Abstract
MexR is a MarR family protein that negatively regulates multidrug efflux systems in the human pathogen Pseudomonas aeruginosa. The mechanism of MexR-regulated antibiotic resistance has never been elucidated in the past. We present here that two Cys residues in MexR are redox-active. They form intermonomer disulfide bonds in MexR dimer with a redox potential of −155 mV. This MexR oxidation leads to its dissociation from promoter DNA, derepression of the mexAB–oprM drug efflux operon, and increased antibiotic resistance of P. aeruginosa. We show computationally that the formation of disulfide bonds is consistent with a conformation change that prevents the oxidized MexR from binding to DNA. Collectively, the results reveal that MexR is a redox regulator that senses peroxide stress to mediate antibiotic resistance in P. aeruginosa.
Keywords: antibiotic resistance regulation, thiol modification, MarR, oxidative stress, transcription regulation
Approximately 10% of all hospital-acquired infections are the result of Pseudomonas aeruginosa, a Gram-negative, opportunistic human pathogen. This pathogen infects immunosuppressed patients and causes high fatality rates in patients hospitalized with cancer, cystic fibrosis, and burns. P. aeruginosa is unique in that it possesses intrinsic resistance to a variety of antimicrobial agents. This intrinsic resistance results from a low outer membrane permeability and expression of specific drug efflux pumps such as those coded by the mexAB–oprM operon (1, 2). The tripartite pumps expressed from this operon couple the inner and outer membranes for extrusion of a range of antibiotics, including tetracycline, chloramphenicol, quinolones, novobiocin, macrolides, trimethoprim, and β-lactams (1–5). The MexR protein was found to be a negative regulator of this efflux system (Fig. 1A) (6). Expression of MexR represses mexAB–oprM and mexR itself, which is located upstream of mexA and is transcribed in the opposite direction. Mutation of mexR has been shown to activate the mexAB–oprM operon and confer the mutant strain increased resistance toward a range of different antibiotics (7–10). Some clinical and laboratory isolated multidrug-resistant (MDR) strains have been found to carry mutations in the mexR gene (5, 11). This regulation has also been suggested to link to bacterial virulence in P. aeruginosa (12). For example, the hyperexpression of the MexAB–OprM multidrug efflux system might lead to reduced expression of LasR–LasI-dependent virulence factors (13, 14).
Fig. 1.
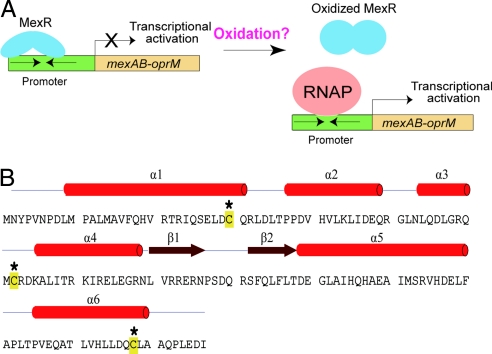
P. aeruginosa MexR. (A) MexR is a transcriptional repressor of the mexAB–oprM multidrug efflux operon. Oxidation stress may serve as a signal to activate MexR. (B) Sequence and secondary structure of MexR with 3 Cys residues highlighted.
MexR contains 147 aa (Fig. 1B) and functions as a transcriptional repressor. It is a member of the MarR family transcriptional regulators and forms a stable homodimer (15, 16). A commonly accepted mechanism for the MarR family proteins involves binding of a small-molecule drug to the dimer protein, which leads to dissociation of the protein from DNA (16, 17). This mechanism was proposed based on a binding assay that shows that salicylate (a simple structural homolog of the fluoro quinolone-type antibiotics) has a Kd of ≈0.5–1 mM to MarR (15). A footprinting experiment indicates that salicylate, at 5 mM, can cause MarR dissociation from its promoter site (15). The crystal structure of Escherichia coli MarR was obtained with two bound salicylates while 250 mM salicylate was used in the crystallization conditions (17).
The effect of high concentrations of salicylate on MarR is well established; however, the low Kd values measured between MarR and salicylate-type molecules cast doubts on the physiological importance of the direct binding mechanism. The mar regulon also responds to chloraphenicol, tetracycline, fluoroquinolones, and various other agents. It is hard to envision that a simple protein like MarR (142 aa) or MexR is able to recognize such a diverse range of structures under physiological conditions. Thus, alternative signaling pathways may exist for the MarR-type regulators. In fact, the signals and regulation mechanisms for most MarR family proteins are unknown.
The crystal structure of MexR was solved recently (18). Multiple conformations were observed from the structure. Based on the observed conformational flexibility, it was proposed that MexR responds to an unknown signal and changes its DNA affinity by reorienting its DNA-binding helices (18). We have been interested in elucidating the molecular mechanism of the MarR family proteins, in particular, the physiological signals that trigger responses of these proteins. Our recent work on MgrA, a MarR homolog that regulates virulence and antibiotic resistance in Staphylococcus aureus, showed that this protein uses a thiol oxidation-based mechanism to sense peroxide stress (19). Oxidation of a key Cys residue in MgrA led to dissociation of this protein from promoter DNA and activation of the mgrA regulon. We have also established that peroxide stress activates antibiotic resistance of S. aureus to fluoroquinoline and vancomycin through MgrA-mediated redox-sensing pathways. A recent study from Collins and coworkers (20) also suggested that many antibiotics exert their bactericidal effects by generating oxidative stress through hydroxyl radical formation. These results led us to test whether oxidative stress also serves as a signal to activate MexR.
