Abstract
Ecological responses to on-going climate change are numerous, diverse, and taxonomically widespread. However, with one exception, the relative roles of phenotypic plasticity and microevolution as mechanisms in explaining these responses are largely unknown. Several recent studies have uncovered evidence for temporal declines in mean body sizes of birds and mammals, and these responses have been interpreted as evidence for microevolution in the context of Bergmann's rule—an ecogeographic rule predicting an inverse correlation between temperature and mean body size in endothermic animals. We used a dataset of individually marked red-billed gulls (Larus novaehollandiae scopulinus) from New Zealand to document phenotypic and genetic changes in mean body mass over a 47-year (1958–2004) period. We found that, whereas the mean body mass had decreased over time as ambient temperatures increased, analyses of breeding values estimated with an “animal model” approach showed no evidence for any genetic change. These results indicate that the frequently observed climate-change-related responses in mean body size of animal populations might be due to phenotypic plasticity, rather than to genetic microevolutionary responses.
Keywords: adaptation, animal model, quantitative genetics
One of the major challenges faced by conservation and evolutionary biologists is to understand the role of microevolution in the population responses to the rapid, ongoing global change (1). Evidence for phenotypic responses to global warming is numerous, diverse, and taxonomically widespread (2–5). Examples include advances in phenology (2, 6, 7), poleward shifts in distribution ranges (4, 8), and changes in population mean phenotypes such as body size (9). Although adaptation to climate change is often evoked to explain phenotypic time trends (e.g., refs. 7, 10, and 11), the available evidence for evolutionary responses sensu stricto is still scarce (refs. 12, 13; but see refs. 14–16).
A large number of correlative studies have found evidence for declines in mean body size of birds (10, 11) and mammals (17–19) over time. These results have been interpreted as evidence for animal adaptation to a warming climate in the framework of Bergmann's rule. The original formulation of the Bergmann's rule applies to geographic variation (20) and predicts an increasing mean body size with increasing latitude as an energetic adaptation to a colder environment. This is because large body size reduces the surface-to-volume ratio and thereby the loss of energy due to conduction. By inference, Bergmann's rule predicts a decreased body size as an adaptive, genetically based response to global warming (9). However, no study has examined whether there is a genetic basis for this interpretation. The decrease in mean body size in a population can also occur as a plastic response to changes in environmental conditions, such as changes in food abundance, food quality, or prevalence of parasites. Hence, decreased mean body size over time might not represent adaptation to global warming but might be a sign of deteriorating environmental conditions. We assessed the value of these contrasting explanations by investigating changes in mean phenotypic and genetic body mass of red-billed gulls in New Zealand using a long-term dataset of individually marked birds collected over a 47-year period (1958–2004).
