 10.1038/sj.emboj.7600244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Luo J, et al. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. [DOI] [PubMed] [Google Scholar]
- 52.Vaziri H, et al. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. [DOI] [PubMed] [Google Scholar]
- 53.Langley E, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cheng HL, et al. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc Natl Acad Sci USA. 2003;100:10794–10799. doi: 10.1073/pnas.1934713100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. 2000;408:307–310. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 56.Vogelstein B, Kinzler KW. Cancer genes and the pathways they control. Nat Med. 2004;10:789–799. doi: 10.1038/nm1087. [DOI] [PubMed] [Google Scholar]
- 57.Luo J, et al. Deacetylation of p53 modulates its effect on cell growth and apoptosis. Nature. 2000;408:377–381. doi: 10.1038/35042612. [DOI] [PubMed] [Google Scholar]
- 58.Chen WY, et al. Tumor suppressor HIC1 directly regulates SIRT1 to modulate p53-dependent DNA-damage responses. Cell. 2005;123:437–448. doi: 10.1016/j.cell.2005.08.011. [DOI] [PubMed] [Google Scholar]
- 59.Heltweg B, et al. Antitumor activity of a small-molecule inhibitor of human silent information regulator 2 enzymes. Cancer Res. 2006;66:4368–4377. doi: 10.1158/0008-5472.CAN-05-3617. [DOI] [PubMed] [Google Scholar]
- 60.Ota H, et al. Sirt1 inhibitor, Sirtinol, induces senescence-like growth arrest with attenuated Ras-MAPK signaling in human cancer cells. Oncogene. 2006;25:176–185. doi: 10.1038/sj.onc.1209049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.