Here, we show that two Cys residues in MexR are redox-active and form intermonomer disulfide bonds in MexR dimer under mild oxidation conditions. The disulfide-linked MexR dissociates from promoter DNA that activates the mexAB–oprM operon. The discovery not only provides a mechanistic understanding of an antibiotic resistance regulation in P. aeruginosa but also has significant implications for redox regulation in human pathogens in general.
Results
Oxidation Leads to Dissociation of MexR from DNA.
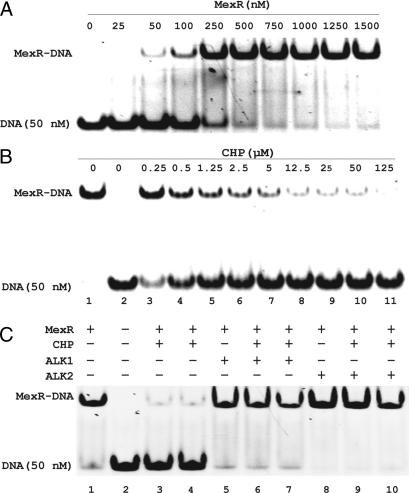
We first cloned, expressed, and purified MexR and examined its DNA-binding activity under reduced and oxidized conditions. Electrophoretic mobility shift assay (EMSA) was performed with MexR and a duplex DNA probe containing the promoter sequence recognized by MexR (21). Under reduced conditions, MexR formed a tight complex with DNA (Fig. 2A). Treatment of DNA-bound MexR (1 μM based on dimer concentration) with various concentrations (0.25–125 μM) of cumene hydroperoxide (CHP) for 60 min led to dissociation of MexR from DNA as shown in Fig. 2B. Even stoichiometric amounts of CHP (0.5–2.5 μM) dissociated a large portion of MexR from DNA (Fig. 2B). This important finding indicates that oxidation of MexR significantly weakens its DNA affinity, which may lead to derepression of the drug efflux systems as proposed in Fig. 1A.
Fig. 2.
Dissociation of oxidized MexR from DNA. (A) EMSA of MexR with a DNA probe (50 nM) that contains MexR-binding sequence showing formation of a MexR–DNA complex. (B) Oxidation of MexR (1 μM per dimer) with CHP (0–125 μM) led to dissociation of the protein from DNA. Lane 1, 1 μM MexR formed a complex with DNA; lane 2, DNA probe alone; lanes 3–11, oxidation of MexR with varying concentrations of CHP for 60 min before being applied to the shift assay. (C) Cys alkylation blocks oxidation-induced dissociation of MexR (1 μM per dimer) from DNA (50 nM). Lane 1, MexR–DNA complex; lane 2, DNA alone; lanes 3 and 4, oxidized MexR by CHP (80 and 800 μM) did not bind DNA; lane 5, MexR, alkylated with 80 μM phenyl vinyl sulfonate (ALK1), still bound DNA; lanes 6 and 7, MexR, alkylated with phenyl vinyl sulfonate (80 μM) for 50 min before treating with CHP (80 and 800 μM) for 90 min, still bound DNA; lane 8, same as lane 5 except with iodoacetamide (ALK2) as the alkylator; lanes 9 and 10, same as lanes 6 and 7 except with ALK2.
Three Cys residues, Cys-30, Cys-62, and Cys-138, exist in the sequence of MexR. To probe whether Cys oxidation is responsible for the observed dissociation of MexR from DNA, we pretreated 1 μM MexR with two Cys alkylators, phenyl vinyl sulfonate and iodoacetamide, for 50 min before mixing with 80 or 800 μM CHP for 90 min. Alkylation of Cys residues completely blocked the CHP-mediated dissociation of MexR from DNA (Fig. 2C), indicating that Cys oxidation is responsible for the observed MexR modification by CHP.
In the MgrA (19) and OhrR (22) cases, the protein uses a single Cys residue to sense peroxide stress (15, 23, 24). This Cys residue, located at the protein dimerization interface, is readily oxidized by peroxides. This Cys oxidation disrupts a hydrogen-bonding network at the protein dimer interface and abolishes the affinity of the protein for DNA. The crystal structure of MexR shows that its three Cys residues are not engaged in interactions between two monomers like that observed in MgrA (19).
Biochemical Assays for MexR Oxidation.
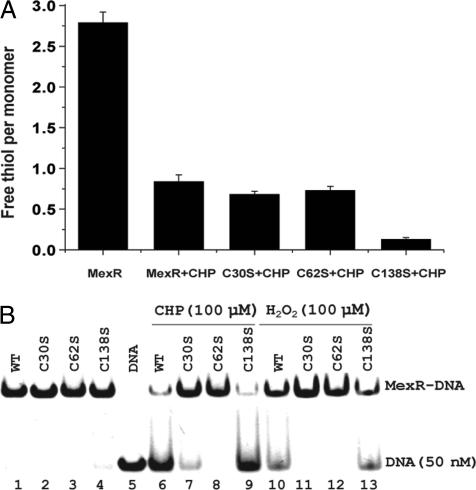
Next, biochemical experiments were performed on the wild-type MexR and three mutants, C30S, C62S, and C138S, with each Cys mutated to Ser, respectively. The mutant proteins were expressed and purified by following the same procedure as the wild-type MexR. Cys oxidation was quantified by measuring the free thiol content per MexR monomer by using the 5,5-dithiobis(2-nitrobenoic acid) (DTNB) assay (Fig. 3A) (25). The reduced wild-type MexR showed three thiols per monomer, as expected. After treating with 3 equivalents (per MexR monomer) of CHP, only one thiol per monomer was observed. Both C30S and C62S mutants showed one thiol per monomer after CHP treatment, indicating that both Cys-30 and Cys-62 are susceptible to oxidation. The C138S mutant exhibited close to 0 equiv of free thiol after treating with CHP, indicating that Cys-138 was not oxidized in the wild-type MexR.