Results and Discussion
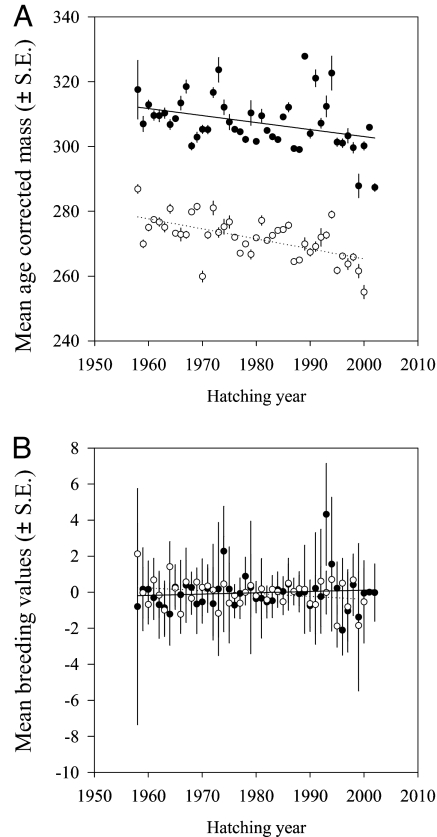
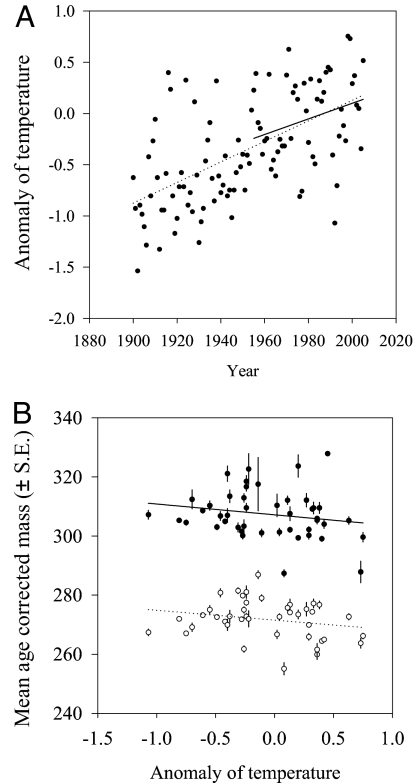
Over the duration of the study (1958–2004), we found that mean age-corrected body mass has been decreasing in the red-billed gull population (b = −0.28; SE = 0.05) (Table 1 and Fig. 1A) in both sexes, corresponding to a rate of 0.1 SD-units per generation. As expected under Bergmann's rule, this decline is significantly correlated with increased temperature in New Zealand over the same period (b = −3.57; SE = 1.98) (Table 1 and Fig. 2). However, analysis of breeding values, estimated with the “animal model” approach (21) and using hatching year as a covariate to avoid bias toward phenotypic trend (22), showed no corresponding change (b = −0.003; SE = 0.02) (Table 1 and Fig. 1B) or correlation with temperature (b = 0.165; SE = 0.44) (Table 1). Consequently, the decreased mean body mass and the absence of change at the genotypic level suggest that the observed trend is mostly, if not entirely, of environmental origin.
Table 1.
GLMM analyses of age-corrected mass and breeding values for mass
| Environmental effect | Age-corrected body mass (NObservations = 3,417/NHatching years = 45) |
Breeding values for body mass (NObservations = 3,283/NHatching years = 43) |
||||
|---|---|---|---|---|---|---|
| df | F | P | df | F | P | |
| Time trend | ||||||
| Hatching year | 1/43 | 27.26 | <0.0001 | 1/41 | 0.005 | 0.946 |
| Sex | 1/3,370 | 14,413.13 | <0.0001 | 1/3,238 | 0.056 | 0.812 |
| Sex:Hatching year | 1/3,370 | 1.57 | 0.210 | 1/3,238 | 0.036 | 0.849 |
| Temperature anomaly | ||||||
| Anomaly | 1/43 | 4.89 | 0.032 | 1/41 | 0.789 | 0.380 |
| Sex | 1/3,370 | 14,408.46 | <0.0001 | 1/3,238 | 0.042 | 0.837 |
| Sex:Anomaly | 1/3,370 | 0.02 | 0.896 | 1/3,238 | 0.098 | 0.754 |
Breeding values for mass are estimated from an animal model containing individual and permanent environment effects. Time trends in phenotypic and breeding values were assessed against the hatching year of the birds. Shown also is the effect of temperature anomaly on body mass.
Fig. 1.
Time trends in mean body mass (in g) (A) and breeding values (B) for female (open symbols and dotted lines) and male (filled symbols and solid lines) red-billed gulls as a function of hatching year (1958–2002). Trends for phenotypic values are statistically significant (P < 0.0001) both in males and females, whereas breeding values have not changed over time (P ≥ 0.9; see text for details of statistical tests.).
Fig. 2.
Associations with time, temperature anomaly, and body mass of red-billed gulls in New Zealand. (A) Temperature anomaly in New Zealand as a function of time (ρ = 0.30, n = 45, P = 0.04). (B) Mean (phenotypic) body mass of male (solid dots and line) and female (open dots and dotted line) red-billed gulls as a function of temperature anomaly. The relationship between mass and temperature is significant (P < 0.05, see text for details of statistical tests). Temperature anomaly data were obtained from figure 6 of ref. 45.
