Abstract
H5N1 influenza viruses have spread extensively among wild birds and domestic poultry. Cross-species transmission of these viruses to humans has been documented in over 380 cases, with a mortality rate of ≈60%. There is great concern that a H5N1 virus would acquire the ability to spread efficiently between humans, thereby becoming a pandemic threat. An H5N1 influenza vaccine must, therefore, be an integral part of any pandemic preparedness plan. However, traditional methods of making influenza vaccines have yet to produce a candidate that could induce potently neutralizing antibodies against divergent strains of H5N1 influenza viruses. To address this need, we generated a consensus H5N1 hemagglutinin (HA) sequence based on data available in early 2006. This sequence was then optimized for protein expression before being inserted into a DNA plasmid (pCHA5). Immunizing mice with pCHA5, delivered intramuscularly via electroporation, elicited antibodies that neutralized a panel of virions that have been pseudotyped with the HA from various H5N1 viruses (clades 1, 2.1, 2.2, 2.3.2, and 2.3.4). Moreover, immunization with pCHA5 in mice conferred complete (clades 1 and 2.2) or significant (clade 2.1) protection from H5N1 virus challenges. We conclude that this vaccine, based on a consensus HA, could induce broad protection against divergent H5N1 influenza viruses and thus warrants further study.
The highly pathogenic H5N1 influenza viruses have caused outbreaks in poultry and wild birds since 2003 (1). These viruses have infected not only avian species but also over 383 humans, of which 241 cases proved to be fatal (http://www.who.int/csr/disease/avian_influenza/country/cases_table_2008_05_28/en/index.html). To date, the human cases have largely been infected by close contact with sick poultry, and the viruses isolated from them still show characteristics of avian influenza viruses (2). Nonetheless, serious concerns have been raised about the possibility of an avian influenza virus evolving to become transmissible among people, resulting in a global influenza pandemic (3, 4). In light of such a threat, new prophylactic and therapeutic strategies to combat human infections by H5N1 viruses are essential for influenza pandemic preparedness.
Over the past 60 years, vaccination has been the most effective method to protect the population against influenza infection (5). Conventional influenza vaccines can be divided into inactivated vaccines and live attenuated influenza vaccines. Virus-based influenza vaccines need to be amplified in the allantoic cavity of specific-pathogen-free (SPF) embryonated hens' eggs, with or without inactivation followed by purification. Inactivated influenza vaccines are safe and well-tolerated. When injected into muscle, they can induce significant protective neutralizing antibodies, with a clinical efficacy of 60–90% in children and adults (6). The live attenuated vaccine, on the other hand, is administered intranasally and can induce local neutralizing immunity and a cell-mediated immune response (7). Although effective, current egg-based vaccine strategies require a long timeline and a large supply of SPF eggs that could be threatened during an influenza pandemic that also affects poultry. Several approaches have been investigated to improve the vaccine manufacturing capacity. For example, reverse genetics has been used to generate reassortant viruses comprised of hemagglutinin (HA) and neuraminidase (NA) from target viruses and internal proteins from strain A/Puerto Rico/8/34 (8). Based on this technology, several groups, such as the Novartis Corporation and Baxter Biosciences, have developed cell-based strategies that use Vero or Madin-Darby Canine Kidney (MDCK) cells to amplify the viruses. Such cell-based production methods allow for faster and more flexible start-up of vaccine manufacturing (9, 10). Influenza vaccines based on inactivated virions have been shown to confer protection against H5N1 infection in animals. For example, inactivated H5N2 vaccines adjuvanated with oil emulsion have been widely used in chickens to protect against H5N1 viruses (11). A similar approach using H5N3 viruses, however, induced only limited protection in mice (12). Some clinical trials have shown that vaccines based on inactivated H5N1 virions can elicit serum-neutralizing antibodies against the homologous virus, but with limited activity against divergent viruses (10, 13).
In addition to virus-based vaccines, other approaches have been used to induce protective immunity against the key structural proteins of H5N1 viruses. Some of the promising approaches include recombinant protein vaccines (14), adenovirus-based technologies (15, 16), and DNA plasmids (17). These strategies, especially plasmid DNA vaccines, allow for easier manipulation and faster production when compared with traditional influenza vaccines. DNA vaccines, however, have not been as immunogenic as the traditional vaccines and thus show insufficient protection against virus infection (18). The main reason for this suboptimal immune response is inadequate gene delivery and gene expression when the DNA vaccine is given intramuscularly. Recent animal studies suggest that this obstacle could be overcome by the use of in vivo electroporation (EP), which results in higher transfection efficiency and protein expression (19).