Fig. 3.
Biochemical assays for MexR oxidation. (A) Quantification of free thiols in MexR in the reduced and oxidized protein. The reduced MexR contains three thiols per monomer. One equiv of thiol per monomer remained after treating MexR with 3 equiv of CHP for 1 h at room temperature. Both the C30S and C62S mutants showed 1 equiv of thiol remaining after the same CHP treatment; however, almost no thiol remained for the C138S mutant after it was treated with CHP. (B) EMSA (with 50 nM DNA) with various forms of MexR (0.5 μM). The wild-type (WT), C30S, C62S, and C138S all bound DNA when freshly purified. Treating these proteins with CHP (100 μM) or H2O2 (100 μM) for 40 min led to dissociation of the wild-type (lanes 6 and 10) and C138S mutant (lanes 9 and 13) from DNA, indicating that Cys-30 and Cye62 are involved in oxidation sensing. Lane 5 is control with DNA only.
EMSAs were conducted with the mutant proteins. The wild-type and all three mutant MexR proteins bind to the DNA probe used in Fig. 2A (21). However, treatment of DNA-bound MexR with either CHP or H2O2 led to dissociation of only the wild-type and C138S mutant proteins from DNA (Fig. 3B). The organic hydroperoxide CHP was more effective in causing dissociation of MexR from DNA than H2O2. This result confirms that both Cys-30 and Cys-62 are engaged in oxidation sensing.
Disulfide Bond Formation Between Cys-30 and Cys-62 in the Oxidized MexR.
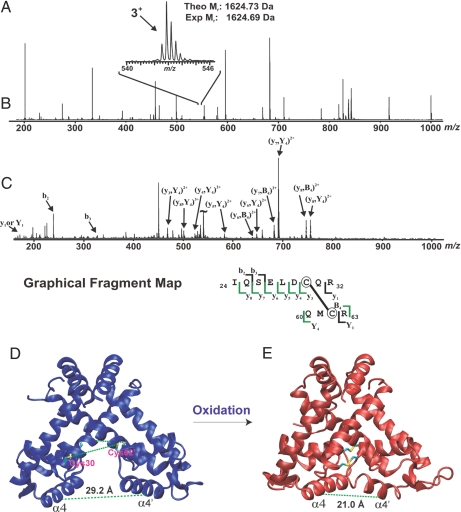
A close examination of the MexR structure eludes that Cys-30 from one monomer is close to Cys-62 from the other monomer (18). The two sulfurs atoms are ≈12–15 Å away from each other in a conformation that favors DNA binding. Because formation of disulfide bond has been observed in many oxidation-sensing/defense proteins (26–33), We wondered whether oxidation could generate an intermonomer disulfide bond in MexR. To reveal the nature of Cys oxidation in MexR, we used mass spectrometric mapping of the oxidized MexR. Purified MexR (35 μM) was oxidized by 350 μM CHP at 37°C for 1.5 h. Any remaining free thiol was blocked by treatment with excess iodoacetamide. The protein sample was run on a nonreducing, denaturing SDS/polyacrylamide gel. Formation of a covalently linked dimer band was clearly visualized on the gel with oxidized MexR [supporting information (SI) Materials and Methods and Fig. S1]. The dimer band was excised and in-gel-digested by trypsin. There was a clear 3+ charge state peak (m/z 542) corresponding to the disulfide-containing peptide of interest (cross-link between Cys-30 and Cys-62; theoretical molecular mass, 1,624.73 Da) by electrospray ionization (ESI)–quadrupole (Q)-TOF mass spectrometry (Fig. 4 A–C). This result provides strong evidence that oxidation of MexR results in the formation of disulfide bonds of the protein between Cys-30 and Cys-62.
Fig. 4.
Mass spectrometric mapping of disulfide bond in MexR and the simulated structure of oxidized MexR. (A) ESI-Q-TOF mass spectrum (m/z 200–1,000) of an unfractionated tryptic digestion. (Inset) The 3+ charge state (m/z 540–546) corresponding to the disulfide-containing peptide of interest (theoretical molecular mass, 1,624.73 Da). (B) MS/MS fragmentation of the 3+ charge state (m/z 542). (C) Graphical fragment map correlating the fragmentation ions to the sequence of the disulfide-containing peptide. The disulfide-linked cysteines are circled. (D) Structure of the reduced MexR dimer; the “open” CD dimer from PDB 1LNW (α4 and α4′ are the two DNA-binding helices). (E) Computationally predicted “closed” form with the disulfide bonded Cys-30–Cys-62 indicated.