Lack of expected changes in predicted breeding values could occur because they are easily biased if the information content of the underlying pedigrees is low (22). This is unlikely to explain our results for four reasons. First, such a bias should shift the breeding values toward phenotypic values rather than away from them. Second, 3 years of data with 100 individuals measured per year would already give good estimates of genetic parameters (23). Our pedigree contains >40 years of data and >100 individuals for >5 years. Third, although correcting for hatching year in the animal model could reduce the likelihood of detecting a time trend in breeding values, this is an unlikely problem in our analyses. In other words, when the connectedness of the pedigree across the fixed effects is low, i.e., when there are observations of an individual and its relatives in only a subset of years, the genetic component of the variation among years may become absorbed into the year effect instead of the breeding values (22). However, the mean lineage length in our pedigree was 2.15 (SD 0.97, maximum 7), corresponding to 22 years, which is approximately half of the study period. Hence, the substantial proportion of time covered by the observations should prevent an “over correction” of the data masking a trend in the breeding values. Fourth, the accuracy of breeding values (correlation between true and predicted values, assessed by the square-root of the ratio between variance in breeding values and additive genetic variance) (24) was good (0.6) and well within the range of typical values from other studies (e.g., r = 0.55 in ref. 25, r = 0.65 in ref. 26). However, we note that the small difference between heritability (h2 ± SE.: males, h2 = 0.33 ± 0.07; females, h2 = 0.27 ± 0.07) and reliability (0.62 = 0.36) suggests that the information contained by breeding values may be limited and that the analyses of breeding values may be subject to low statistical power. This means that we cannot rule out the possibility that part of the observed change in phenotypic means over time might be of genetic rather than environmental origin.
The interpretation that the decline in mean body mass over time is due to environmental rather than genetic effects is further supported by two lines of evidence. First, the analyses of natural selection on body mass show no evidence for directional (or quadratic) selection on body mass in either sex whether using lifetime production of recruits (Table 2) or fledglings (Table 2) as estimates of fitness. Likewise, no evidence for directional selection through survival was observed [supporting information (SI) Table S1]. Hence, although body mass is heritable in both sexes, no evolutionary response of mass is to be expected. Second, in analyzing changes in traits indicative of the degree of environmental stress experienced by the population, we found a significant decline in survival probability over the study period (posterior mean estimate and 95% confidence interval for change in annual survival probability from 1968 to 2004 in males: −0.042 [−0.085; 0.000), P = 0.026; in females: −0.063 (−0.104; −0.025), P = 0.001], supporting the interpretation that the environmental conditions in the study population have been deteriorating over time. Consequently, the breeding population size of red-billed gulls has declined since 1983 (b = −141.70, SE = 45.95, F1, 20 9.51, P = 0.006) from 5,888 to 3,737 breeding pairs.
Table 2.
Selection on body mass of breeding male and female red-billed gulls
| Parameter | N | Standardized directional selection differential |
Standardized quadratic selection differential |
||||
|---|---|---|---|---|---|---|---|
| Estimate ± SE | t | P | Estimate ± SE | t | P | ||
| Reproductive success | |||||||
| Males | 338 | −0.003 ± 0.08 | −0.043 | 0.97 | −0.051 ± 0.05 | −0.943 | 0.35 |
| Females | 213 | 0.107 ± 0.12 | 0.872 | 0.38 | 0.016 ± 0.09 | 0.173 | 0.86 |
| Fledging success | |||||||
| Males | 346 | 0.032 ± 0.045 | 0.722 | 0.471 | −0.052 ± 0.030 | −1.73 | 0.085 |
| Females | 213 | 0.104 ± 0.063 | 1.645 | 0.101 | 0.015 ± 0.044 | 0.343 | 0.732 |
Standardized selection differentials were estimated regressing relative estimate of lifetime reproductive success and lifetime fledging success against standardized mass according to the procedures described by Lande and Arnold (40).
Our results question the interpretation of temporal “Bergmann clines” as adaptive responses to a warming climate. In contrast, they indicate that body size declines can simply be environmentally induced responses to some form of environmental stress. Unlike some other seabird species, such as the black-legged kittiwake (Rissa tridactyla) where a decline in fish stocks has affected feeding conditions (e.g., ref. 27), there has not been a commensurate decline in the availability of euphausiids, the main food of the red-billed gull, during the breeding season to account for the decline in the size of the bird (28). At the same time, a reduction in the abundance of food outside of the breeding season, when the bird is a more generalist feeder may have been a contributing factor. However, in the context of global change, many other factors, such as changes in habitat quality and interspecific interactions, may account for this decline. If so, this also means that temporal Bergmann clines could perhaps be viewed as warning signals, rather than comforting examples of microevolutionary adaptation in response to climate change. In more general terms, our results demonstrate the danger of drawing evolutionary inference from purely phenotypic data, a conceptual point also brought home by two case studies where phenotypic and genetic changes occurred in opposite directions (29, 30).