The influenza virus is comprised of 11 proteins, and the HA is no doubt the major target for protective immunity. Antibodies against this surface glycoprotein can provide protection by blocking virus attachment and entry (20). Influenza viruses, however, are continuously evolving. The influenza proteins, including HA, can either change gradually through point mutations (antigenic drift) or change abruptly through reassortment with another divergent virus (antigenic shift) (21). As a result, the immunity generated against one HA is only protective against another virus strain that shares an antigenically related HA (22). The influenza vaccines thus need to be annually updated owing to antigenic changes of circulating strains.
In this study, we test the hypothesis that a consensus HA (CHA5) based on available H5N1 sequences incorporated into a DNA vaccine (pCHA5) can induce cross-protection against divergent H5N1 influenza viruses. Our results show that pCHA5 delivered intramuscularly via in vivo EP induced both humoral and cell-mediated immune responses without the use of adjuvants. In addition, sera from mice immunized with pCHA5 displayed robust and broad neutralizing activity against a panel of viruses that have been pseudotyped with HAs from divergent H5N1 viruses. Virus challenge experiments confirm that pCHA5 can protect mice from mortality and morbidity caused by several significantly divergent strains of H5N1 viruses. Altogether, our findings suggest that the EP delivery of pCHA5 represents one vaccine approach against H5N1 viruses worthy of further investigation. Such a strategy may be applicable to vaccine development for seasonal influenza as well.
Results
Design of a Prototype HA-Based DNA Vaccine.
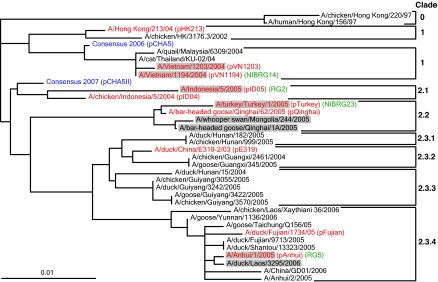
The HA sequences of circulating H5N1 viruses fell into phylogenetic clades and subclades previously designated by the World Health Organization (WHO). We hypothesized that a consensus HA-based vaccine would confer cross-protection against various H5N1 strains. A consensus HA (CHA5) was deduced from 467 HA sequences of H5N1 influenza viruses available in the GenBank in early 2006. Fig. 1 shows the phylogenetic position of CHA5, situated between clades 1 and 2.
Fig. 1.
Phylogenetic analysis of the H5N1 HA protein sequences. The circulating avian flu H5N1 viruses have been grouped into two major clades. The CHA5 sequence (blue) was generated in early 2006 and the CHA5II (blue) was generated in December 2007. The HAs used in this study are shown in red, with the plasmid names indicated in the first set of parentheses. The WHO-suggested vaccine strains are highlighted in gray. The reassortant influenza viruses used for HI and virus-challenge experiments are shown in green.
pCHA5 Conferred High-Level Expression of a Functional Protein.
Our preliminary studies showed that the original (unmodified) nucleotide sequence of influenza HA does not express the protein well in 293T cells (data not shown). The causes of the nonexpression include poor mRNA export to the cytoplasm (23) and inefficient translation because of differences in codon usage (24). Therefore, to improve HA expression in mammalian cells, the nucleotides of CHA5 were optimized with human codons (25), and the percentage of G/C was subsequently increased from 42% to 58% to provide better mRNA stability, processing, and nucleocytoplasmic transport (23). This HA gene was commercially synthesized and then inserted into the pVAX vector to create a consensus HA-based DNA vaccine (pCHA5). It was confirmed that pCHA5 expressed HA of the proper molecular weight in 293T cells [see supporting information (SI) Fig. S1A]. The function of CHA5 was subsequently verified by its ability to mediate hemadsorption of chicken red blood cells (cRBC) when expressed on transfected cells (see SI Materials and Methods and Fig. S1B).
pCHA5 Induced High-Antibody Titers and Elicited Cell-Mediated Immunity in BALB/c Mice.
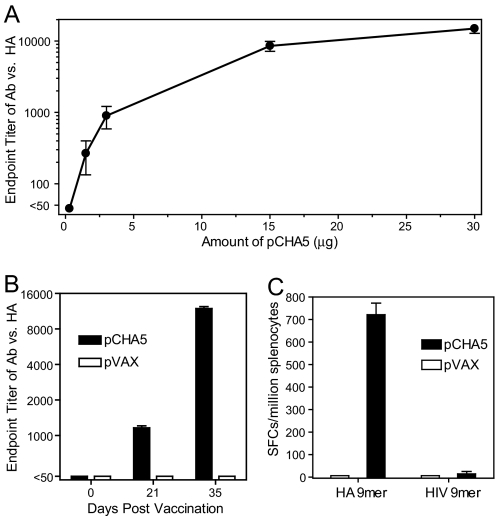
To assess the immunogenicity of pCHA5, female BALB/c mice were immunized with pCHA5 intramuscularly followed by EP at weeks 0 and 3. ELISA analysis of sera obtained 2 weeks after vaccination revealed a dose-dependent response in HA-specific antibodies (Fig. 2A). Two vaccinations with only 1.5 μg of pCHA5 elicited detectable HA-specific antibodies, with the endpoint titer between 1:100 and 1:400. The endpoint antibody titer reached ≈1:10,000 when 15 μg of pCHA5 was administered. One injection of 30 μg of pCHA5 induced a specific antibody titer between 1:400 and 1:1,600, whereas two injections generated a titer >1:10,000 (Fig. 2B).
