Abstract
Splice-site mutations in the beta-globin gene can lead to aberrant transcripts and decreased functional beta-globin, causing beta-thalassemia. Triplex-forming DNA oligonucleotides (TFOs) and peptide nucleic acids (PNAs) have been shown to stimulate recombination in reporter gene loci in mammalian cells via site-specific binding and creation of altered helical structures that provoke DNA repair. We have designed a series of triplex-forming PNAs that can specifically bind to sequences in the human beta-globin gene. We demonstrate here that these PNAs, when cotransfected with recombinatory donor DNA fragments, can promote single base-pair modification at the start of the second intron of the beta-globin gene, the site of a common thalassemia-associated mutation. This single base pair change was detected by the restoration of proper splicing of transcripts produced from a green fluorescent protein-beta-globin fusion gene. The ability of these PNAs to induce recombination was dependent on dose, sequence, cell-cycle stage, and the presence of a homologous donor DNA molecule. Enhanced recombination, with frequencies up to 0.4%, was observed with use of the lysomotropic agent chloroquine. Finally, we demonstrate that these PNAs were effective in stimulating the modification of the endogenous beta-globin locus in human cells, including primary hematopoietic progenitor cells. This work suggests that PNAs can be effective tools to induce heritable, site-specific modification of disease-related genes in human cells.
Keywords: beta-thalassemia, gene correction, triplex-forming oligonucleotides, gene targeting
Mutations in the beta-globin gene that affect any stage in beta-globin biogenesis can cause beta-thalassemia. Identified mutations include single base pair changes that lead to frameshift mutations or changes in canonical sequences that affect mRNA stability and processing (1). As monogenic disorders, beta-thalassemia and sickle cell anemia have attracted substantial efforts directed at gene therapy by gene replacement, and there has been ongoing progress in this regard. In one approach specific to the thalassemias in which the genetic defect affects mRNA splicing, antisense oligonucleotides have been used to manipulate the splice site choice in beta-globin premRNA to prevent aberrant splicing. Restoration of proper beta-globin splicing has been demonstrated in human erythroid cells derived from beta-thalassemic patients, and in transgenic mouse models containing splicing mutations in the beta-globin gene (2, 3).
In this study, we use an antigene approach to correct a thalassemia-causing splice-site mutation at the level of chromosomal DNA in cultured cells, generating heritable, site-specific modification of the beta-globin gene. We have used peptide nucleic acids (PNAs), a class of triplex-forming molecules shown to be effective at provoking recombination and repair at chromosomal sites near PNA binding sites (4). PNAs contain standard nucleobases linked to a peptide-like backbone, and their advantages include resistance to nucleases and proteases, and increased stability of the PNA/DNA duplex compared with DNA/DNA. PNAs can bind DNA by strand invasion into the duplex, forming a so-called “D-loop” (5, 6). At homopurine/homopyrimidine stretches, the purine strand of the DNA duplex can be bound by two PNAs linked together (bis-PNA) to form a PNA/DNA/PNA triplex. One PNA segment of the bis-PNA serves as the Watson–Crick complementary strand to the purine strand of the target site, whereas the other PNA employs Hoogsteen bonding to the newly formed DNA/PNA duplex to form a PNA/DNA/PNA structure. There has been considerable interest in bis-PNAs both in antisense and antigene applications (4, 7–10).
A common beta-thalassemia mutation occurs at the first position of intron 2 in the human beta-globin gene (designated IVS2–1). A G→A mutation at this position disrupts normal splicing and causes thalassemia. We have identified several polypurine sequences in the vicinity of the IVS2–1 mutation that would be suitable sites for bis-PNA-mediated triple-helix formation. To test the ability of bis-PNAs to induce recombination and thereby direct sequence change at the IVS2–1 position of the beta-globin gene, we have designed a cellular assay that allows quantification of site-specific gene modification at IVS2–1. In this assay, IVS2 has been inserted into the green fluorescent protein (GFP) gene, such that GFP expression requires correct IVS2 splicing. Cells were engineered to contain a single copy of this GFP/IVS2 fusion gene with a G→A mutation at IVS2–1 that abrogates GFP fluorescence. GFP fluorescence can be restored by bis-PNA-stimulated correction of this G→A mutation by homologous recombination.
We report here that bis-PNA binding at homopurine sites within the IVS2 sequence can stimulate correction of the beta-globin IVS2–1 G→A mutation by short (51 nt) single-stranded donor DNAs in our cellular assay. The gene correction is shown to be stimulated by bis-PNAs designed to bind 35, 64, 194, 512, and 830 base pairs away from the position of the splice-site mutation. We also show that the gene correction is influenced by cell cycle phase, with the highest activity observed in cells in S phase. Finally, we demonstrate that bis-PNAs can provoke targeted modification within the endogenous beta-globin locus in human cells, including human primary hematopoietic progenitor cells, highlighting the potential of these DNA-reactive molecules to provoke heritable and long-term changes at disease-related sites in the human genome. To our knowledge, this demonstration of endogenous gene modification in human CD34+ cells using bis-PNAs has been previously undescribed.
Results
Development of Cellular Assay to Quantify Gene Conversion in the Beta-Globin Gene.