Most microevolutionary studies of climate change responses have overlooked or failed to find evidence for genetic adaptation (31). Yet the knowledge about the genetic vs. plastic basis of observed responses is important, both in terms of devising conservation practices and management strategies (e.g., refs. 31 and 32) as well as for our understanding of temporal phenotypic trends in the context of Bergmann's rule (9). However, disentangling genetic from plastic responses to human impacts has proven challenging (12, 31). Fisheries induced responses to size selective harvesting provide a case in point. Despite a wealth of phenotypic data for which time trends match expected response to selection from fishing, hard evidence for genetically based changes in life history traits from the wild is still lacking (33–35). As the results of this study show, the same applies to the temporal trends in vertebrate body size commonly interpreted to reflect adaptive responses to climate change in the context of Bergmann's rule.
Methods
Data.
Data on red-billed gulls (Larus novaehollandiae scopulinus) were collected between 1958 and 2004 from a colony breeding in Kaikoura, New Zealand. Each year, breeding attempts were monitored from egg-laying until fledging of the young, and all parents and young were banded with individually numbered metal bands. A pedigree of seven generations and 16,520 individuals was based on these records. Because generation length of red-billed gulls—defined as the average age of breeding individuals—is ≈10 years, the data hence covered roughly on average four generations. Mass was recorded for 3,417 breeding individuals of known age, and 871 individuals were measured twice or more.
As a precaution against the possibility that body condition may change faster than body size, we tested for time trend in tarsus length. Mean tarsus length at phenotypic level decreased with time (estimate ± SE: −0.376 ± 0.068, F1, 42 = 66, P < 0.001) but not at level of breeding values (0.009 ± 0.016, F1, 41 = 0.34, P = 0.56). Hence, the data on linear measurements corroborates the results obtained using body mass. Because (i) more body mass than tarsus length measurements were available, and (ii) Bergmann's rule deals explicitly with volume (and hence mass) rather than with linear measurement of body size per se, only results based on body mass are reported.
Age-corrected mass, used in the estimation of time trends and selection analyses, was obtained by using mixed model predictions, with individual identity as a random effect and age as a covariate for which polynomials up to the second degree were fitted. The age at which mass is predicted was arbitrarily set to 5 years. Individuals were followed 1958–2004, and hence, with respect to hatching year, records were available for birds hatched in 2002 or before. Because only a subset of the individuals in the population were followed through their lifetimes, the sample of birds we used in the selection analyses (see below) was significantly smaller than the sample of birds of known age for which we had mass measurements.
Quantitative Genetic Analysis.
Predicted breeding values and heritabilities were estimated with the aid of an animal model, which is a mixed model restricted maximum likelihood (REML) procedure using the software ASReml (36). Animal models were fitted separately for females and males to break down individuals' phenotypic mass into components of additive genetic value and other random and fixed effects (37, 38). In all models, individual's age and hatching year were fitted as categorical fixed effects. Because permanent environmental (PE) effects and maternal effects can inflate estimates of additive genetic variance, we used a model selection approach to estimate the significance of these effects. Both effects were nonsignificant (see Table S2) but PE was kept in the final model to account for the nonindependence of repeated measurements of same individuals in different years.
The narrow sense heritability (h2) was estimated as the ratio of the additive variance (VA) to the total phenotypic variance after accounting for fixed effects (VP): h2 = VA/VP. Best linear unbiased predictors (BLUP) of individual breeding values (i.e., the expected effect of the genes that an individual passes on to its offspring) were quantified. Changes in BLUP estimates of breeding values in different generations or years reflect changes in the genetic composition of the population resulting from selection, genetic drift, or inbreeding (38). Because the annual size of the Kaikoura breeding population was large, varying annually from 2,315 to 9,212 pairs, the occurrence of genetic drift was unlikely. Likewise, there was no inbreeding in this population. Therefore, changes in breeding values over time can be taken as evidence of a response to selection (29).
Analyses of Time Trends.
Testing for time trends in both age-corrected mass measures and breeding values were conducted by using generalized linear mixed models, where hatching year was fitted both as a linear fixed effect to estimate time trend and as a random effect to account for the nonindependence of observations within the same year of birth. Analyses performed by using yearly mean values weighted by the inverse of their standard deviations gave qualitatively similar results. These analyses were run with the R statistical package (39).
