Abstract
Two proteins, STIM1 in the endoplasmic reticulum and Orai1 in the plasma membrane, are required for the activation of Ca2+ release-activated Ca2+ (CRAC) channels at the cell surface. How these proteins interact to assemble functional CRAC channels has remained uncertain. Here, we determine how many Orai1 and STIM1 molecules are required to form a functional CRAC channel. We engineered several genetically expressed fluorescent Orai1 tandem multimers and a fluorescent, constitutively active STIM1 mutant. The tandem multimers assembled into CRAC channels, as seen by rectifying inward currents and by cytoplasmic calcium elevations. CRAC channels were visualized as fluorescent puncta in total internal reflection microscopy. With single-molecule imaging techniques, it was possible to observe photo-bleaching of individual fluorophores and to count the steps of bleaching as a measure of the stoichiometry of each CRAC channel complex. We conclude that the subunit stoichiometry in an active CRAC channel is four Orai1 molecules and two STIM1 molecules. Fluorescence resonance energy transfer experiments also showed that four Orai1 subunits form the assembled channel. From the fluorescence intensity of single fluorophores, we could estimate that our transfected HEK293 cells had almost 400,000 CRAC channels and that, when intracellular Ca2+ stores were depleted, the channels clustered in aggregates containing ≈1,300 channels, amplifying the local Ca2+ entry.
Keywords: endoplasmic reticulum, fluorescence resonance energy transfer
Calcium can be released from intracellular stores [i.e., endoplasmic reticulum (ER)] in response to receptor activation. If the stores become depleted, Ca2+ release-activated Ca2+ (CRAC) channels of the plasma membrane are opened and continue the Ca2+ response (1). Ca2+ entry through CRAC channels is critical for many cellular responses, such as T cell proliferation (2, 3) or release of proinflammatory cytokines from mast cells (4, 5). However, because the unitary conductance of CRAC channels is <1% of that for voltage-gated calcium channels (6), their downstream effectors need to be in close proximity to the sites of Ca2+ entry. The molecular identity of the CRAC channels has long been a mystery, but two essential components, the ER Ca2+ sensor STIM1 (7, 8) and the plasma membrane channel pore-forming subunit Orai1/CRACM1, were identified recently through genomic screens (9–11). When CRAC channels are induced by store depletion, fluorescently labeled STIM1 and Orai1 migrate in their respective membranes and aggregate in tightly packed coclusters where the membranes come close together (12–14 nm) (6, 12–14). Presumably, aggregation intensifies the Ca2+ signal locally as in the immune synapse of T cells activated by antigen (6, 15, 16).
Functional ion channels often form from subunits assembled into multimers. Thus, Orai1 proteins associate with each other in vivo (3, 17, 18), but the stoichiometry for forming a CRAC channel is controversial. A recent study (19) adopted techniques previously used to determine the composition of mammalian potassium channels (20); they recorded macroscopic currents from HEK293 cells coexpressing STIM1 with preassembled tandem multimeric Orai1 constructs and a dominant negative Orai1 E106Q mutant. All of the tandem Orai1 constructs showed small but appropriate CRAC-like currents; however, only cells transfected with Orai1 4 tandems plus STIM1 showed CRAC current that was insensitive to coexpression of a monomeric dominant negative Orai1 mutant. This was taken as evidence for a tetrameric assembly of Orai1 subunits. Relatively low expression of STIM1 and variable expression of Orai1 from different vectors might complicate the interpretation of these important experiments. In contrast to the electrophysiological data, biochemical analysis showed a predominance of Orai1 dimers compared with tetramers in quiescent cells and in cells in which Ca2+ stores had been depleted (3). Therefore, the stoichiometry of CRAC channel needs further study.