To quantify the ability of PNAs to induce gene correction at the human beta-globin locus, we generated a Chinese hamster ovary (CHO) reporter cell line containing a single copy of the GFP gene interrupted by the second intron of the human beta-globin gene, such that GFP expression requires proper splicing of this intronic sequence (8). A G→A mutation at position 1 of this intron (GFP/IVS2–1G→A) abolishes the normal donor splice site while activating a cryptic splice site, resulting in a GFP mRNA that retains an additional 47 nt of intron and yields no GFP protein expression (Fig. 1A). This GFP/IVS2–1G→A construct was stably transfected into CHO cells to create a reporter cell line (CHO-GFP/IVS2–1G→A). The insertion was directed into a single, predefined locus as previously described (11). A similar cell line, containing the GFP gene interrupted by a wild-type beta-globin intron 2 (GFP/IVS2wt), was also made. These CHO-GFP/IVS2wt cells exhibited high-level GFP expression in >95% of the cells as observed by FACS and fluorescence microscopy, whereas CHO-GFP/IVS2–1G→A cells demonstrated undetectable levels of background fluorescence (data not shown). Southern blots confirmed the integration of a single copy of the GFP/IVS2 constructs in the respective cell lines (data not shown).
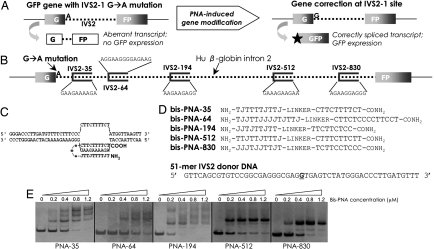
Fig. 1.
Strategy for targeted correction of beta-globin gene IVS2–1 mutation in mammalian cells. (A) Schematic of the cellular assay, in which intron 2 of the beta-globin gene, mutated at position 1, interrupts the GFP sequence. In cell lines made from this construct, this mutation leads to an aberrant transcript. GFP expression in these cells requires reversion of this IVS2–1 G→A mutation, via PNA-stimulated gene correction, thereby restoring proper splicing. (B) Relative positions and sequences of the homopurine bis-PNA binding sites in intron 2 of the beta-globin gene. (C) Putative strand invasion complex formed by bis-PNA-35 at the beta-globin IVS2–35 site, resulting in a PNA/DNA/PNA triplex structure. (D) Sequences of the bis-PNAs used in the current study. The sequence of the 51-mer IVS2 donor DNA corresponds to the nontemplate strand (single base pair change underlined). J, pseudoisocytidine. (E) Gel shift assays of the bis-PNAs binding to their respective target sites in a plasmid construct.
Bis-PNA Molecules for Targeted Correction of Beta-Globin IVS2–1.
Next, we identified homopurine sequences within the beta-globin IVS2 that can serve as binding sites for formation of PNA/DNA/PNA triple helices, based on the strategy that PNA-mediated triplex formation can stimulate recombination by altering local DNA topology and triggering DNA repair activity (4, 12). We identified five such sites within human beta-globin IVS2, ranging from IVS2–35 (and therefore, 34 base pairs from our mutation site of interest) to a sequence beginning at IVS2–830 (829 bp from the IVS2–1 mutation) (Fig. 1B). Bis-PNAs with ethylene glycol-containing linkers were designed to form PNA/DNA/PNA triplex-invasion complexes at these sites (Fig. 1C). For the bis-PNAs used in this study (Fig. 1D), cytosines in the Hoogsteen-bonding PNA strand were substituted with pseudoisocytosine to eliminate the requirement for N3 protonation of cytosine, thereby enhancing complex formation at neutral pH (13). Lysines were also conjugated to the PNA molecules to increase solubility and to enhance strand invasion by the molecules (14).
To assess bis-PNA binding to target sites, gel shift assays were carried out. Increasing concentrations of the various bis-PNAs were incubated with plasmids containing the relevant PNA binding sites (Fig. 1E). As shown in Fig. 1E, the PNAs can strand invade into and bind to their target sites in the plasmid substrate in vitro at concentrations as low as 200 nM. The multiple bands seen in the gel shift assay result from structural or stoichiometric isomers formed by bis-PNAs binding to duplex DNA (15). These isomers are formed at high PNA concentrations in vitro, but nonetheless provide evidence of high-affinity binding. PNAs of the reverse orientation, in contrast, showed no evidence of binding to IVS2 (data not shown).
Induced Genomic Correction at IVS2–1 by PNAs and Donor DNAs.
Next, the ability of bis-PNAs to stimulate gene correction in conjunction with short (51-mer), single-stranded “donor” oligonucleotides containing the desired reverting sequence change at IVS2–1 was tested in the CHO-GFP/IVS2–1G→A cell assay (Fig. 2A). The cells were mock transfected, transfected with IVS2 donor DNA alone, or transfected with IVS2 donor DNA plus bis-PNAs as indicated. Because PNAs and DNAs are differentially charged at physiologic pH, electroporation rather than cationic lipid comixture was chosen as the method of transfection. The cells were allowed 48 h to recover, after which they were inspected by fluorescence microscopy and analyzed by FACS for GFP expression, indicating correction at the IVS2–1G→A mutation and expression of a functional gene product. In initial experiments with asynchronous CHO cells, overall correction frequencies were low (data not shown). However, in cells synchronized in S phase, cotransfection of bis-PNA-35 with the donor DNA yielded correction at this site at an average frequency of 0.16% (Fig. 2A), more than three times that generated by transfection of donor DNA alone. Restoration of GFP fluorescence in the CHO-GFP/IVS2–1G→A reporter cell line was also stimulated using bis-PNAs targeting more distant sites in the IVS2 sequence of beta-globin. Bis-PNAs targeting IVS2–64, IVS2–194, IVS2–512, and IVS2–830 were all able to provoke correction of the IVS2–1 mutation at frequencies >0.1% (Fig. 2A), indicating that bis-PNA binding can provoke DNA metabolism over distances of several hundred base pairs.