Time trends in survival were estimated separately for each sex from capture-mark-recapture data by using a Bayesian hierarchical model (40) in OpenBUGS software (http://mathstat.helsinki.fi/openbugs) (41). We included the 4,603 color-banded individuals of known age in the analysis. Band loss was not a problem as all color-marked individuals had a stainless steel numbered band in the combination and none of these metal bands were lost due to band wear even after 20–25 years. Annual survival probability and annual probability of capture after the marking of the individual were modeled on logit-normal scale as a function of mean, year and age of the individual. Age was treated as a random effect in both cases. Year was treated as a random effect for capture probability and both as a random effect and a covariate for survival. The inclusion of year as both random effect and covariate allowed us to estimate the time trend in survival and the variation around it. We used vague normal priors (mean 0, variance 10) for the mean and the year as a covariate and noninformative priors [gamma(0.001, 0.001)] for the precision of random effects. Three chains of 20,000 iterations were run with a burn-in period of 10,000 iterations. Convergence and mixing were monitored by eye.
Selection Analyses.
Selection on body mass was assessed with standard regression methodology by regressing a relative estimate of fitness (see below) against standardized (age-corrected) mass (42). Because red-billed gulls are long-lived (average lifespan = 13 years but potentially up to 30 years), we used reproductive success (number of offspring recruited into the breeding population) at an age of 15 years as a proxy for lifetime reproductive success (LRS) to obtain a reasonable sample size and to avoid favoring individuals who died young. This was justified because (i) 76% of females and 84% of males had died by the age of 15 and because (ii) the correlation with this proxy and true LRS for a subset of birds known to have completed their life and died after they were 15 was very strong (males: n = 70; r = 0.86, P < 0.0001; females: n = 41; r = 0.92, P < 0.0001). Hence, the selection analyses were restricted to birds born before 1985 so that offspring had still 5 years to be recruited and observed as breeders in the population (age at first reproduction in males and females is 3.3 and 4.2 years, respectively). This process resulted in a dataset of 551 individuals for which mass measurements were available. Sample size is smaller in females than in males because we had to exclude from the analysis all of the females that had been breeding in a female-female pair at some point in their life.
Because lifetime reproductive success estimated as the number of recruited offspring in the population can confound both parental and offspring fitness (43), we also ran selection analysis by using lifetime fledging success and yearly survival as fitness estimates. Selection analyses were made with the software package R (39).
Weather Data.
We used temperature anomaly estimates (calculated as the annual differences from the 1971–2000 average) at the country level as a temperature proxy because outside of the breeding season red-billed gulls move around widely, but also because many studies have shown that larger scale estimates of climate can be better predictors of biological phenomena than local weather (44). These data were obtained from the New Zealand meteorological center website (45).
Supplementary Material
Acknowledgments.
We thank Bob O'Hara for help with statistical analyses; Phillip Gienapp, Alastair J. Wilson, and two anonymous referees for comments on the manuscript; and the numerous people who have helped with data collection and management over the years. Our research was supported by the Finnish Ministry of Education, the Research Foundation of the University of Helsinki, the Emil Aaltonen Foundation, and the Academy of Finland.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. D.R. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/cgi/content/full/0800999105/DCSupplemental.