CRAC channel function depends on its ER component, STIM1, which interacts with Orai1 to open the channel. This kind of activation mechanism is unprecedented, so determining the stoichiometry between STIM1 and Orai1 is essential to understanding CRAC channel function. Overexpressing Orai1 alone in HEK293 cells without also expressing exogenous STIM1 actually depresses store-operated calcium entry and the CRAC channel current that can be induced (17, 21, 22), and cotransfecting with STIM1 and Orai1 vectors in a ratio of 10:1 yields “monster” CRAC currents as high as 30 pA/pF (23). Expressing different ratios of STIM1 and Orai1 vectors yields different amplitudes of CRAC currents (unpublished data). However, because the relationship between the ratio of cDNA transfected and the molecular stoichiometry in the expressed channels is unclear, the number of STIM1 molecules involved in opening a CRAC channel remains to be resolved.
Here, we resolve these problems by using single-molecule imaging. By counting bleaching steps of fluorescent EGFPs tagged onto various Orai1 or STIM1 constructs expressed in HEK293 cells, we confirmed that the Orai1 channel is a homotetramer and found evidence that the molecular stoichiometry for STIM1 is 2 per functional complex. Additional experiments with FRET between Orai1 subunits gave the same answer. Based on fluorescence intensity profiles of Orai1 clusters, we also estimated the assembled CRAC channel density and numbers after ER calcium store depletion.
Results and Discussion
Orai1 Tandem Constructs Are Active.
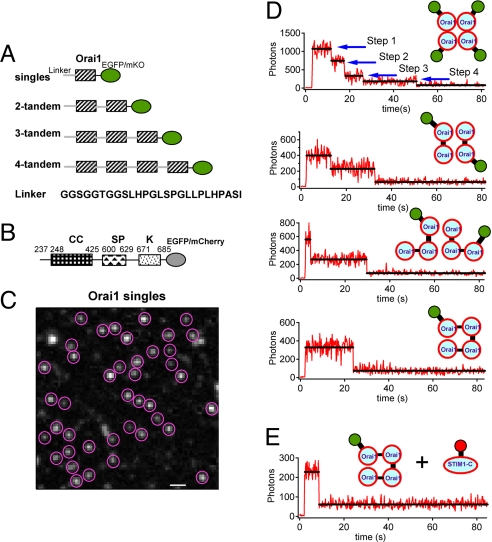
We constructed several fluorescent forms of Orai1 and STIM1 (Fig. 1 A and B). Single Orai1 and three tandem multimeric forms were tagged with one EGFP at their cytosolic C termini (Fig. 1A). Coexpression of STIM1 with any of the Orai1 constructs (DNA transfection ratio of 5:1) reconstituted 10–60 pA/pF CRAC currents in HEK293 cells when Ca2+ stores were depleted with BAPTA [supporting information (SI) Fig. S1], indicating that all chimeras can mediate large CRAC currents in living cells. The current magnitude varied widely among cells. Each of the exogenously expressed tandem Orai1 constructs also generated fluorescence at the cell membrane seen with total internal reflection (TIRF) microscopy (Fig. S2A) and aggregated into large, bright clusters when ER Ca2+ was depleted with thapsigargin (TG) (Figs. 3B and S2 B and C), as has been described before for Orai1 singles (6, 13, 14). The shorter the construct, the more total light was seen in quiescent cells under TIRF microscopy (Fig. S2A), indicating that, with multiple tandem Orai1 units in the construct, fewer molecules reached the cell surface.
Fig. 1.
Bleaching steps for EGFP attached to tandem multimeric constructs of Orai1 reveal a tetrameric complex of Orai1 proteins. (A) Schematic representation of the fluorescent Orai1 constructs used in this study in which each shaded box represents a full-length Orai1. Orai1 multimers were assembled in tandem using the linker sequence GGSGGTGGSLHPGLSPGLLPLHPASI. (B) A fluorescent, constitutively active C-terminal construct (STIM1-C) made from WT STIM1 (residues 237–685) by deletion of the endoplasmic reticulum and transmembrane domains and fusion with EGFP or mCherry at the C terminus. (C) A representative image (EGFP channel) taken using TIRF microscopy shows Orai1 puncta on the plasma membrane of a fixed cell. The puncta enclosed with red circles were chosen for later single-molecule bleaching analysis. These cells also co-express STIM1-mOrange. The image resolution is 160 nm/pixel, and the scale bar represents 1 μm. (D) HEK293 cells were co-transfected with different EGFP-tagged Orai1 tandem multimers and STIM1-mOrange. Representative time courses of EGFP emission for these different constructs are shown after background correction. The y axis plots photons collected per 200-ms sampling period for comparison. (E) Fluorescent puncta of cells cotransfected with Orai1 4-tandems and the truncated, constitutively active STIM1-C-mCherry usually bleach in a single step.