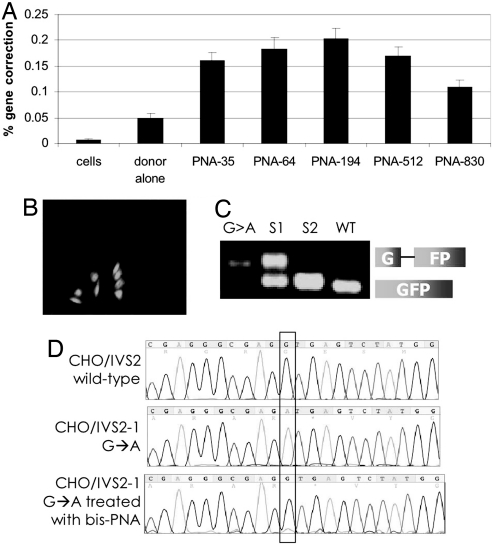
Fig. 2.
bis-PNAs induce correction of the IVS2–1 mutation by donor DNAs in the CHO-GFP/IVS2–1G→A reporter cell assay. (A) Gene correction was measured as the percentage of transfected cells expressing GFP by FACS analysis. CHO-GFP/IVS2–1G→A cells, synchronized in S phase, were transfected with no oligonucleotides (cells), with IVS2 donor DNA (donor alone), or with the indicated bis-PNA plus IVS2 donor DNA. Bar graph represents data collected from at least three independent experiments, and the error bars indicate standard errors. (B) Fluorescence micrograph of PNA and donor DNA-treated cells 48 h after transfection showing GFP-expressing CHO-GFP/IVS2–1G→A cells in a field of predominantly GFP-negative, uncorrected cells. (C) RT-PCR analysis of RNA harvested from GFP-positive CHO-GFP/IVS2–1G→A cells that were isolated by FACS. The IVS2–1 splicing mutation yields a longer RT-PCR product. G > A: untreated CHO-GFP/IVS2–1G→A cells; S1 and S2: first and second sort, respectively, of CHO-GFP/IVS2–1G→A cells treated with bis-PNA-35 and IVS2 donor DNA; WT: CHO-GFP/IVS2wt cells with wild-type and thus properly spliced transcript. Sorted CHO-GFP/IVSG→A cells that were treated with bis-PNA and IVS2 donor DNA show increasing proportions of RT-PCR product matching that of CHO-GFP/IVS2wt cells, indicating restoration of correct splicing. (D) Genomic DNA from sorted GFP-expressing cells maintained in culture for 1 month was harvested and sequenced to verify genomic modification at the target site. Chromograms are from untreated CHO-GFP/IVS2wt cells containing the wild-type intron (Top), untreated CHO-GFP/IVS2–1G→A cells with the IVS2–1 G→A mutation (Middle), and CHO-GFP/IVS2–1G→A cells treated with bis-PNA-35 and IVS2 donor DNAs, and sorted by FACS for GFP-positive cells (Bottom).
As further evidence that proper splicing was restored by PNA-induced gene correction, we used reverse-transcriptase (RT)-PCR to analyze RNA extracted from the cells that had acquired GFP fluorescence after treatment with bis-PNA and IVS2 donor DNA (Fig. 2B). The GFP-expressing cells were sorted by two rounds of FACS resulting in a stepwise enrichment of the GFP-positive cell population. The RT-PCR analysis of the sorted cells (Fig. 2C, lanes 2 and 3) was carried out in comparison with CHO cells containing the GFP gene with the wild-type beta-globin intron (Fig. 2C, lane 4, WT), and with untreated CHO cells containing the GFP gene with the mutated intron (Fig. 2C, lane 1, G > A). Note that the CHO-GFP/IVS2–1G→A cells with the mutant splice site have a larger RT-PCR product consistent with incorrect splicing (i.e., use of an aberrant splice site almost 50 nt from the IVS2–1 site). The CHO-GFP/IVS2wt cells with the wild-type intron yield a smaller RT-PCR product, indicative of correct splicing out of the entire intron. The sorted cell populations (Fig. 2C, lanes 2 and 3) show progressive increases in the proportion of the correct (smaller) RT-PCR product, in keeping with the restoration of the wild-type splice site sequence at the IVS2–1 position, and consistent with the observed, acquired GFP expression by microscopy and FACS. In addition, genomic DNA was extracted and sequenced from the sorted GFP-fluorescent cells that had been in culture for 1 month to demonstrate the presence and persistence of the expected single base pair change at the genomic level (Fig. 2D).
Treatment of cells with the lysomotropic agent chloroquine has been reported to enhance transfection of PNAs into cells (16, 17). Accordingly, we found that chloroquine treatment after electroporation augmented correction frequencies to >0.4%, whereas background fluorescence in mock-transfected cells did not change, consistent with an increase in effective delivery of the bis-PNAs and DNAs (Fig. 3A).