Fig. 2.
HA-specific antibody responses and IFN-γ-secreting CD8+ T cell response to pCHA5 vaccinations. (A) BALB/c mice were vaccinated with increasing doses of pCHA5 by IM/EP. The mice were immunized on days 0 and 21, and the antiserum was collected on day 35 for ELISA analysis of HA-specific antibodies. (B) BALB/c mice were vaccinated with 30 μg of pVAX or pCHA5 on days 0 and 21 by IM/EP. The antiserum was collected on days 21 and 35 for analysis of HA-specific antibodies. (C) The splenocytes of immunized mice were obtained after two immunizations and assessed by an ELISpot assay using a specific peptide (HA 9mer) or an irrelevant peptide (HIV 9mer). P < 0.0001, pCHA5 vs. pVAX.
In addition to humoral response, cell-mediated immunity was also evaluated by using the ELISpot assay to monitor the ability of splenocytes of immunized mice to secrete cytokines after in vitro restimulation with a peptide homologous to a known CD8+ T cell epitope. As shown in Fig. 2C, the splenocytes from pCHA5-immunized mice were restimulated by a HA-specific peptide but not by an irrelevant HIV-1-specific 9mer. The number of IFN-γ-secreting cells increased from <10 to ≈750 spot-forming cells per million cells by immunization with pCHA5.
Antisera to pCHA5 Demonstrated Broad Neutralization Activity Against HA from Various H5N1 Viruses.
As a first step in assessing the potential of pCHA5 for broad-spectrum heterologous protection, the cross-reactivity of immunized sera to HAs from various H5N1 strains was evaluated by using the hemagglutination inhibition (HI) assay, where cross-reactivity is defined as a fourfold higher result than that of naïve sera (≦1:10) (22). Fig. 3A shows that the HI titers of the pCHA5 antiserum were 1:640 against NIBRG14 (clade 1) and NIBRG23 (clade 2.2) and perhaps slightly lower against RG5 (clade 2.3.4), but only 1:160 against RG2 (clade 2.1). To evaluate the cross-reactivity of the pCHA5 antiserum against additional HAs, we modified the traditional HI assay by using HA-expressing cells. The plasmids encoding HAs from a variety of H5N1 viruses were thus created by mutagenizing CHA5 at specific sites. Each individual plasmid was transfected into 293T cells, and the protein expression was confirmed by using immunoblot analysis (data not shown). After the cells were incubated with cRBCs in the presence of antiserum, it appeared that the pCHA5 antiserum caused a >50% reduction in hemadsorption to HA from clade 1 and clade 2.2 viruses (Fig. 3B). For HA from clade 2.1 and clade 2.3 viruses, varying degrees of reduction was observed.
Fig. 3.
Inhibition of hemagglutination and HA hemadsorption by antisera obtained from mice after receiving two injections of pCHA5. (A) The HI titer was determined with reassortant H5N1 viruses in the presence of CHA5 antisera. (B) Human 293T cells were transfected with pCHA5 or other HA genes from different H5N1 strains for 48 h and then incubated with cRBC in the presence or absence of pCHA5 antisera for 30 min. The absorbed red blood cells were quantitated by using the amount of hemoglobin detected by OD540. The percent of reduction was calculated as (ODw/o antiserum − ODw/antiserum)/(ODw/o antiserum − ODbackground) × 100%. Data were obtained from three independent experiments and presented as mean ± SEM.
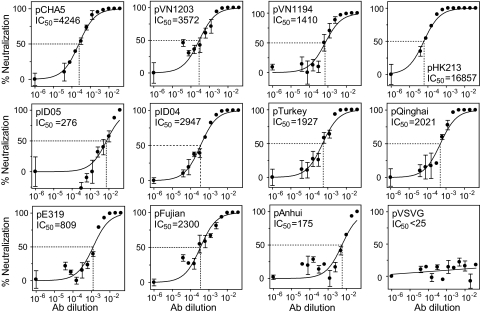
Development of an HA-Pseudotyped Virus Assay and Its Use in Evaluating the Neutralizing Activity of Antisera.