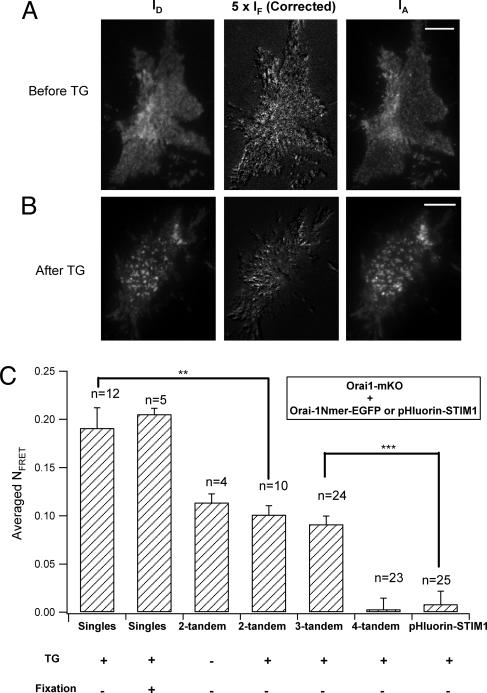
Fig. 3.
FRET between Orai1 singles and tandem multimers in living cells observed under TIRF microscopy. (Scale bars, 10 μm.) (A) Images IA and ID (see Methods) for a representative resting cell coexpressing Orai1-mKO and two-tandem Orai1-EGFP, excited by the 488-nm laser. The corrected FRET image was obtained from the raw IF image by subtracting the EGFP fluorescence bleedthrough to the red channel and the direct excitation of mKO by the 488-nm laser and then scaling by a factor of 5 to improve visibility. Most of the fluorescent regions of the cell show appreciable Orai1-Orai1 FRET. (B) Similar images for cells stimulated with TG (1 μM) for 10 min. Orai1 has assembled into aggregates on the plasma membrane. (C) Summaries of mean NFRET (see Methods) between Orai1-mKO and different EGFP-tagged vectors. No FRET signal was detected when cells were co-expressing Orai1-mKO and EGFP-tagged four tandem (**, P < 0.01; ***, P < 0.001).
Orai1 Forms Tetramers in Quiescent Cells.
When quiescent cells were visualized by using TIRF microscopy at high gain, the exogenously expressed Orai1 constructs gave a punctate appearance of fluorescence on the surface membrane (Fig. 1C). We suggest that many such puncta represent single CRAC channel complexes. Applying a defined continuous laser power to the cells induced stepwise loss of this fluorescence by photobleaching (Fig. 1D). For each of the chosen fluorescent spots, we could count the number of steps of bleaching to determine the number of fluorescing EGFP molecules it contained, as described for other ion channels (24, 25). High expression of the constructs often led to two or more apparent channel complexes lying within a diffraction-limited spot, which complicated interpretation of the results. Therefore, we used HEK293 cells cotransfected for as little as 2–5 h with Orai1 tandem constructs and STIM1. Cells were then fixed to prevent fluorescence fluctuation induced by the lateral movement of fusion proteins. During TIRF data collection on the fixed cells, we selected a field of 10 × 10 μm with between 20 and 200 fluorescence puncta, and the fluorescence intensity time course for each acceptable punctum was extracted as described in Methods.