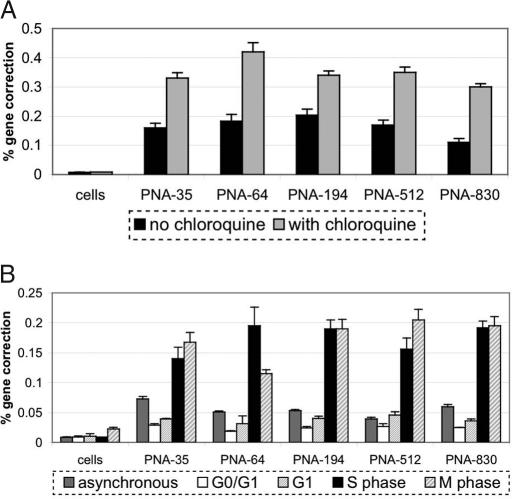
Fig. 3.
bis-PNA induction of gene correction at IVS2–1 is enhanced by chloroquine treatment and is affected by cell cycle phase. (A) CHO-GFP/IVS2–1G→A cells in S phase were electroporated with bis-PNAs and IVS2 donor DNAs and treated with or without chloroquine (as indicated) posttransfection. Two days later, the cells were analyzed by FACS to detect GFP expression, representing a gene correction event. (B) Cells were transfected with IVS2 donor DNA and the indicated bis-PNAs during various cell cycle stages (followed by chloroquine treatment), and then analyzed by FACS as above. Each experiment was performed in triplicate and standard errors are shown.
These results were obtained in cells transfected during S phase. To further explore the cell-cycle dependence of gene correction induced by bis-PNAs in the CHO-GFP/IVS2–1G→A reporter cells, we investigated bis-PNA activity in other stages of the cell cycle. We synchronized cells in various cell cycle stages using established CHO cell synchronization protocols (18), and transfected cells in each phase by electroporation, followed by chloroquine treatment. As shown in Fig. 3B, the introduction of bis-PNAs and IVS2 donor DNAs into cells synchronized in the G0/G1 or G1 stages stimulates gene correction at frequencies similar to that provoked by bis-PNA treatment of an asynchronous population. Compared with frequencies of gene correction in cells in G0, G1, or log-growth phases, treatment in S-phase cells yields higher levels of PNA-induced gene correction (Fig. 3B), consistent with previous data on favorable chromatin accessibility during S phase (18). Treatment of cells arrested in M phase also resulted in increased gene correction frequencies, similar to that seen with S phase targeting. This is not unexpected in light of published work indicating that the disintegration of the nuclear envelope during M phase results in increased gene transfer efficiencies regardless of transfection method (19–21).
Targeted Modification at the Endogenous Beta-Globin Locus.
We next sought to extend the results from the GFP reporter system to the endogenous beta-globin locus using two erythroid cell lines: human erythroleukemic K562 cells and mouse bone marrow cells containing the human beta-globin locus (beta-YAC BMCs) (22). We designed a 50-mer donor oligonucleotide homologous to the human beta-globin gene, except for a 6-bp change at positions 21–27 of the donor (corresponding to the last position of exon 2 to +6 of intron 2 of beta-globin) that was designed to abrogate proper splicing of the second intron and thereby introduce a thalassemia-like mutation (sequence change shown in Fig. 4A). We found that treatment of K562 or beta-YAC BMCs with this HBB donor DNA alone does lead to low-level genomic modification that is specifically detected by an allele-specific forward primer [Fig. 4B and see supporting information (SI) Fig. S1 for validation of assay specificity]. As controls, mock transfected cells show no PCR amplification using mutant-specific forward primers, indicating a lack of gene modification in these cells, whereas the use of wild-type-specific primers yields amplification products from all treated cell populations as expected, because only a small proportion of the HBB donor DNA-treated cells would be predicted to undergo a sequence change.
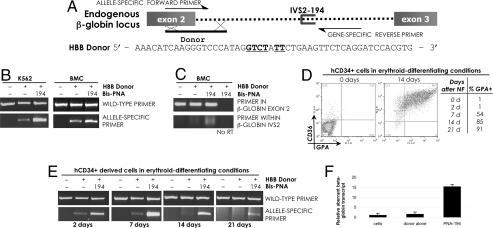
Fig. 4.
PNA-stimulated modification of the endogenous beta-globin gene in human cells. (A) Schematic of bis-PNA-194 designed to bind within intron 2 of the endogenous beta-globin gene, and of a single-stranded HBB donor DNA molecule designed for modification of the beta-globin gene at the exon 2/intron 2 boundary. The modification is a 6 nt substitution (underlined) that introduces a splicing mutation in the beta-globin gene. The HBB donor DNA sequence corresponds to the template strand of the target site. (B) K562 cells synchronized in S phase, or beta-YAC BMCs, were transfected with bis-PNA-194 and HBB donor DNA. Genomic DNAs from the transfected cells were analyzed by allele-specific PCR to detect the presence of the sequence change introduced by HBB donor DNA. (C) RT-PCR of RNA harvested from transfected BMCs, using primers that flank the human beta-globin IVS2 region (Upper), or that anneal to the 5′ portion of the IVS2 region (Lower). Beta-YAC BMCs treated with bis-PNA-194 and HBB donor DNA show evidence of beta-globin transcripts that retain a portion of IVS2; such transcripts are absent in mock transfected BMCs. “No RT” indicates the lack of reverse transcriptase enzyme in the RT-PCR. (D) Human CD34+ cells were nucleofected (NF) with bis-PNA-194 and HBB donor DNA and then placed in erythroid-differentiating conditions for up to 21 days. Initially <2% of these NF cells were positive for CD36 or GPA; by 21 days postnucleofection, >90% of the nucleofected cells were GPA-positive. (E) Genomic DNAs from cells harvested at the indicated times posttransfection were analyzed by allele-specific PCR to detect the introduced modification in the beta-globin gene. (F) Analysis of RNA prepared from human CD34+-derived cells maintained and differentiated in culture 21 days after transfection of bis-PNA-194 and donor DNA. Quantitative RT-PCR using primers that anneal to the IVS2 sequence reveal the presence of an aberrant beta-globin transcript in the bis-PNA- and HBB donor DNA-treated cells. Aberrant transcript levels were normalized to total beta-globin transcript in all samples.