The neutralizing activity of the pCHA5 antiserum was further assessed, more quantitatively, by using a HIV pseudotype virus that carries a luciferase reporter gene and HA as its envelope protein. The entry of the HA-pseudotyped viruses into target cells was efficient, with luminescence values between 50,000 and 200,000 relative luminescence units. The IC50 of pCHA5 antiserum was well over a dilution of 1,000 against many divergent HAs, except for those from A/Indonesia/5/05 (clade 2.1), A/Duck/China/E319 (clade 2.3.2), and A/Anhui/1/05 (clade 2.3.4) (Fig. 4). The pCHA5 antiserum was clearly able to block the entry of the pseudotype virus enveloped with HA from all clade 1 and clade 2.2 H5N1 viruses and some of the H5N1 viruses in clades 2.1 and 2.3.
Fig. 4.
Neutralization of the infectivity of HA-pseudotyped viruses by antisera obtained from mice after receiving two injections of pCHA5. The IC50 is defined as the reciprocal of the antiserum dilution at which virus entry is 50% inhibited (dashed line). Data were collected from three independent experiments and presented as mean ± SEM.
pCHA5 Protected Mice from Lethal Challenges of Reassortant H5N1 Viruses.
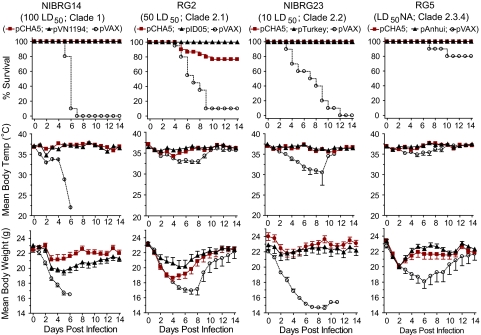
The benchmark of an influenza vaccine is protection against a lethal virus challenge. Again, BALB/c mice were immunized with two doses of pCHA5 or a plasmid encoding the homologous HA. After resting for 2 weeks, the immunized mice were intranasally challenged with a genetically modified H5N1 virus: NIBRG14, RG2, NIBRG23, or RG5. Immediately after virus challenge, all mice experienced a decrease in body temperature and weight (Fig. 5). The mice vaccinated with pCHA5 or the homologous HA gradually recovered after day 6 after a higher challenge dose (NIBRG14 and RG2) and after day 3 after lower challenge doses (NIBRG23 and RG5). The control mice vaccinated with the empty pVAX vector, however, continued to lose weight until they died. The only exception was the RG5 challenge, as the pathogenicity of this virus was too low to cause significant mortality in mice. Nonetheless, the data showed that pCHA5 did indeed protect mice from virus-induced morbidity, with complete protection observed against NIBRG14 and NIBRG23 and partial protection observed against RG2 (Fig. 5). The superiority of the DNA vaccine carrying the homologous HA was only seen in challenge experiments using RG2.
Fig. 5.
Vaccine protection against lethal challenges by H5N1 reassortant viruses. BALB/c mice were immunized with two injections of the consensus HA-based DNA vaccine (pCHA5), the homologous HA plasmid, or the control plasmid (pVAX). The immunized mice were intranasally challenged with the reassortant NIBRG14 (clade 1), RG2 (clade 2.1), NIBRG23 (clade 2.2), or RG5 (clade 2.3.4) (n = 10 per group). After virus challenge, survival, body weight, and body temperature were recorded for 14 days.
Discussion
Our goal was to investigate whether a consensus-HA-based DNA vaccine, pCHA5, given by EP delivery, can provide protection in mice against lethal challenges of H5N1 influenza viruses. A major obstacle in vaccine development against viruses such as HIV-1 and influenza is the extent of genetic diversity. For example, a clade-1 whole-virus vaccine raised a significant neutralizing antibody response against the homologous strain, but much less so against heterologous strains (10). One way to minimize the sequence diversity between a vaccine strain and circulating viruses is to create an artificial sequence to “centralize” the immunogenicity of the vaccine antigen (18, 26, 27). One approach is to construct, computationally, a consensus sequence that resides toward the middle of the viral phylogenetic tree. Another is to construct an “ancestral” sequence. Both approaches have been shown to elicit broader, more cross-reactive antibody responses than an antigen derived from a single strain (28). Initially we generated an HA ancestral sequence, but it was obviously biased toward the H5N1 viruses in clade 1, which have not been predominant in recent years (data not shown). We therefore deliberately favored the consensus approach to centralize the HA sequence. The deduced consensus sequence (CHA5) is indeed more centrally located in the phylogenetic tree (Fig. 1). We thus decided to use pCHA5 as our prototype HA DNA vaccine. A concern of using this consensus approach is that such an artificial sequence may not yield a structurally and functionally intact protein. However, this concern was alleviated when CHA5 was found to exhibit hemadsorption activity (see SI Materials and Methods and Fig. S1).