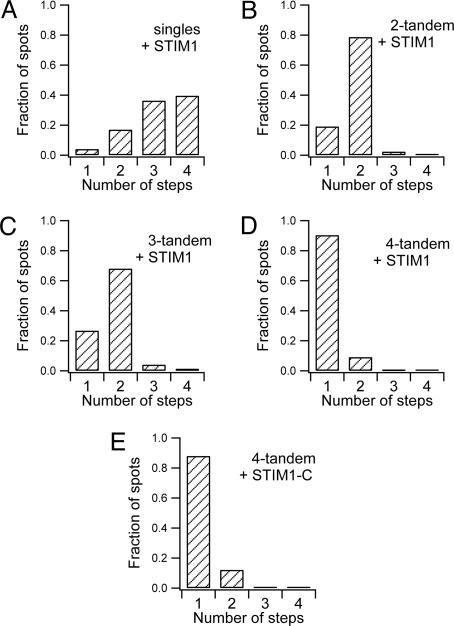
First, we considered the bleaching steps observed with expressed Orai1 singles (Fig. 1D). The first few points are taken before the laser has been turned on, and all points in the figure are corrected with a time-dependent background correction. Of the analyzed puncta, 40% (49 of 124, from three cells) bleached in four steps, 36% (45 of 124) bleached in three steps, and the remaining 21% bleached in one or two steps (Fig. 2A). In addition, four puncta (3%) bleached in five steps. These data are consistent with formation of a tetrameric channel in which a small fraction of the Orai1 monomers are nonfluorescent because they are endogenous unlabeled Orai1 molecules or they are fusion proteins carrying immature EGFP, similar to what has been proposed in oocytes (25). For example, if the abundance of nonfluorescent subunits was 0.20, the binomial distribution predicts 41% of four-step bleaches, 41% of three-step bleaches, and 18% of one- and two-step bleaches.
Fig. 2.
Distribution of EGFP bleaching steps seen with TIRF in fixed HEK293 cells expressing Orai1 single or tandem constructs. (A) Frequency of one-step and multistep bleaching events in cells cotransfected with STIM1-mOrange and Orai1-EGFP singles. (B and C) With either two- (B) or three-tandem (C) Orai1 tagged with EGFP, a large number of fluorescence spots bleached in two steps. (D and E) When four Orai1 molecules were linked to a EGFP, nearly all fluorescence spots on the plasma membrane bleached in a single step, regardless of whether the cells had been cotransfected with STIM1-mOrange (D) or with the constitutively active STIM1-C-mCherry (E). Except in E, three experiments (cells) were done for each construct.
As a further test, we examined the bleaching steps for tandem Orai1 constructs. When fluorescent Orai1 two tandems were coexpressed with STIM1, the majority of bleaching events (79%, 103 of 131 events, from three cells) were two-step processes, and only a minority were single-step processes (25 of 131 events; Figs. 1D and 2B). Similarly, when fluorescent Orai1 three tandems were coexpressed with STIM1, the majority of bleaching events still were two-step events (Figs. 1D and 2C). As has been proposed for other ion channels (20), two three tandems might contribute a subset of their six subunits to form one functional tetramer. Finally, we turned to Orai1 four tandems. Nearly all individual four-tandem spots were bleached in a single step (90%, 130 of 144 events, from three cells; Figs. 1D and 2D). Hence, the experiments with singles and those with tandems agree that the diffraction-limited spots on the plasma membrane in fixed quiescent HEK293 cells must contain Orai1 tetramers.
We noted earlier that, the more Orai1 units were linked together in a tandem construct, the less EGFP appeared at the cell surface. Perhaps this reflects a homeostatic mechanism that limits the number of functional CRAC channels. Thus, compared with Orai1 singles, only 25% as many four tandems would be needed to make the same number of channels. In addition, with transfections lasting only a few hours, cells may not be able to make, fold and mature as many longer tandem constructs as singles. In SI Text and Fig. S3, we also show that the variance of the fluorescence signal is proportional to the number of unbleached EGFP molecules remaining. These fluctuations arise primarily from noisy optical properties of EGFP rather than from instrument noise or counting statistics.
Active CRAC Channels Are Orai1 Tetramers as Well.