Cotransfection of bis-PNA-194 in conjunction with HBB donor DNA into K562 cells or into beta-YAC BMCs was found to yield increased genomic modification at the beta-globin locus, compared with the effect of HBB donor DNA alone, as evidenced by the increased levels of amplification products using the primers specific for the introduced mutation (Fig. 4B). The specificity of the bis-PNA effect was demonstrated by the use of a control bis-PNA that does not bind the beta-globin gene; when transfected with HBB donor DNA, this control bis-PNA fails to stimulate gene modification beyond that mediated by donor DNA alone (Fig. S1C). We chose bis-PNA-194 for these studies because it induced the highest relative frequencies of gene correction in our CHO reporter assay. The K562 cells were transfected during S phase and treated with chloroquine posttransfection to enhance oligonucleotide uptake and delivery, whereas the beta-YAC BMCs were transfected as an asynchronous population. The PCR-based analysis performed here is not quantitative in the manner of the FACS analysis of GFP expression in the CHO reporter cells, however, and so precise quantitative conclusions cannot be drawn at this point. Nonetheless, although we cannot assign an absolute value to the frequency of modification, we can conclude that PNA-stimulated gene targeting can be achieved at the endogenous beta-globin locus in erythroid cells.
We then hypothesized that the PNA and HBB donor DNA-induced 6-bp mutation at the exon 2/intron 2 border would confer a change in beta-globin mRNA splicing, because the modification would remove a canonical donor splice site at this position and might thereby activate downstream, cryptic splice sites. Such aberrantly spliced beta-globin mRNAs would therefore retain a portion of IVS2 sequence. As beta-globin mRNA is undetectable in K562 cells, we used the beta-YAC BMC line previously demonstrated to follow human adult-pattern globin gene expression and regulation (22). The beta-YAC BMCs express human wild-type beta-globin mRNA, as shown by RT-PCR in Fig. 4C (Top). However, we were able to detect a new species of mRNA in the bis-PNA-194 and HBB donor treated cells that retained a portion of IVS2, as determined by use of primers that anneal specifically to the 5′ end of IVS2 (Fig. 4C, Bottom). This alternatively spliced beta-globin transcript was not detected in mock-transfected BMCs, and no amplification was seen when reverse transcriptase was omitted from the reaction, indicating that this RT-PCR product did not result from genomic DNA amplification. These results show that the PNA-stimulated genomic modification at the IVS2–1 splice site is also detectable at the transcript level, yielding a variantly spliced beta-globin mRNA in the beta-YAC BMCs.
For eventual clinical application, primary human hematopoietic stem and progenitor cells would be the appropriate targets for the bis-PNA-mediated beta-globin gene correction strategy discussed here. Therefore, we tested the ability of bis-PNA-194 and HBB donor DNA to introduce the 6-bp splice site mutation into the endogenous beta-globin gene of human CD34+ progenitor cells, harvested from peripheral blood in nonthalassemic donors. Less than 2% of the CD34+ cell population expressed mature erythroid (glycophorin A, CD36), myeloid (CD14, CD15), or megakaryocyte (CD41) markers at the time of transfection with bis-PNA-194 and HBB donor DNA (Fig. 4D, Fig. S2, and data not shown). Forty-eight hours after transfection and growth in cell expansion conditions, the treated CD34+ cells were then separated into three pools: (i) those grown in erythroid differentiation conditions, (ii) those grown in neutrophil differentiation conditions, or (iii) those harvested for genomic DNA analysis. As shown in Fig. 4E, gene modification can be detected in genomic DNA harvested from these cells 2 days after transfection with bis-PNA-194 and donor DNA. Moreover, at 21 days after the single administration of bis-PNA and donor DNA, when the majority of the cells have acquired specific lineage markers due to exposure to appropriate cytokines, the genomic modification is still detectable in the differentiated cells (erythroid lineage in Fig. 4E and neutrophil lineage in Fig. S2). At 21 days posttreatment, the erythroid-differentiated cells were found to express an aberrant splice form of beta-globin mRNA, as detected using quantitative RT-PCR with an IVS2-specific primer (Fig. 4F). Thus, the targeted gene modification was detected in both erythroid and neutrophil populations derived from the initial CD34+ cells, indicating that bis-PNA and donor DNA can be used for targeted gene modification in primary human hematopoietic cells. Because the initially treated CD34+ cells were predominantly a progenitor cell population, and some of the corrected cells were able to differentiate down the erythroid or neutrophil pathways, the data presented here suggest that the genomic DNA of individual myeloerythroid progenitors can undergo targeted modification using this approach. Alternatively, it is also possible that separate myeloid and erythroid progenitor cells may have been targeted. Regardless, the results demonstrate that primary human hematopoietic cells can be modified at the beta-globin locus by bis-PNAs and donor DNAs, and that the cells can give rise to differentiated, lineage-specific cell populations. In addition, the induced gene modification was seen to persist for up to 3 weeks and yielded a detectable change at the transcript level, further suggesting that functional gene modification of human progenitor cells can be achieved by PNA-mediated targeting.