We further probed formation of intermolecular cross-link between the two MexR monomers from in vivo samples. A mexR mutant strain Lys-1491 of P. aeruginosa PAO1 (8) complemented with a FLAG-tagged mexR cloned in a shuttle vector pMMB67HE (ATCC number 37623) was used (34). Cells were treated with 0, 0.15, or 0.5 mM CHP for 1 h before harvesting. Proteins extracts were separated by nonreducing, denaturing SDS/PAGE, and MexR bands were visualized by Western blotting with an anti-FLAG antibody. Oxidation activates expression of the FLAG-tagged MexR inside P. aeruginosa because this protein serves as the repressor of its own transcript (Fig. S2). This observation agrees with the oxidation activation mechanism of MexR. Importantly, we also observed increased formation of covalently linked MexR dimer in samples treated with CHP, confirming the intermonomer disulfide bond formation in MexR inside bacteria.
Computational Analysis of the Oxidized MexR.
The MexR C138S mutant was treated with glutathione disulfide GSSG, and the reaction mixture was subjected to separation by size exclusion chromatography. MexR protein elutes as a dimer, and the oxidized form elutes slower than the reduced form (Fig. S3), suggesting a conformational change of the protein upon oxidation. To determine how formation of an intermonomer disulfide bond between cysteines 30 and 62 could impact DNA binding, we constructed a model of the oxidized dimer as described in the SI Materials and Methods. Results obtained from simulations that started with the CD dimer (Fig. 4D) in Protein Data Bank (PDB) ID code 1LNW (18) are shown in Fig. 4E; similar results were obtained from the other three available structures of MexR and from alternate representations of the solvent (data not shown). For the case shown, formation of the disulfide bond decreases the spacing between the DNA binding α4-helices (as measured by the Cα–Cα distance for the Arg-73 residues in the α4-helices of the two monomers, marked by the dashed green line in Fig. 4 D and E) from 29.2 Å to 21.0 Å. Such a conformational change would prevent the DNA binding α4-helices from both binding in the major groove on the same face of the duplex DNA (18), thus significantly reducing the affinity of MexR for DNA.
Redox Potential of MexR.
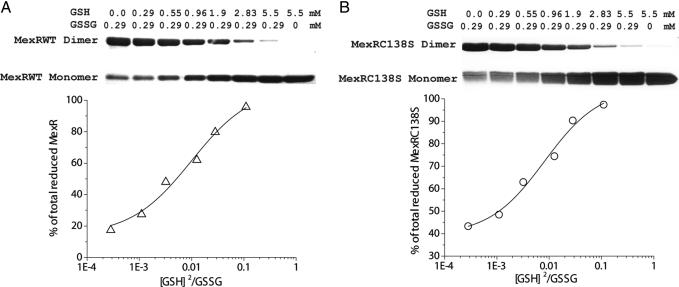
Oxidation of MexR is reversible as excess DTT retained formation of the MexR–DNA complex by EMSA (Fig. S4). The intermonomer disulfide bond in oxidized MexR can be reversed to free cystines by treatment with reducing agents, resembling that of E. coli OxyR (27, 30, 32, 33). We used a redox titration method (27) with a combination of GSH/GSSG to measure the redox potential of MexR. Both the wild-type and C138S mutant MexR were used in the experiment. The protein was incubated with defined concentrations of GSH/GSSG for 2 h to reach equilibrium. The reaction was quenched by addition of excess iodoacetamide to alkylate free thiols. The ratio of the oxidized versus reduced MexR was determined by denaturing SDS/PAGE that separates the covalently linked dimers from reduced monomers (Fig. 5A). These titration data were best fit by assuming a concerted two-monomer oxidation and reduction (Reaction 1), which is consistent with the observation that MexR is a dimer in solution. The equilibrium constant was obtained based on the titration data (2.7:1, GSH:GSSG ratio for 50% oxidation of MexR; Fig. 5A) and Reaction 1. The redox potential was calculated to be approximately −155 mV according to the Nernst equation (35). The measured redox potential is ≈100 mV higher than the estimated cytosol redox potentials for common bacteria (e.g., −280 mV for E. coli) (36), which is consistent with a role of MexR as an oxidation sensor inside P. aeruginosa (36). This value is also very much comparable with the redox potential measured for OxyR (−185 mV), the oxidative stress sensor in E. coil (27). It should be noted that this redox potential of MexR was measured in the absence of DNA. DNA binding could change the potential, which will be studied in the future.
Fig. 5.
Redox potential measurement for MexR. (A) Various concentrations of GSH and GSSG were incubated with 25 μM freshly purified MexR and C138S mutant protein at pH 7.4 and 37°C for 2 h. Reaction was stopped by adding excess iodoacetamide (19 mM). Protein samples were analyzed by nonreducing, denaturing SDS/PAGE. (B) Fitting of the titration data according to Reaction 1. Triangles (wild-type MexR) and circles (C138S mutant) correspond to experimental data, and the solid line is the theoretic fit. The redox potential was determined to be around −155 mV.
Activation of the mexAB–oprM Operon by Oxidation.