References
- 1.Ferriere R, Dieckmann U, Couvet D. Evolutionary Conservation Biology. Cambridge, UK: Cambridge Univ Press; 2004. [Google Scholar]
- 2.Parmesan C, Yohe G. A globally coherent fingerprint of climate change impacts across natural systems. Nature. 2003;421:37–42. doi: 10.1038/nature01286. [DOI] [PubMed] [Google Scholar]
- 3.Root TL, MacMynowski DP, Mastrandrea MD, Schneider SH. Human-modified temperatures induce species changes: Joint attribution. Proc Natl Acad Sci USA. 2005;102:7465–7469. doi: 10.1073/pnas.0502286102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Walther GR, Berger S, Sykes MT. An ecological ‘footprint’ of climate change. Proc R Soc London B. 2005;272:1427–1432. doi: 10.1098/rspb.2005.3119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Adger N, et al. Intergovernmental Panel on Climate Change. Cambridge, UK: IPCC) (Cambridge Univ Press; 2007. Climate Change 2007: Climate Change Impacts, Adaptation and Vulnerability. [Google Scholar]
- 6.Root TL, et al. Fingerprints of global warming on wild animals and plants. Nature. 2003;421:57–60. doi: 10.1038/nature01333. [DOI] [PubMed] [Google Scholar]
- 7.Jonzen N, et al. Rapid advance of spring arrival dates in long-distance migratory birds. Science. 2006;312:1959–1961. doi: 10.1126/science.1126119. [DOI] [PubMed] [Google Scholar]
- 8.Parmesan C. Ecological and evolutionary responses to recent climate change. Annu Rev Ecol Evol Syst. 2006;37:637–669. [Google Scholar]
- 9.Millien V, et al. Ecotypic variation in the context of global climate change: Revisiting the rules. Ecology Letters. 2006;9:853–869. doi: 10.1111/j.1461-0248.2006.00928.x. [DOI] [PubMed] [Google Scholar]
- 10.Yom-Tov Y. Global warming and body mass decline in Israeli passerine birds. Proc R Soc London Ser B. 2001;268:947–952. doi: 10.1098/rspb.2001.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yom-Tov Y, et al. Recent changes in body weight and wing length among some British passerine birds. Oikos. 2006;112:91–101. [Google Scholar]
- 12.Gienapp P, Leimu R, Merilä J. Responses to climate change in avian migration time—microevolution or phenotypic plasticity? Clim Res. 2007;35:25–35. [Google Scholar]
- 13.Holt RD. The microevolutionary consequences of climate change. Trends Ecol Evol. 1990;14:96–101. doi: 10.1016/0169-5347(90)90088-U. [DOI] [PubMed] [Google Scholar]
- 14.Reale D, McAdam AG, Boutin S, Berteaux D. Genetic and plastic responses of a northern mammal to climate change. Proc R Soc London Ser B. 2003;270:591–596. doi: 10.1098/rspb.2002.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Franks SJ, Sim S, Weis AE. Rapid evolution of flowering time by an annual plant in response to a climate fluctuation. Proc Natl Acad Sci USA. 2007;104:1278–1282. doi: 10.1073/pnas.0608379104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bradshaw WE, Holzapfel CM. Genetic shift in photoperiodic response correlated with global warming. Proc Natl Acad Sci USA. 2001;98:14509–14511. doi: 10.1073/pnas.241391498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Millien V. Relative effects of climate change, isolation and competition on body-size evolution in the Japanese field mouse, Apodemus argenteus. J Biogeogr. 2004;31:1267–1276. [Google Scholar]
- 18.Smith FA, Betancourt L, Brown JH. Evolution of body size in the woodrat over the past 25,000 years of climate change. Science. 1995;270:2012–2014. [Google Scholar]
- 19.Yom-Tov Y, Yom-Tov S. Climatic change and body size in two species of Japanese rodents. Biol J Linn Soc London. 2004;82:263–267. [Google Scholar]
- 20.Bergmann C. Ueber die verhältnisse der wärmeökonomie der thiere zu ihrer grösse. Gottinger Studien. 1847;3:595–708. [Google Scholar]
- 21.Kruuk LEB. Estimating genetic parameters in natural populations using the “animal model””. Philos Trans R Soc London Ser B. 2004;359:873–890. doi: 10.1098/rstb.2003.1437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Postma E. Implications of the difference between true and predicted breeding values for study of natural selection and micro-evolution. J Evol Biol. 2006;19:309–320. doi: 10.1111/j.1420-9101.2005.01007.x. [DOI] [PubMed] [Google Scholar]