Our initial subunit counting was limited in that it was done only in quiescent cells. During depletion of Ca2+ stores in the ER, STIM1 and Orai1 colocalize to form large and densely packed aggregates that correlate with the opening of CRAC channels (Fig. S2B) (6, 13, 14). Individual bleaching steps could not be resolved in such dense and bright aggregates, so it remains unclear whether Orai1 tetramers represent the functional channel pore, or perhaps some higher-order complex of Orai1 oligomers forms during CRAC channel activation. To address this question, we cotransfected cells with Orai1 four tandems (coupled to EGFP) and with the cytoplasmic C terminus of STIM1 (STIM1-C; Fig. 1B), a constitutively active activator of Orai1 (26). As expected for cells with activated CRAC channels, the resting cytoplasmic Ca2+ concentrations ([Ca2+]i) were much higher than in nontransfected cells (Fig. S4) (26). Fortunately, TIRF microscopy showed that these Orai1 four tandems activated by STIM-C did not gather into larger aggregates. They formed single puncta or small clusters, so we were again able to use single-molecule EGFP bleaching measurements to study subunit compositions. As shown in Figs. 1E and 2E, the majority of the puncta recorded (88%, 80 of 91 events, from two cells) exhibited single-step behavior implying just one four tandem per punctum, whereas only a few puncta bleached in two steps. From these data, we conclude that four Orai1 monomers form the unitary pore complex in both resting and STIM1-C-activated CRAC channels.
Stoichiometry of Orai1 in Live Cells.
A question in our bleaching work is whether fixation of the cells for single-molecule TIRF may have distorted the apparent stoichiometry or altered the cellular distribution of CRAC channels (27). To confirm our conclusions by a different method and in living cells, we used FRET. EGFP was the donor, and monomeric Kushibara Orange (mKO) (28) was the FRET acceptor. We used a sensitized emission method based on taking three images (29) (Fig. 3 A and B). Robust FRET was seen in cells coexpressing Orai1-EGFP singles and Orai1-mKO singles under TIRF illumination (Fig. 3C), showing molecular proximity of Orai1 subunits in their assembly, as suggested by biochemical experiments (3, 17, 18), our bleaching results and recent FRET observations (30). Fixation of cells with paraformaldehyde without mounting in antifading solution did not affect the cellular localization of EGFP or mKO, and the FRET efficiency was the same in these fixed cells as in the live ones (27) (Fig. 3C), suggesting that fixation does not change channel stoichiometry. Furthermore, TG treatment induced an aggregation of Orai1 on the surface membrane (live cells) without altering the FRET efficiency (Fig. 3B), as also reported (30). This suggests that preexisting Orai1 oligomers do not change their stoichiometry during CRAC channel activation. A negative control experiment used STIM1 labeled at its ER luminal N terminus with EGFP-derived pHluorin. No FRET with Orai1-mKO was detected after store depletion despite the perfect colocalization of pHluorin-STIM1 and Orai1-mKO (Fig. 3C), consistent with the orientation of these fluorescent tags on opposite sides of the membrane.
Using this FRET technique, we found that the stoichiometry of Orai1 subunits in CRAC channels of living cells is tetrameric, as it is in fixed cells. The approach was to ask whether Orai1-mKO could join in channel complexes formed by different tandem Orai1 multimers tagged with EGFP. As expected, mKO singles could interact (FRET) with the two- and three-tandem versions of Orai1-EGFP (Fig. 3), indicating that these tandems alone were not the fully assembled form of an active channel. Conversely, no FRET signal was detected in cells coexpressing Orai1-mKO and four-tandem Orai1-EGFP, presumably because of efficient assembly of a channel pore by the four-tandem constructs alone leaving no spot for a fifth subunit. This latter result is quite analogous to the electrophysiological finding that monomeric dominant negative Orai1 subunits could not join and suppress current in channels formed by four-tandems (19). Our experiments on living cells give the same answers as those in fixed cells and show that fixation, which was necessary to stop the molecular motions in single-molecule TIRF experiments, need not disturb protein stoichiometry.
Two STIM1 Molecules Combine with One CRAC Channel.