Previous studies from other laboratories have established MexR as the negative regulator of the mexAB–oprM operon (5, 6, 8, 11). Results presented here imply that oxidation of MexR inside P. aeruginosa would activate expression of this operon. The expression levels of mexB, oprM, and mexA were analyzed in PAO1 (wide-type), mexR mutant, and mexR mutant complemented with pMXR1–mexR (6). The wild-type PAO1 cells were also grown to exponential phase and treated with various concentrations of H2O2 or CHP for 30 min to probe the expression levels of the mexAB–oprM operon. Total RNAs were isolated and subjected to Northern blotting. The results show that mexB, oprM, and mexA were constitutively activated in mexR mutant and repressed in mexR mutant complemented with pMXR1–mexR (Figs. S5 and S6). These three genes were activated under peroxide stress conditions in PAO1 (Fig. S5), thus supporting the oxidation activation mechanism for MexR.
Antibiotic Sensitivity Assay.
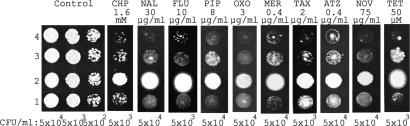
To provide further in vivo support to our proposed mechanism for MexR, we examined antibiotic sensitivity of four P. aeruginosa strains: wild-type PAO1 with empty plasmid pAK1900 as control, mexR mutant PAO1 with empty pAK1900, mexR mutant complemented with wild-type mexR in pAK1900 (pMXR1), and mexR mutant complemented with mexRC30SC62S (C30S/C62S double mutant) in pAK1900. Nine different antibiotics (NAL, nalidixic acid; FLU, Flumequine; PIP, pipemidic acid; OXO, oxolinic acid; MER, meropenem; TAX, cefotaxime; ATZ, aztreonam; NOV, novobiocin; TET, tetracycline) shown to be cleared through the mexAB–oprM efflux pump system (7–10) were chosen as well as CHP. As shown in Fig. 6, the mexR mutant strain was more resistant to all nine antibiotics and CHP than PAO1. These phenotypes could be complemented with wild-type mexR encoded in a shuttle plasmid expressed in the mexR mutant strain. Importantly, increased bacterial susceptibility to all of these antibiotics and CHP was observed when the shuttle plasmid encoding mexRC30SC62S was expressed in the mexR mutant strain compared with both wild-type PAO1 and complementary experiments with wild-type mexR. In addition, Northern blot analysis showed that transcripts of mexB and oprM had little change in the mexR mutant complemented with mexRC30SC62S when treated with CHP or antibiotics; they were activated in the mexR mutant complemented with wild-type mexR when treated with CHP or antibiotics (Fig. S5). These results firmly establish the thiol oxidation-based sensing and regulation mechanism for the MexR-mediated antibiotic resistance in P. aeruginosa.
Fig. 6.
Plate sensitivity assay. Four P. aeruginosa strains were treated by CHP and different antibiotics. Row 1, wide-type PAO1 with pAK1900 (the empty plasmid); row 2, mexR mutant with pAK1900; row 3, mexR mutant complemented by mexR in pAK1900; row 4, mexR mutant complemented with mexRC30SC62S in pAK1900. The control plate had no antibiotics.
Discussion
The MarR family of regulatory proteins plays important roles in antibiotic resistance and virulence regulation in bacteria (15–17, 19). Elucidating activation mechanisms of these proteins is important for understanding their regulation pathways and designing small molecules that may modulate their functions. Our previous work on MgrA, a MarR homolog in S. aureus, has demonstrated that peroxide stress serves as the signal to trigger the MgrA-mediated virulence and antibiotic resistance regulation (19). The MgrA/OhrR type proteins use a thiol oxidation-based sensing and regulation mechanism (19, 24, 37–41). A sole Cys residue is oxidized to sulfenic acid, which could be further modified inside a microbe to form mixed disulfides with small-molecule thiols (32, 42). This Cys oxidation disrupts the DNA-binding activity of the protein and activates the corresponding regulon. A recently discovered subfamily of OhrR was found to use the disulfide bond formation mechanism to sense organic hydroperoxide (37, 43), but its sequence homolog identified in P. aeruginosa (GenBank accession no. AAG06237) is different from MexR and has not been characterized.
As shown in this work, MexR forms intermonomer disulfide cross-link upon oxidation. Biochemical and cell-based experiments indicate that Cys-30 and Cys-62 are engaged in oxidation sensing and formation of intermonomer disulfide bonds. The oxidized MexR dissociates from promoter DNA that leads to activation of the mexAB–oprM operon. This protein senses peroxide stress formed inside bacterium upon challenging with antibiotics (20) and activates drug efflux defensive system. The measured redox potential for MexR supports its role as an oxidative stress sensor inside bacterial cells. Computational simulation of the disulfide cross-linked MexR shed further light into the detailed mechanism. The crystal structure of MexR has been solved in different conformations (18). In one conformation the protein dimer orients its two DNA-binding helices in a perfect spacing to engage in interaction with the major groove on the same face of a B-form duplex DNA. In a different conformation, the two DNA-binding helices were reoriented, and the spacing between them was reduced, and this form was deemed unfavorable for DNA binding. Starting from the DNA-binding conformation of MexR (Fig. 4D), we built in intermonomer disulfide bonds. The simulated cross-linked structure shows that the two DNA-binding helices realign, and the spacing between them is significantly reduced (Fig. 4E). As a result, the conformation is not expected to be DNA binding-competent.