Discussion
The work reported here demonstrates that triplex-forming bis-PNAs can stimulate site-specific modification within human beta-globin gene sequences in mammalian cells, including human hematopoietic progenitor cells. In CHO cells carrying a thalassemia-associated mutation at position IVS2–1, we demonstrate targeted correction of the mutation at not only the genomic level, but also the transcript and protein levels. Comparison of cells transfected with bis-PNAs and donor DNAs while in various phases of the cell cycle revealed that the highest levels of induced gene correction were achieved in cells transfected while in S phase. This is consistent with studies by Majumdar et al., suggesting that the open chromatin structure in S-phase cells affords increased accessibility of the chromosomal target site to DNA-binding molecules such as PNAs and DNA TFOs (18). In addition, unwound duplexes with partial single-stranded regions (as occur at replication forks) may be particularly conducive to PNA/DNA/PNA triplex formation by bis-PNAs. When cell cycle synchronization was also combined with postelectroporation treatment of cells with chloroquine, correction frequencies of up to 0.4% were obtained. This enhancement by chloroquine, which disrupts lysosomes and is thought to thereby release trapped molecules, indicates that the bioavailability of transfected PNAs and DNAs is a critical factor.
Targeted gene modification was achieved not only in the reporter construct in CHO cells but also at the exon 2/intron 2 boundary within the human beta-globin gene in K562 cells, beta-YAC BMCs, and human CD34+ progenitor cells, in the latter two cases causing a detectable change in beta-globin splicing. Moreover, we demonstrate that the PNA-mediated gene modification can persist for at least a month in the CHO cells and at least 3 weeks in cells derived from the human CD34+ cells. In addition, the PNA- and donor DNA-modified CD34+ cell population was able to give rise to both erythroid and myeloid lineages in which the beta-globin gene modification was still detectable. This persistence in CD34+-derived cells supports the feasibility of a transplantation therapy based on ex vivo treatment of mobilized peripheral blood progenitor cells in individuals with thalassemia.
Interestingly, bis-PNAs targeting polypurine sites as far away as 830 bp from the IVS2–1 mutation site were effective at stimulating gene correction by the 51 nt donor DNAs. The ability of triplex-forming bis-PNAs, and of triplex-forming DNA oligonucleotides (11), to stimulate recombination with donor DNAs has been shown to depend, in part, on the nucleotide excision repair pathway (4, 23). Triplex structures are recognized by the XPA/RPA (24) and XPC/hRad23 (25) damage recognition complexes, suggesting that triplexes constitute a helical alteration that can provoke DNA repair. In the case of the bis-PNAs used here, we tested whether knockdown of XPA could affect the stimulation of gene modification by bis-PNAs. We found that K562 cells, with levels of XPA reduced by siRNA knockdown, showed lower levels of induced gene modification in the bis-PNA-194 plus HBB donor DNA sample (Fig. S3), in concert with our previous mechanistic studies on triplex-provoked repair. However, the full mechanism(s) by which triplexes stimulate recombination, and by which they can do so over distances of several hundred base pairs [as reported here with bis-PNAs and previously with DNA TFOs (26)] remain to be elucidated.
Going forward, as evidence accumulates that site-specific gene targeting can be stimulated in primary human cells, additional efforts will be needed to optimize techniques to enhance the targeting of disease-related genes in progenitor cells of various relevant tissues. We provide important evidence here that human hematopoietic progenitor cells can be substrates for gene targeting by PNAs.
Materials and Methods
Oligonucleotides.
The sequences of the bis-PNAs used in this study are shown in Fig. 1D. Bis-PNAs with 8-amino-2,6-dioxaoctanoic acid linkers were either synthesized by us or purchased from Bio-Synthesis (Lewisville TX) or Applied Biosystems (Framingham MA). All bis-PNAs, purified by HPLC, had three terminal lysines on each end except for bis-PNA-512 and bis-PNA-830, which had three lysines attached to the central linker. Donor oligonucleotides 50 nt and 51 nt in length were synthesized by Midland Certified Reagent (Midland TX), 5′ and 3′ end protected by three phosphorothioate internucleoside linkages at each end and purified by reversed-phase HPLC.
Electrophoretic Mobility Shift Assays.
To test for formation of bis-PNA triplex-invasion complexes, plasmid DNA engineered to contain the IVS2 sequence (see SI Methods for plasmid construction) was incubated with increasing concentrations of bis-PNA in TE buffer (pH 7.4) with 10 mM KCl, and digested with flanking restriction enzymes to release a ≈110-bp fragment containing the plasmid/bis-PNA complexes. The products were separated by nondenaturing PAGE and visualized by silver staining.
Cell Lines.