DNA vaccines have certain advantages over conventional influenza vaccines. Plasmid DNAs are readily amenable to modification once the circulating strain is identified. Large-scale production of DNA plasmids, albeit challenging, is definitely feasible. Although numerous studies have shown them to be promising, safe, and efficacious, DNA vaccines have not reached the market, and only a few are in clinical trials (29). One of the main problems of DNA vaccines has been inefficient gene delivery and expression. It is believed that the level of gene expression in a vaccinated host correlates with the induced immune response, i.e., low-level antigen expression elicits a low immune response. Although expression can be greatly improved by gene design and optimization, delivery remains a bottleneck for DNA vaccine development. It has been shown that DNA vaccine delivered by a cationic lipid can elicit a robust immune response (30). Another way to increase the cellular uptake of the DNA vaccine and thus to enhance antigen expression and immunogenicity is to administer a quick electrical field to muscle tissue where a DNA vaccine has been injected (31, 32). Because EP can efficiently augment humoral and cellular immune response, another advantage of EP is its vaccine dose-sparing effect. In fact, in vivo EP has reached the stage of human trials with a good tolerability profile to date (19).
It is generally thought that DNA vaccines can generate significant immune responses, but the responses are not as good as those elicited by virus-based vaccines or adjuvanted protein vaccines (29). In our study, however, the nonadjuvanted pCHA5 vaccine delivered by EP elicited robust humoral and cellular immune responses. The potential for broad-spectrum protection was first evaluated by an HI assay as well as by a neutralization assay using HA-pseudotyped viruses. As has been reported (33), results from such in vitro assays do correlate with in vivo protection against lethal virus challenges. Indeed, we observed that antisera to pCHA5 had broad activity in HI assays when tested against H5N1 viruses, although the activity against clades 2.1 and 2.3 was weaker or variable (Fig. 3). Similarly, antisera to pCHA5 showed substantial virus neutralization against many divergent H5N1 pseudotype viruses, but again with lower activity against some viruses in clade 2.1 and 2.3 (Fig. 4). This trend is reflected in the virus challenge experiments as well.
Our results show that immunization with pCHA5 protected mice from H5N1 viruses from clade 1 and clade 2.2 and to a lesser degree against the clade 2.1 and clade 2.3 viruses. Sequence analyses indicated that pCHA5 generally differs by only 3–5 amino acids from the HA of H5N1 viruses in clade 1 and clade 2.1, but by 16–18 amino acids from the HA of H5N1 viruses in clades 2.2 and 2.3. Nevertheless, the broad protection profile of pCHA5 demonstrated in our study compares quite favorably with the extent of protection observed for other H5N1 influenza vaccine candidates reported to date (18, 30, 34, 35).
H5N1 clade 2.3 viruses have been detected more frequently in southeast Asia in recent years (36). We know that fewer HA sequences from clade 2.3 viruses were available when CHA5 was designed in early 2006. We have since redesigned a new consensus sequence using all 1192 full-length sequences available by the end of 2007. As shown in Fig. 1, this second-generation consensus HA (CHA5 II) is now closer to clade 2 compared with the original CHA5. We are currently evaluating the immunogenicity and protection profile of the new DNA vaccine, pCHA5 II.
We have found that a consensus HA-based DNA vaccine delivered by EP can elicit robust cross-protective immune responses. However, it is difficult to separate the contribution of the EP administration from that of the consensus sequence approach. Intramuscular injection of a DNA vaccine generally elicits antibody titers 10-fold lower than EP delivery of the same vaccine (19, 32, 37). In our unpublished studies, we have noted that a nonconsensus H5N1 DNA vaccine induced less cross-protective neutralizing antibodies than pCHA5, suggesting that the consensus sequence is indeed contributing to the breadth of protection observed in this study.
Taken together, our strategy of employing the codon-optimized HA DNA vaccine and the EP delivery system has proven to be effective in eliciting significant protective immunity in mice against divergent strains of H5N1 viruses. Influenza viruses continuously undergo antigenic changes so that a consensus HA-based strategy is unlikely to provide a one-for-all solution that is universally desired. Nevertheless, DNA vaccines can be modified readily and rapidly to adjust to changes in the circulating viral populations. Others have reported that a DNA vaccine can protect ferrets and rhesus macaques from H5N1 virus challenge (30, 38). We believe the vaccine strategy described herein warrants further evaluation as a potentially fast, affordable, and stable prophylactic approach to combating H5N1 influenza viruses.
Materials and Methods
Viruses.