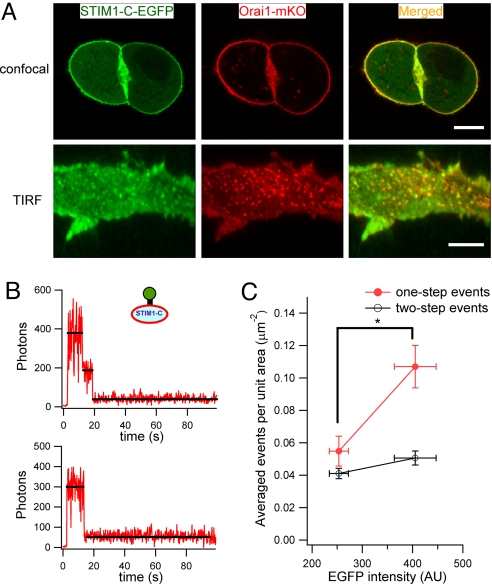
Next, we attempted to determine the number of STIM1-C molecules in the fluorescent puncta using a single-molecule imaging strategy similar to that used for Orai1. In live HEK293 cells transfected with both STIM1-C-EGFP and monomeric Orai1-mKO, most of the STIM1-C molecules localized with Orai1 at the cell surface under confocal microscopy and in the small coclusters seen in TIRF microscopy (Fig. 4A). Because these STIM1-C-expressing cells also exhibited elevated resting [Ca2+]i relative to nontransfected cells (as in Fig. S4 but done with different fluorophores), we believe that some of this surface STIM1-C interacts closely with Orai1 and opens CRAC channels (26, 30, 31). In recent work, activation of CRAC channels was seen to induce an increase in FRET observed both between STIM1 and STIM1 (30, 32) and between STIM1 and Orai1 (30). Thus, in the activation of CRAC channels, several STIM1 molecules join in an oligomeric complex with each other, and they in turn form a close molecular complex with Orai1. To be able to resolve fluorescent puncta of constitutively active STIM1-C, we picked transfected cells expressing a low amount of STIM1-C and containing only their endogenous Orai1. We measured the number of bleaching steps for 298 puncta. Of these, 56% bleached in one step (Fig. 4B), 42% in two steps, 1.7% in three steps, and none in four steps. The one- and two-step events occurred at mean densities of 0.05 ± 0.009 events/μm2 and 0.04 ± 0.003 events/μm2. We then analyzed another subset of the same transfected cells that expressed more STIM1-C (Fig. 4C). The total fluorescence of STIM1-C was 60% higher, and the density of one-step bleaches almost doubled, but the number of two-step bleaches hardly increased. Conversely, if we increased the amount of Orai1 in the cells by using an HEK293 cell line stably expressing Orai1–3× FLAG-SBP-CBP, and transfecting them with STIM1-C-EGFP (mean expression 274 ± 14 counts), the number of two-step bleaches increased (0.07 ± 0.007 events/μm2, n = 7, P < 0.01) and the number of one-step bleaches was unchanged (0.05 ± 0.007, events/μm2). We interpret these phenomena in a model with two membrane pools of overexpressed STIM1-C. One pool is productively interacting in a stoichiometric manner with the limited number of endogenous Orai1 complexes present and thereby activates functional CRAC channels. This pool should not increase when more STIM1-C is expressed because it depends on Orai1 complexes, but it would increase in the cells expressing more Orai1. It therefore corresponds to the two-step bleaches. The other pool represents monomers that possibly have a nonspecific interaction via their polycationic C terminus with acidic lipids of the plasma membrane. This pool should increase when more STIM1-C is expressed and corresponds to one-step bleaches. Based on that model, we suggest that two STIM1 molecules can activate a CRAC channel.
Fig. 4.
Two STIM1-C proteins bind to Orai1 complexes. (A) Upper shows a representative confocal image captured in the middle focal plane of live HEK293 cells cotransfected with STIM1-C-EGFP and Orai1-mKO. Bottom shows colocalization of these two proteins at the membrane under TIRF illumination again in live cells. (Scale bars, 10 μm.) (B) Representative fluorescence steps in TIRF microscopy for HEK293 cells transfected with STIM1-C-EGFP and no exogenous Orai1. Puncta bleached either in two steps (Upper) or one step (Lower). (C) Comparison of one- and two-step bleaches in subpopulations of cells from the same dish expressing low (253 ± 19 arbitrary units; n = 3) or higher (405 ± 41; n = 3) amounts of STIM1-C-EGFP. The higher expressers had significantly more one-step bleach events occurring per μm2 (P < 0.05) but no change in two-step events.