CHO cell lines were generated using the CHO-Flp-In System (Invitrogen, Carlsbad CA) to contain a single copy of the EGFP gene, interrupted by the thalassemic IVS2–1G→A intron or its wild-type equivalent. Refer to SI Methods for details. The CHO cells were maintained in Ham's F12 media with 10% fetal bovine serum (FBS). K562 (American Type Culture Collection CCL-243, Manassas VA) and beta-YAC BMC (22) cells were maintained in RPMI with 10% FBS. Human CD34+ stem and progenitor cells were obtained from granulocyte colony-stimulating factor (G-CSF)-mobilized peripheral blood of normal healthy donors (Yale Center of Excellence in Molecular Hematology, Yale University, New Haven, CT), thawed from cryopreservation and maintained in StemSpan Serum-Free Expansion Media supplemented with StemSpan CC100 cytokine mixture (StemCell Technologies, Vancouver, Canada).
Cell Transfection.
Cells in 100 μl or 250 μl of complete media were electroporated or nucleofected (Amaxa Biosystems, Gaithersburg MD) in the presence of 12 μM donor DNA and 0 or 8 μM bis-PNA, as indicated. Electroporation conditions are provided in SI Methods. Cells were in logarithmic growth or synchronized at various cell cycle stages using established protocols (see SI Methods) at the time of transfection. Where indicated, the cells were exposed to 100 μM chloroquine in the media at 4 h after electroporation for a total of 4-h chloroquine treatment. Forty-eight hours after transfection, cells were collected for genomic DNA and/or RNA analysis. CD34+-derived cells were grown separately in erythroid- or neutrophil-differentiating conditions for up to 21 days (see SI Methods for culture conditions) and harvested for genomic DNA analysis by allele-specific PCR (see below), or stained with PE- or FITC-conjugate antibodies directed against lineage-specific markers for FACS analysis (see SI Methods).
RNA Extraction and RT-PCR.
RNA was isolated from CHO cells, beta-YAC BMCs, or human CD34+ cells using the Absolutely RNA Miniprep Kit (Stratagene, La Jolla CA). The RNA was treated with DNaseI, and cDNA was made using the SuperScript III RT-PCR kit (Invitrogen). Primers that anneal within the GFP sequence, flanking the IVS2 sequence, were used for CHO cell RNA analysis. For BMC and CD34+ cell beta-globin RT-PCR, primers were designed to anneal to exon sequences that flank IVS2. To detect transcripts that retain IVS2, a sense primer annealing to IVS2 sequence was used. RT-PCR reactions that omitted RT enzyme were used as controls to demonstrate lack of genomic contamination. Quantitative PCR was performed using Brilliant SYBR Green QPCR on an Mx3000P real-time PCR machine (Stratagene).
Sequence Analysis of Genomic DNA.
Corrected GFP-expressing CHO-GFP/IVS2–1G→A cells were isolated using a Becton Dickinson FACSVantage SE and maintained in F12 media with 10% FBS for 1 month. Genomic DNA was purified using the Wizard Genomic DNA Purification kit (Promega, Madison WI), and the GFP-IVS2 region was amplified by PCR using primers that flank the IVS2–1 site. The resulting PCR products were gel purified and sequenced.
Allele-Specific PCR.
Genomic DNA was harvested from K562 cells, BMCs, or CD34+-derived cells as above. Equal amounts of genomic DNA were subjected to allele-specific PCR, in which the 3′ end of the forward primer corresponds to the wild type or mutated sequence as introduced by the donor DNA (primer sequences and PCR conditions available upon request).
Supplementary Material
Acknowledgments.
We thank Bernard Forget (Yale University School of Medicine, New Haven, CT) for helpful discussions and members of the Glazer Lab for their assistance. The hCD34+ cells were obtained from the Yale Center of Excellence in Molecular Hematology (National Institutes of Health grant DK072442). This work is supported National Institutes of Health grants R01CA64186 and R01HL082655 (to P.M.G.) and MSTP training grant 5T32GM07205 (to J.Y.C.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0711793105/DCSupplemental.