- 23.Quinn JL, Charmantier A, Garant D, Sheldon BC. Data depth, data completeness, and their influence on quantitative genetic estimation in two contrasting bird populations. J Evol Biol. 2006;19:994–1002. doi: 10.1111/j.1420-9101.2006.01081.x. [DOI] [PubMed] [Google Scholar]
- 24.Falconer DS, Mackay TFC. Introduction to Quantitative Genetics. New York: Longman; 1996. [Google Scholar]
- 25.Gienapp P, Postma E, Visser ME. Why breeding time has not responded to selection for earlier breeding in a songbird population. Evolution. 2006;60:2381–2388. [PubMed] [Google Scholar]
- 26.Charmantier A, Perrins C, McCleery RH, Sheldon BC. Evolutionary response to selection on clutch size in a long-term study of the mute swan. Am Nat. 2006;167:453–465. doi: 10.1086/499378. [DOI] [PubMed] [Google Scholar]
- 27.Frederiksen M, Wanless S, Rothery P, Wilson LJ. The role of industrial fisheries and oceanographic change in the decline of North Sea black-legged kittiwakes. J Appl Ecol. 2004;41:1129–1139. [Google Scholar]
- 28.Mills J. A., et al. The impact of climate fluctuation on food availability and reproductive performance of the planktivorous red-billed gull Larus novaehollandiae scopulinus. J Anim Ecol. 2008 doi: 10.1111/j.1365-2656.2008.01383.x. in press. [DOI] [PubMed] [Google Scholar]
- 29.Merilä J., Kruuk LEB, Sheldon BC. Cryptic evolution in a wild bird population. Nature. 2001;412:76–79. doi: 10.1038/35083580. [DOI] [PubMed] [Google Scholar]
- 30.Garant D, Kruuk LEB, McCleery RH, Sheldon BC. Evolution in a changing environment: A case study with great tit fledging mass. Am Nat. 2004;164:E115–E129. doi: 10.1086/424764. [DOI] [PubMed] [Google Scholar]
- 31.Gienapp P, et al. Climate change and evolution: Disentangling environmental and genetic responses. Mol Ecol. 2008;17:167–178. doi: 10.1111/j.1365-294X.2007.03413.x. [DOI] [PubMed] [Google Scholar]
- 32.Jorgensen C, et al. Ecology-Managing evolving fish stocks. Science. 2007;318:1247–1248. doi: 10.1126/science.1148089. [DOI] [PubMed] [Google Scholar]
- 33.Kuparinen A, Merilä J. Detecting and managing fisheries-induced evolution. Trends Ecol Evol. 2007;22:652–659. doi: 10.1016/j.tree.2007.08.011. [DOI] [PubMed] [Google Scholar]
- 34.Kuparinen A, Merilä J. The role of fisheries-induced evolution. Science. 2008;320:47–48. [PubMed] [Google Scholar]
- 35.Browman HI, Law R, Marshall CT. The role of fisheries-induced evolution. Science. 2008;320:47. doi: 10.1126/science.320.5872.47b. [DOI] [PubMed] [Google Scholar]
- 36.Gilmour AR, Gogel BJ, Cullis BR, Thompson R. ASReml User Guide Release 2.0. VSN International Ltd. Hempstead, UK: Hemel; 2006. [Google Scholar]
- 37.Merilä J, Sheldon BC, Kruuk LEB. Explaining stasis: Microevolutionary studies in natural populations. Genetica. 2001;112:199–222. [PubMed] [Google Scholar]
- 38.Lynch M, Walsh B. Genetics and Analysis of Quantitative Traits. Sunderland, MA: Sinauer; 1998. [Google Scholar]
- 39.R Development Core Team. Vienna: R Foundation for Statistical Computing; 2006. R: A language and environment for statistical computing. [Google Scholar]
- 40.Gelman A., Carlin J. B., Stern H. S., Rubin D. B. Bayesian Data Analysis. London: Chapman & Hall; 2004. [Google Scholar]
- 41.Thomas A, O'Hara R, Ligges U, Sturtz S. Making BUGS open. R News. 2007;6:12–17. [Google Scholar]
- 42.Lande R, Arnold SJ. The measurement of selection on correlated characters. Evolution. 1983;37:1210–1226. doi: 10.1111/j.1558-5646.1983.tb00236.x. [DOI] [PubMed] [Google Scholar]
- 43.Wolf JB, Wade MJ. On the assignment of fitness to parents and offspring: Whose fitness is it and when does it matter? J Evol Biol. 2001;14:347–356. [Google Scholar]
- 44.Hallett TB, et al. Why large-scale climate indices seem to predict ecological processes better than local weather. Nature. 2004;430:71–75. doi: 10.1038/nature02708. [DOI] [PubMed] [Google Scholar]
- 45.Wrat D, Salinger J, Bell R. Climate Variability and Change: Past Climate Variations over New Zealand. Auckland: National Climate Centre; 2006. [Accessed November 9, 2006]. Available at www.niwascience.co.nz/ncc/clivar/pastclimate#temperature. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.