The attenuated reassortant H5N1 influenza viruses A/Vietnam/1194/2004/NIBRG14 and A/turkey/Turkey/01/2003/NIBRG23 were procured from the reference collection of the National Institute for Biological Standards and Control. A/Indonesia/5/2005/RG2 was obtained from the Center for Disease Control (CDC) in Indonesia. A/Anhui/1/2005/RG5 was provided by the U.S. CDC. All viruses were cultivated in the allantoic cavity of SPF embryonated eggs, titered in MDCK cells, and expressed as 50% tissue culture infective dose (TCID50). The 50% lethal dose (LD50) in mice was determined for each virus before use in challenge experiments.
Vaccine and Plasmid Construction.
All 467 full-length HA sequences from H5N1 viruses available in early 2006 were downloaded from the NCBI database and aligned by the ClustalW algorithm from the BioEdit program (version 7.0.9; Tom Hall, Ibis Biosciences, Carlsbad, CA) (39). The most conserved amino acid at each position was chosen to create a consensus HA (CHA5). The codons of CHA5 were optimized for mammalian expression by using human codons, and the G/C percentage of the resultant sequence was increased to 58% from 42% (Entelechon). The optimized coding sequence was then synthesized (Blue Heron Biotech) and built into the pVAX expression vector (Invitrogen) to create pCHA5, a prototype HA-based DNA vaccine. By using pCHA5 as a template, HAs from various H5N1 viruses (Fig. 1) were constructed by site-directed mutagenesis (Multi Site-Directed Mutagenesis Kit, Stratagene). To produce the coating antigen for determination of HA-specific antibodies, the polybasic cleavage site of CHA5 was changed to RER and amino acids 1–523 of CHA5 were fused to a human Fc (CHAedc-Fc). The sequence of CHAedc-Fc was built into the pVAX vector for expression in mammalian cells. Furthermore, the NA gene from influenza virus A/Vietnam/1194/2004 was also optimized, synthesized, and cloned into pVAX (pNA) for use in the production of HA-pseudotyped viruses.
Phylogenetic Analysis of HA Genes.
HA genes from WHO-recommended H5N1 vaccine strains and other representative H5N1 viruses were obtained from GenBank (NCBI). The HA genes were aligned as described, and the aligned sequences were used to generate a phylogenetic tree by using the default setting in the CustalW2 program provided by the European Molecular Biology Laboratory (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The tree was presented by the Phylip method and explored by TreeView 1.6 (Rod Page, University of Glasgow, Glasgow, UK).
Vaccination.
Five- to six-week-old female BALB/c mice were immunized with endotoxin-free pCHA5 or other HA constructs, prepared with GenElute HP Endotoxin-Free Plasmid Maxiprep Kit (Sigma-Aldrich). The immunization was performed at weeks 0 and 3 by an intramuscular administration of plasmids, followed immediately by the application of electrical stimulation (TriGrid Delivery System, Ichor Medical Systems) (19, 32). The spacing of the TriGrid electrode array was 2.5 mm, and the electrical field was applied at an amplitude of 250 V/cm of electrode spacing for six pulses totaling 40 msec duration applied over a 400 msec interval (i.e., a 10% duty cycle). After immunization, the mice were housed in the SPF animal facility at the Institute of Cell Biology, Academia Sinica, Taiwan. Two weeks after the second immunization, the immunized mice were bled for HA-specific antibody analysis and for neutralization assay or challenged with lethal doses of the viruses to assess vaccine efficacy. All animal experiments were evaluated and approved by the Institutional Animal Care and Use Committee of Academia Sinica.
Virus Challenge Experiments.
Two weeks after the second immunization, the immunized mice were anesthetized and intranasally challenged with a reassortant H5N1 virus (NIBRG14, RG2, NIBRG23, or RG5) in a final volume of 50 μl. The challenge doses were 250 TCID50 (100 LD50) for NIBRG14, 2 × 105 TCID50 (50 LD50) for RG2, 30,000 TCID50 (10 LD50) for NIBRG23, and 2 × 106 TCID50 for RG5. After infection, the mice were observed daily for 14 days, and survival and clinical parameters such as body weight and temperature were recorded. The challenge experiments were performed under biosafety level-2-plus enhancement conditions.
Statistical Analysis.
The animal experiments to evaluate immune responses were repeated at least three times (n = 3 per group), and the virus challenge studies were done at least twice (n = 10 per group). The response of each mouse was counted as an individual data point for statistical analysis. Data obtained from animal studies and pseudotyped virus neutralization assays were examined by using one-way ANOVA from GraphPad; differences were considered significant at P < 0.05.
Supplementary Material
Acknowledgments.
We thank the Taiwan CDC for providing the RG2 virus, Drew Hannaman and Ichor Medical Systems for help with the EP instrument, and Shih-Chi Wang for his assistance with mouse immunizations. This work was supported by the Taiwan Pandemic Influenza Vaccine Research and Development Program of the Taiwan Centers for Disease Control.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806901105/DCSupplemental.