In summary, we have shown that S. aureus, a Gram-positive pathogen, uses an oxidation-sensing mechanism to regulate antibiotic resistance and virulence (19). P. aeruginosa is a Gram-negative pathogen, and yet, peroxide stress also serves as a signal to activate the antibiotic resistance regulator MexR. Oxidative stress could serve as a general signal for other MarR-type proteins. These results agree with the observation that many antibiotics produce oxidative stress inside bacteria (20). Our study also shows that thiol-based oxidation sensing and regulation are prevalent in human pathogens to counter antibiotic treatments.
Materials and Methods
Construction, Expression, and Purification of MexR.
The mexR gene was amplified by PCR from genomic DNA of P. aeruginosa PAO1 and subcloned into pET-30a vector (Novagen) between NdeI and XhoI sites. The three mutations, MexRC30S, MexRC62S, and MexRC138S, were obtained by using QuikChange II site-directed mutagenesis kit (Stratagene).
E. coli BL21(DE3) cells harboring the constructed plasmid were grown in LB medium containing kanamycin (30 μg/ml) at 37°C with shaking at 250 rpm. Cells were grown to an A600 nm of 0.6 and induced with 1 mM IPTG for an additional 4 h at 30°C. Cells were harvested by centrifugation and stored at −80°C. All subsequent steps were performed at 4°C. A cell pellet (0.5 liter) was suspended in 15 ml of lysis buffer [10 mM Tris (pH 6.8), 10 mM 2-mercaptoethanol (BME), 10% glycerol], disintegrated by sonication, and centrifuged at 17,500 × g for 30 min. Proteins in the supernatant were loaded onto an SFF column (bed volume 20 ml; Amersham Bioscience), equilibrated with buffer A [10 mM Tris (pH 6.8), 10 mM BME], washed with 100 mM NaCl in buffer A, and eluted with 250 mM NaCl in buffer A. Peak fractions were applied to a heparin column equilibrated with buffer A, washed with 300 mM NaCl in buffer A, and eluted with 600 mM NaCl in buffer A. The fractions containing MexR were concentrated and loaded onto a Superdex-200 gel filtration column with a running condition of 10 mM Tris (pH 7.4), 100 mM NaCl, and 5 mM BME. The purified MexR fractions were collected and concentrated for further experiments (>95% purity as estimated by SDS/PAGE).
EMSA.
A 28-mer duplex DNA (5′-ATTTTAGTTGACCTTATCAACCTTGTTT-3′ annealed to its complementary strand) containing the mexRAB–oprM promoter sequence was used for the assay by using 8% nondenaturing polyacrylamide gel made with 10 mM Tris buffer containing 50 mM KCl, 5 mM MgCl2 and 10% glycerol. Gels were run in 0.5× TB [50 mM Tris, 41.5 mM borate (pH 8.0)] at 85 V at room temperature and stained in a 10,000-fold diluted SYBR Gold nucleic acid staining solution (Molecular Probes) for 5 min. The DNA bands were visualized with UV light at 254 nm.
Redox Titration.
Samples of purified MexR (25 μM) were incubated with different ratios of GSH/GSSG (0–5.5 mM for GSH; 0.29–0 mM for GSSG); 100 mM GSH and GSSH stock solutions were made by using 1 M Tris·HCl (pH 7.4) in 50 mM Tris·HCl (pH 7.4) with 100 mM NaCl at 37°C for 2 h. Free thiols were then alkylated by treating with 19 mM iodoacetamide for 30 min at 37°C. The protein samples were analyzed by nonreducing, denaturing SDS/PAGE. Intensities of protein bands were measured by ImageJ 1.40 version (National Institutes of Health), and the ratios of the oxidized versus reduced protein were used for calculating the redox potential (27, 36).
Other Procedures.
Detailed procedures are available in SI Materials and Methods.
Supplementary Material
Acknowledgments.
We thank Dr. Satoshi Nishida and the reviewers for suggestions. The strains of Pseudomonas aeruginosa PAO1, mexR mutant PAO1, and plasmids pAK1900 and pMXR1 were generous gifts from Dr. Keith Poole from Queen's University, Kingston, ON, Canada. This work was supported by the W. M. Keck Foundation (to C.H.), National Institute of Allergy and Infectious Diseases/National Institutes of Health Grant R01 AI074658 (to C.H.), National Science Foundation Award MCB-0547854 (to A.R.D.), and the University of Chicago.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/cgi/content/full/0803391105/DCSupplemental.