In SI Text, we determine the fluorescence intensity of single fixed EGFP molecules in our TIRF microscope and use that to count the number of CRAC channels. We estimate there that, in the clusters formed upon ER calcium-store depletion (i.e., TG), complete CRAC channels consisting of four Orai1 molecules would be ≈34 nm apart, too far for FRET interactions between channels. Macroscopically, we also estimate 1,300 channels in each TG-induced cluster and 370,000 channels per cell. If aggregates of CRAC channels normally contain 1,300 channels per cluster, for example at the immune synapse, then despite the low conductance per channel, the cluster would indeed generate a large but focused calcium signal to drive physiological responses. The total number of CRAC channels per cell agrees well with previous calculations based on current density and single-channel conductance analysis (33). It should be remembered, however, that our calculations are for HEK293 cells that have been transfected with exogenous Orai1 and STIM1.
In summary, we find that single-molecule fluorescence bleaching methods (24, 25) and FRET can be successfully applied in mammalian cells to CRAC channels. We show by these two methods that four Orai1 monomers assemble to form a tetrameric pore of the CRAC channel, in full agreement with recent electrophysiological work (19). We also suggest by one method that pairs of STIM1 molecules are associated with the CRAC channels when they are activated. Finally we reach a self-consistent count of the number of CRAC channels in aggregates formed after activation of the CRAC system by depletion of intracellular Ca2+ stores.
Methods
Solutions and Chemicals.
For measurements of intracellular Ca2+ concentration ([Ca2+]i), we used standard extracellular Ringer solution containing: 145 mM NaCl, 4.5 mM KCl, 10 mM CaCl2, 1 mM MgCl2, 10 mM d-glucose, and 5 mM Hepes (pH 7.4, adjusted with NaOH). For cells fixed with paraformaldehyde, we used a PBS solution that contained: 137 mM NaCl, 2.7 mM KCl, 4.3 mM Na2HPO4, and 1.47 mM KH2PO4 (pH 7.4, adjusted with NaOH). In patch–clamp experiments, the standard external solution contained: 145 mM NaCl, 4.5 mM KCl, 10 mM CaCl2, 1 mM MgCl2, 10 mM d-glucose, 10 mM tetraethylammonium, and 5 mM Hepes (pH 7.4, adjusted with NaOH). The pipette solution contained: 140 mM Cs-glutamate, 8 mM MgCl2, 12 mM BAPTA, and 10 mM Hepes (pH 7.2, adjusted with CsOH). Stock solutions of TG were prepared in DMSO at a concentration of 2 mM. External solutions were applied by using a gravity-driven perfusion system, and the local solution was completely exchanged in <2 seconds. Fura-2/AM was purchased from Invitrogen. Unless otherwise specified all reagents and chemicals were from Sigma-Aldrich.
Imaging of Single Molecules.
The imaging experiments of Figs. 1, 2, 4 B and C, S2, S3, S5, and S6 were done with fixed cells. Transfected cells were viewed under either a confocal microscope (Fig. 4) or a TIRF microscope, as described in ref. 34. EGFP was excited with a 488-nm laser, and ≈6 mW laser power was applied to the rear pupil of an oil objective (Zeiss 100×, N.A. = 1.45). The TIRF image of Orai1-mKO was excited with a 532-nm laser. To reduce background noise and maximize emission, photons were detected by using a back-illuminated electron-multiplying charge-coupled device (EMCCD) camera (Andor iXon DV-897 BV) and high-quality filters (Chroma, 49002 ET-GFP). In addition, a laser blocker absorbed the reflected laser beam at the rear pupil of the objective. We set the gain of our EMCCD camera at 300 throughout all single-molecule imaging experiments and at 3 to collect images of cells emptied of their ER Ca2+ store (Fig. S2). These settings were in the linear dynamic range of the EMCCD camera. In addition, we measured the illumination field of the TIRF microscope to evaluate its heterogeneity. Because the intensity at the edge of the excitation field was at least 85% of that at the center, we treated the illumination as a relatively homogeneous field. The output of the camera is measured in counts from its 14-bit analog-to-digital (A/D) converter. We used the manufacturer's equation to convert from counts to photons (i.e., photoelectrons) collected during the 200-ms sampling period:
where P is the number of photons, S is the observed fluorescence intensity (in counts), TA/D is the conversion factor to convert EMCCD counts to photoelectrons (10.5 electrons per A/D count at unity gain), and G is the EMCCD gain.