References
- 1.Li KS, et al. Genesis of a highly pathogenic and potentially pandemic H5N1 influenza virus in eastern Asia. Nature. 2004;430:209–213. doi: 10.1038/nature02746. [DOI] [PubMed] [Google Scholar]
- 2.Ungchusak K, et al. Probable person-to-person transmission of avian influenza A (H5N1) N Engl J Med. 2005;352:333–340. doi: 10.1056/NEJMoa044021. [DOI] [PubMed] [Google Scholar]
- 3.Beigel JH, et al. Avian influenza A (H5N1) infection in humans. N Engl J Med. 2005;353:1374–1385. doi: 10.1056/NEJMra052211. [DOI] [PubMed] [Google Scholar]
- 4.Kuiken T, et al. Host species barriers to influenza virus infections. Science. 2006;312:394–397. doi: 10.1126/science.1122818. [DOI] [PubMed] [Google Scholar]
- 5.Osterholm MT. Preparing for the next pandemic. N Engl J Med. 2005;352:1839–1842. doi: 10.1056/NEJMp058068. [DOI] [PubMed] [Google Scholar]
- 6.Neuzil KM, et al. Immunogenicity and reactogenicity of 1 versus 2 doses of trivalent inactivated influenza vaccine in vaccine-naive 5–8-year-old children. J Infect Dis. 2006;194:1032–1039. doi: 10.1086/507309. [DOI] [PubMed] [Google Scholar]
- 7.Couch RB, Keitel WA, Cate TR. Improvement of inactivated influenza virus vaccines. J Infect Dis 176 Suppl. 1997;1:S38–S44. doi: 10.1086/514173. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann E, et al. Rescue of influenza B virus from eight plasmids. Proc Natl Acad Sci USA. 2002;99:11411–11416. doi: 10.1073/pnas.172393399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ghendon YZ, et al. Development of cell culture (MDCK) live cold-adapted (CA) attenuated influenza vaccine. Vaccine. 2005;23:4678–4684. doi: 10.1016/j.vaccine.2005.04.039. [DOI] [PubMed] [Google Scholar]
- 10.Ehrlich HJ, et al. A clinical trial of a whole-virus H5N1 vaccine derived from cell culture. N Engl J Med. 2008;359:2573–2584. doi: 10.1056/NEJMoa073121. [DOI] [PubMed] [Google Scholar]
- 11.Swayne DE, Lee CW, Spackman E. Inactivated North American and European H5N2 avian influenza virus vaccines protect chickens from Asian H5N1 high pathogenicity avian influenza virus. Avian Pathol. 2006;35:141–146. doi: 10.1080/03079450600597956. [DOI] [PubMed] [Google Scholar]
- 12.Lu X, et al. Cross-protective immunity in mice induced by live-attenuated or inactivated vaccines against highly pathogenic influenza A (H5N1) viruses. Vaccine. 2006;24:6588–6593. doi: 10.1016/j.vaccine.2006.05.039. [DOI] [PubMed] [Google Scholar]
- 13.Bresson JL, et al. Safety and immunogenicity of an inactivated split-virion influenza A/Vietnam/1194/2004 (H5N1) vaccine: Phase I randomised trial. Lancet. 2006;367:1657–1664. doi: 10.1016/S0140-6736(06)68656-X. [DOI] [PubMed] [Google Scholar]
- 14.Treanor JJ, et al. Safety and immunogenicity of a recombinant hemagglutinin vaccine for H5 influenza in humans. Vaccine. 2001;19:1732–1737. doi: 10.1016/s0264-410x(00)00395-9. [DOI] [PubMed] [Google Scholar]
- 15.Gao W, et al. Protection of mice and poultry from lethal H5N1 avian influenza virus through adenovirus-based immunization. J Virol. 2006;80:1959–1964. doi: 10.1128/JVI.80.4.1959-1964.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoelscher M, et al. Development of adenoviral-vector-based pandemic influenza vaccine against antigenically distinct human H5N1 strains in mice. Lancet. 2006;367:475–481. doi: 10.1016/S0140-6736(06)68076-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kong WP, et al. Protective immunity to lethal challenge of the 1918 pandemic influenza virus by vaccination. Proc Natl Acad Sci USA. 2006;103:15987–15991. doi: 10.1073/pnas.0607564103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laddy DJ, et al. Immunogenicity of novel consensus-based DNA vaccines against avian influenza. Vaccine. 2007;25:2984–2989. doi: 10.1016/j.vaccine.2007.01.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Luxembourg A, Evans CF, Hannaman D. Electroporation-based DNA immunization: Transition to the clinic. Exp Opin Biol Ther. 2007;7:1647–1664. doi: 10.1517/14712598.7.11.1647. [DOI] [PubMed] [Google Scholar]
- 20.Lee CW, Senne DA, Suarez DL. Development and application of reference antisera against 15 hemagglutinin subtypes of influenza virus by DNA vaccination of chickens. Clin Vaccine Immunol. 2006;13:395–402. doi: 10.1128/CVI.13.3.395-402.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nicholson KG, Wood JM, Zambon M. Influenza. Lancet. 2003;362:1733–1745. doi: 10.1016/S0140-6736(03)14854-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.World Health Organization. Antigenic and genetic characteristics of H5N1 viruses and candidate H5N1 vaccine viruses developed for potential use as human vaccines. [Accessed February 2008];2008 Available at http://www.who.int/csr/disease/avian_influenza/guidelines/H5VaccineVirusUpdate20080214.pdf.