References
- 1.Forget BG. Molecular basis of hereditary persistence of fetal hemoglobin. Ann N Y Acad Sci. 1998;850:38–44. doi: 10.1111/j.1749-6632.1998.tb10460.x. [DOI] [PubMed] [Google Scholar]
- 2.Sierakowska H, Sambade MJ, Agrawal S, Kole R. Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc Natl Acad Sci USA. 1996;93:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sazani P, et al. Systemically delivered antisense oligomers upregulate gene expression in mouse tissues. Nat Biotechnol. 2002;20:1228–1233. doi: 10.1038/nbt759. [DOI] [PubMed] [Google Scholar]
- 4.Rogers FA, Vasquez KM, Egholm M, Glazer PM. Site-directed recombination via bifunctional PNA-DNA conjugates. Proc Natl Acad Sci USA. 2002;99:16695–16700. doi: 10.1073/pnas.262556899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peffer NJ, et al. Strand-invasion of duplex DNA by peptide nucleic acid oligomers. Proc Natl Acad Sci USA. 1993;90:10648–10652. doi: 10.1073/pnas.90.22.10648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Footer M, Egholm M, Kron S, Coull JM, Matsudaira P. Biochemical evidence that a D-loop is part of a four-stranded PNA-DNA bundle. Biochemistry. 1996;35:10673–10679. doi: 10.1021/bi960486p. [DOI] [PubMed] [Google Scholar]
- 7.Mologni L, Nielsen PE, Gambacorti-Passerini C. In vitro transcriptional and translational block of the bcl-2 gene operated by peptide nucleic acid. Biochem Biophys Res Commun. 1999;264:537–543. doi: 10.1006/bbrc.1999.1548. [DOI] [PubMed] [Google Scholar]
- 8.Sazani P, et al. Nuclear antisense effects of neutral, anionic and cationic oligonucleotide analogs. Nucleic Acids Res. 2001;29:3965–3974. doi: 10.1093/nar/29.19.3965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhilina ZV, Ziemba AJ, Nielsen PE, Ebbinghaus SW. PNA-nitrogen mustard conjugates are effective suppressors of HER-2/neu and biological tools for recognition of PNA/DNA interactions. Bioconjug Chem. 2006;17:214–222. doi: 10.1021/bc0502964. [DOI] [PubMed] [Google Scholar]
- 10.Kim KH, Nielsen PE, Glazer PM. Site-specific gene modification by PNAs conjugated to psoralen. Biochemistry. 2006;45:314–323. doi: 10.1021/bi051379a. [DOI] [PubMed] [Google Scholar]
- 11.Knauert MP, Kalish JM, Hegan DC, Glazer PM. Triplex-stimulated intermolecular recombination at a single-copy genomic target. Mol Ther. 2006;14:392–400. doi: 10.1016/j.ymthe.2006.03.020. [DOI] [PubMed] [Google Scholar]
- 12.Bacolla A, Jaworski A, Connors TD, Wells RD. Pkd1 unusual DNA conformations are recognized by nucleotide excision repair. J Biol Chem. 2001;276:18597–18604. doi: 10.1074/jbc.M100845200. [DOI] [PubMed] [Google Scholar]
- 13.Egholm M, et al. Efficient pH-independent sequence-specific DNA binding by pseudoisocytosine-containing bis-PNA. Nucleic Acids Res. 1995;23:217–222. doi: 10.1093/nar/23.2.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuhn H, Demidov VV, Frank-Kamenetskii MD, Nielsen PE. Kinetic sequence discrimination of cationic bis-PNAs upon targeting of double-stranded DNA. Nucleic Acids Res. 1998;26:582–587. doi: 10.1093/nar/26.2.582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Krupnik OV, et al. Stability and transformations of bis-PNA/DNA triplex structural isomers. J Biomol Struct Dyn. 2004;21:503–512. doi: 10.1080/07391102.2004.10506944. [DOI] [PubMed] [Google Scholar]
- 16.Shiraishi T, Nielsen PE. Enhanced delivery of cell-penetrating peptide-peptide nucleic acid conjugates by endosomal disruption. Nat Protoc. 2006;1:633–636. doi: 10.1038/nprot.2006.92. [DOI] [PubMed] [Google Scholar]
- 17.Abes S, et al. Endosome trapping limits the efficiency of splicing correction by PNA-oligolysine conjugates. J Controlled Release. 2006;110:595–604. doi: 10.1016/j.jconrel.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 18.Majumdar A, et al. Cell cycle modulation of gene targeting by a triple helix-forming oligonucleotide. J Biol Chem. 2003;278:11072–11077. doi: 10.1074/jbc.M211837200. [DOI] [PubMed] [Google Scholar]
- 19.Yorifuji T, Tsuruta S, Mikawa H. The effect of cell synchronization on the efficiency of stable gene transfer by electroporation. FEBS Lett. 1989;245:201–203. doi: 10.1016/0014-5793(89)80221-2. [DOI] [PubMed] [Google Scholar]
- 20.Escriou V, Carriere M, Bussone F, Wils P, Scherman D. Critical assessment of the nuclear import of plasmid during cationic lipid-mediated gene transfer. J Gene Med. 2001;3:179–187. doi: 10.1002/jgm.174. [DOI] [PubMed] [Google Scholar]
- 21.Golzio M, Teissie J, Rols MP. Cell synchronization effect on mammalian cell permeabilization and gene delivery by electric field. Biochim Biophys Acta. 2002;1563:23–28. doi: 10.1016/s0005-2736(02)00369-3. [DOI] [PubMed] [Google Scholar]
- 22.Blau CA, et al. γ-Globin gene expression in chemical inducer of dimerization (CID)-dependent multipotential cells established from human β-globin locus yeast artificial chromosome (β-YAC) transgenic mice. J Biol Chem. 2005;280:36642–36647. doi: 10.1074/jbc.M504402200. [DOI] [PubMed] [Google Scholar]
- 23.Faruqi AF, Datta HJ, Carroll D, Seidman MM, Glazer PM. Triple-helix formation induces recombination in mammalian cells via a nucleotide excision repair-dependent pathway. Mol Cell Biol. 2000;20:990–1000. doi: 10.1128/mcb.20.3.990-1000.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vasquez KM, Christensen J, Li L, Finch RA, Glazer PM. Human XPA and RPA DNA repair proteins participate in specific recognition of triplex-induced helical distortions. Proc Natl Acad Sci USA. 2002;99:5848–5853. doi: 10.1073/pnas.082193799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thoma BS, Wakasugi M, Christensen J, Reddy MC, Vasquez KM. Human XPC-hHR23B interacts with XPA-RPA in the recognition of triplex-directed psoralen DNA interstrand crosslinks. Nucleic Acids Res. 2005;33:2993–3001. doi: 10.1093/nar/gki610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Knauert MP, et al. Distance and affinity dependence of triplex-induced recombination. Biochemistry. 2005;44:3856–3864. doi: 10.1021/bi0481040. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.