References
- 1.Poole K, Krebes K, McNally C, Neshat S. Multiple antibiotic-resistance in Pseudomonas aeruginosa: Evidence for involvement of an efflux operon. J Bacteriol. 1993;175:7363–7372. doi: 10.1128/jb.175.22.7363-7372.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gotoh N, Tsujimoto H, Poole K, Yamagishi JI, Nishino T. The outer-membrane protein OprM of Pseudomonas Aeruginosa is encoded by oprk of the mexA–mexB–oprK multidrug-resistance operon. Antimicrob Agents Chemother. 1995;39:2567–2569. doi: 10.1128/aac.39.11.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li XZ, Nikaido H, Poole K. Role of MexA–MexB–OprM in antibiotic efflux in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:1948–1953. doi: 10.1128/aac.39.9.1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Köhler T, et al. Multidrug efflux in intrinsic resistance to trimethoprim and sulfamethoxazole in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1996;40:2288–2290. doi: 10.1128/aac.40.10.2288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Srikumar R, Li XZ, Poole K. Inner membrane efflux components are responsible for β-lactam specificity of multidrug efflux pumps in Pseudomonas aeruginosa. J Bacteriol. 1997;179:7875–7881. doi: 10.1128/jb.179.24.7875-7881.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Poole K, et al. Expression of the multidrug resistance operon mexA–mexB–oprM in Pseudomonas aeruginosa: mexR encodes a regulator of operon expression. Antimicrob Agents Chemother. 1996;40:2021–2028. doi: 10.1128/aac.40.9.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ziha-Zarifi I, Llanes C, Köhler T, Pechère JC, Plesiat P. In vivo emergence of multidrug-resistant mutants of Pseudomonas aeruginosa overexpressing the active efflux system MexA–MexB–OprM. Antimicrob Agents Chemother. 1999;43:287–291. doi: 10.1128/aac.43.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Srikumar R, Paul CJ, Poole K. Influence of mutations in the mexR repressor gene on expression of the MexA–MexB–OprM multidrug efflux system of Pseudomonas aeruginosa. J Bacteriol. 2000;182:1410–1414. doi: 10.1128/jb.182.5.1410-1414.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Poole K, Srikumar R. Multidrug efflux in Pseudomonas aeruginosa: Components, mechanisms, and clinical significance. Curr Top Med Chem. 2001;1:59–71. doi: 10.2174/1568026013395605. [DOI] [PubMed] [Google Scholar]
- 10.Köhler T, Pechère JC. In vitro selection of antibiotic resistance in Pseudomonas aeruginosa. Clin Microbiol Infect. 2001;7:7–10. doi: 10.1046/j.1469-0691.2001.00066.x. [DOI] [PubMed] [Google Scholar]
- 11.Masuda N, Sakagawa E, Ohya S. Outer membrane proteins responsible for multiple drug resistance in Pseudomonas aeruginosa. Antimicrob Agents Chemother. 1995;39:645–649. doi: 10.1128/AAC.39.3.645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hirakata Y, et al. Multidrug efflux systems play an important role in the invasiveness of Pseudomonas aeruginosa. J Exp Med. 2002;196:109–118. doi: 10.1084/jem.20020005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evans K, et al. Influence of the MexAB–OprM multidrug efflux system on quorum sensing in Pseudomonas aeruginosa. J Bacteriol. 1998;180:5443–5447. doi: 10.1128/jb.180.20.5443-5447.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pearson JP, Van Delden C, Iglewski BH. Active efflux and diffusion are involved in transport of Pseudomonas aeruginosa cell-to-cell signals. J Bacteriol. 1999;181:1203–1210. doi: 10.1128/jb.181.4.1203-1210.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin RG, Rosner JL. Binding of purified multiple antibiotic resistance repressor protein (MarR) to mar operator sequences. Proc Natl Acad Sci USA. 1995;92:5456–5460. doi: 10.1073/pnas.92.12.5456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Alekshun MN, Levy SB. The mar regulon: Multiple resistance to antibiotics and other toxic chemicals. Trends Microbiol. 1999;7:410–413. doi: 10.1016/s0966-842x(99)01589-9. [DOI] [PubMed] [Google Scholar]
- 17.Alekshun MN, Levy SB, Mealy TR, Seaton BA, Head JF. The crystal structure of MarR, a regulator of multiple antibiotic resistance, at 2.3 angstrom resolution. Nat Struct Biol. 2001;8:710–714. doi: 10.1038/90429. [DOI] [PubMed] [Google Scholar]
- 18.Lim D, Poole K, Strynadka NCJ. Crystal structure of the MexR repressor of the mexRAB–oprM multidrug efflux operon of Pseudomonas aeruginosa. J Biol Chem. 2002;277:29253–29259. doi: 10.1074/jbc.M111381200. [DOI] [PubMed] [Google Scholar]
- 19.Chen PR, et al. An oxidation-sensing mechanism is used by the global regulator MgrA in Staphylococcus aureus. Nat Chem Biol. 2006;2:591–595. doi: 10.1038/nchembio820. [DOI] [PubMed] [Google Scholar]
- 20.Kohanski MA, Dwyer DJ, Hayete B, Lawrence CA, Collins JJ. A common mechanism of cellular death induced by bactericidal antibiotics. Cell. 2007;130:797–810. doi: 10.1016/j.cell.2007.06.049. [DOI] [PubMed] [Google Scholar]