- 23.Goetz RM, Fuglsang A. Correlation of codon bias measures with mRNA levels: Analysis of transcriptome data from Escherichia coli. Biochem Biophys Res Comm. 2005;327:4–7. doi: 10.1016/j.bbrc.2004.11.134. [DOI] [PubMed] [Google Scholar]
- 24.Holm L. Codon usage and gene expression. Nucleic Acids Res. 1986;14:3075–3087. doi: 10.1093/nar/14.7.3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang S, et al. Hemagglutinin (HA) proteins from H1 and H3 serotypes of influenza A viruses require different antigen designs for the induction of optimal protective antibody responses as studies by codon-optimized HA DNA vaccines. J Virol. 2006;80:11628–11637. doi: 10.1128/JVI.01065-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao F, et al. Consensus and ancestral state HIV vaccines. Science. 2003;299:1517–1518. doi: 10.1126/science.299.5612.1515c. [DOI] [PubMed] [Google Scholar]
- 27.Gaschen B, et al. Diversity considerations in HIV-1 vaccine selection. Science. 2002;296:2354–2360. doi: 10.1126/science.1070441. [DOI] [PubMed] [Google Scholar]
- 28.Kothe DL, et al. Ancestral and consensus envelope immunogens for HIV-1 subtype C. Virology. 2006;352:438–449. doi: 10.1016/j.virol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 29.Laddy DJ, Weiner DB. From plasmids to protection: A review of DNA vaccines against infectious diseases. Intl Rev Immunol. 2006;25:99–123. doi: 10.1080/08830180600785827. [DOI] [PubMed] [Google Scholar]
- 30.Lalor PA, et al. Plasmid DNA-based vaccines protect mice and ferrets against lethal challenge with A/Vietnam/1203/04 (H5N1) influenza virus. J Infect Dis. 2008;197:1643–1652. doi: 10.1086/588431. [DOI] [PubMed] [Google Scholar]
- 31.Liu J, Kjeken R, Mathiesen I, Barouch DH. Recruitment of antigen-presenting cells to the site of inoculation and augmentation of human immunodeficiency virus type 1 DNA vaccine immunogenicity by in vivo electroporation. J Virol. 2008;82:5643–5649. doi: 10.1128/JVI.02564-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luxembourg A, et al. Potentiation of an anthrax DNA vaccine with electroporation. Vaccine. 2008 doi: 10.1016/j.vaccine. 2008.03.064. [DOI] [PubMed] [Google Scholar]
- 33.Temperton NJ, et al. A sensitive retroviral pseudotype assay for influenza H5N1-neutralizing antibodies. Influenza Other Resp Diseases. 2007;1:105–112. doi: 10.1111/j.1750-2659.2007.00016.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kistner O, et al. Cell culture (Vero) derived whole virus (H5N1) vaccine based on wild-type virus strain induces cross-protective immune responses. Vaccine. 2007;25:6028–6036. doi: 10.1016/j.vaccine.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hoelscher M, et al. A broadly protective vaccine against globally dispersed clade 1 and clade 2 H5N1 influenza viruses. J Infect Dis. 2008;197:1185–1188. doi: 10.1086/529522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen H, et al. Establishment of multiple sublineages of H5N1 influenza virus in Asia: Implications for pandemic control. Proc Natl Acad Sci USA. 2006;103:2845–2850. doi: 10.1073/pnas.0511120103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang S, et al. The relative immunogenicity of DNA vaccines delivered by the intramuscular needle injection, electroporation and gene gun methods. Vaccine. 2008;26:2100–2110. doi: 10.1016/j.vaccine.2008.02.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Laddy DJ, et al. Heterosubtypic protection against pathogenic human and avian influenza viruses via in vivo electroporation of synthetic consensus DNA antigens. PLoS One. 2008;3:e2517. doi: 10.1371/journal.pone.0002517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hall TA. Bioedit: A user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucl Acid Symp Ser. 1999;41:95–98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.