- 21.Evans K, Adewoye L, Poole K. MexR repressor of the mexAB–oprM multidrug efflux operon of Pseudomonas aeruginosa: Identification of MexR-binding sites in the mexA–mexR intergenic region. J Bacteriol. 2001;183:807–812. doi: 10.1128/JB.183.3.807-812.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuangthong M, Atichartpongkul S, Mongkolsuk S, Helmann JD. OhrR is a repressor of. ohrA, a key organic hydroperoxide resistance determinant in Bacillus subtilis. J Bacteriol. 2001;183:4134–4141. doi: 10.1128/JB.183.14.4134-4141.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong M, Fuangthong M, Helmann JD, Brennan RG. Structure of an OhrR–ohrA operator complex reveals the DNA-binding mechanism of the MarR family. Mol Cell. 2005;20:131–141. doi: 10.1016/j.molcel.2005.09.013. [DOI] [PubMed] [Google Scholar]
- 24.Fuangthong M, Helmann JD. The OhrR repressor senses organic hydroperoxides by reversible formation of a cysteine–sulfenic acid derivative. Proc Natl Acad Sci USA. 2002;99:6690–6695. doi: 10.1073/pnas.102483199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Riddles PW, Blakeley RL, Zerner B. Reassessment of Ellman's reagent. Methods Enzymol. 1983;91:49–60. doi: 10.1016/s0076-6879(83)91010-8. [DOI] [PubMed] [Google Scholar]
- 26.Gilbert HF. Molecular and cellular aspects of thiol disulfide exchange. Adv Enzymol Relat Areas Mol Biol. 1990;63:69–172. doi: 10.1002/9780470123096.ch2. [DOI] [PubMed] [Google Scholar]
- 27.Zheng M, Aslund F, Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science. 1998;279:1718–1721. doi: 10.1126/science.279.5357.1718. [DOI] [PubMed] [Google Scholar]
- 28.Demple B. Signal transduction: A bridge to control. Science. 1998;279:1655–1656. doi: 10.1126/science.279.5357.1655. [DOI] [PubMed] [Google Scholar]
- 29.Hutchison KA, Matic G, Meshinchi S, Bresnick EH, Pratt WB. Redox manipulation of DNA-binding activity and BuGR epitope reactivity of the glucocorticoid receptor. J Biol Chem. 1991;266:10505–10509. [PubMed] [Google Scholar]
- 30.Choi HJ, et al. Structural basis of the redox switch in the OxyR transcription factor. Cell. 2001;105:103–113. doi: 10.1016/s0092-8674(01)00300-2. [DOI] [PubMed] [Google Scholar]
- 31.Poole LB. Bacterial defenses against oxidants: Mechanistic features of cysteine-based peroxidases and their flavoprotein reductases. Arch Biochem Biophys. 2005;433:240–254. doi: 10.1016/j.abb.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 32.Kim SO, et al. OxyR: A molecular code for redox-related signaling. Cell. 2002;109:383–396. doi: 10.1016/s0092-8674(02)00723-7. [DOI] [PubMed] [Google Scholar]
- 33.Lee CJ, et al. Redox regulation of OxyR requires specific disulfide bond formation involving a rapid kinetic reaction path. Nat Struct Mol Biol. 2004;11:1179–1185. doi: 10.1038/nsmb856. [DOI] [PubMed] [Google Scholar]
- 34.Furste JP, et al. Molecular cloning of the plasmid RP4 primase region in a multihost-range TACP expression vector. Gene. 1986;48:119–131. doi: 10.1016/0378-1119(86)90358-6. [DOI] [PubMed] [Google Scholar]
- 35.Aslund F, Berndt KD, Holmgren A. Redox potentials of glutaredoxins and other thiol-disulfide oxidoreductases of the thioredoxin superfamily determined by direct protein–protein redox equilibria. J Biol Chem. 1997;272:30780–30786. doi: 10.1074/jbc.272.49.30780. [DOI] [PubMed] [Google Scholar]
- 36.Schafer FQ, Buettner GR. Redox environment of the cell as viewed through the redox state of the glutathione disulfide/glutathione couple. Free Radic Biol Med. 2001;30:1191–1212. doi: 10.1016/s0891-5849(01)00480-4. [DOI] [PubMed] [Google Scholar]
- 37.Panmanee W, Vattanaviboon P, Poole LB, Mongkolsuk S. Novel organic hydroperoxide-sensing and responding mechanisms for OhrR, a major bacterial sensor and regulator of organic hydroperoxide stress. J Bacteriol. 2006;188:1389–1395. doi: 10.1128/JB.188.4.1389-1395.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chuchue T, et al. ohrR and ohr are the primary sensor/regulator and protective genes against organic hydroperoxide stress in Agrobacterium tumefaciens. J Bacteriol. 2006;188:842–851. doi: 10.1128/JB.188.3.842-851.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mongkolsuk S, et al. The repressor for an organic peroxide-inducible operon is uniquely regulated at multiple levels. Mol Microbiol. 2002;44:793–802. doi: 10.1046/j.1365-2958.2002.02919.x. [DOI] [PubMed] [Google Scholar]
- 40.Lee JW, Soonsanga S, Helmann JD. A complex thiolate switch regulates the Bacillus subtilis organic peroxide sensor OhrR. Proc Natl Acad Sci USA. 2007;104:8743–8748. doi: 10.1073/pnas.0702081104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Oh SY, Shin JH, Roe JH. Dual role of OhrR as a repressor and an activator in response to organic hydroperoxides in Streptomyces coelicolor. J Bacteriol. 2007;189:6284–6292. doi: 10.1128/JB.00632-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pineda-Molina E, et al. Glutathionylation of the p50 subunit of NF-κB: A mechanism for redox-induced inhibition of DNA binding. Biochemistry. 2001;40:14134–14142. doi: 10.1021/bi011459o. [DOI] [PubMed] [Google Scholar]
- 43.Newberry KJ, Fuangthong M, Panmanee W, Mongkolsuk S, Brennan RG. Structural mechanism of organic hydroperoxide induction of the transcription regulator OhrR. Mol Cell. 2007;28:652–664. doi: 10.1016/j.molcel.2007.09.016. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.