Image Analysis.
Usually, an image stack of 500–800 frames was acquired at 5 Hz. We first subtracted the background fluorescence by using the rolling ball method in Image J software (National Institutes of Health; Fig. S6C). Because the noise in the images was high (Fig. S6A), we determined the center of mass of each fluorescence punctum by using a previously developed algorithm written in MATLAB (MathWorks) (25). The algorithm averaged the first 5–10 frames of the EGFP signal (Fig. S6B), then applied a threshold to obtain connected regions of interest (ROIs) above that threshold (Fig. S6D). The threshold was set at 10 times the SD measured in an area with no cells. ROIs smaller than three pixels were regarded as noise and discarded (Fig. S6E). ROIs with areas >15 pixels were segmented by Gaussian fitting, and multiple peaks that were very close were excluded. (For example, the bright spot near the top left corner of Fig. 1A was excluded, because it was large but could not be separated into two spots by our methods.) Centroid analysis or nonlinear Gaussian fitting algorithms were used to find the peaks of ROIs in the averaged image (Fig. S6F, red points). These methods yielded almost identical results to subpixel resolution, as explained (25). Finally, we obtained the summed fluorescence intensity in a square region of 5 × 5 pixels that enclosed a peak. The time course was plotted to identify the bleaching steps of that spot. Subsequently, bleaching transitions were scored manually by one investigator and rescored blindly by another. Apparent variance changes and step size were included in the scoring criteria. Traces receiving inconsistent scoring and traces with erratic behavior of the total fluorescence were rejected. Ambiguities were greatest (80% of traces) in experiments with singles and considerably less (50% of traces) in experiments with tandems, which usually yielded only one or two steps.
FRET Analysis.
FRET was measured on cells expressing different combinations of EGFP (donor)- and mKO (acceptor)-tagged Orai1. The cells were unfixed or fixed, as indicated. Following published methods and notation (29), the two fluorophores were sequentially excited with the 488- and 532-nm lasers, and three images were recorded with pixel intensities: ID at the donor emission with donor excitation (488 nm), IA at acceptor emission with acceptor excitation (532 nm), and IF at the acceptor emission with donor excitation. The normalized FRET (NFRET) in each pixel was defined by the equation:
Here, the terms in α and β correct for cross-excitation and bleed-through, respectively. They are measured in control experiments in which only one fluorophore is expressed. The quantity α is defined as IF/IA in experiments expressing only STIM1-C–EGFP (Fig. S7B). It corrects for EGFP emission that contaminates the red channel (i.e., bleedthrough), and was ≈0.43. The quantity β is defined as IF/ID in experiments expressing only Orai1-mKO (Fig. S7A). It corrects for direct excitation of mKO by the 488-nm laser and was ≈0.34. In the bar graphs, we plot the mean NFRET for all of the pixels in the TIRF footprint of cells.
Data Analysis.
Unless noted otherwise, all current traces were corrected for leak currents. Data were analyzed by using IGOR Pro 5.01 (Wavemetrics). Average results are presented as the mean ± SEM with the number of experiments indicated. Statistical significance was evaluated by using a Student's t test. Asterisks denote statistical significance compared with the control, with P values <0.05, 0.01, and 0.001, respectively.
Supplementary Material
Acknowledgments.
We thank Prof. Roger Y. Tsien (University of California at San Diego, La Jolla, CA) for providing the mOrange and mCherry and William N. Zagotta, Justin W. Taraska, Jill B. Jensen, Tamas Balla, and Ehud Isacoff for commenting on the manuscript. This work was supported by Grants 30670503, 30670504, 30630020, and 30611120531 from the National Science Foundation of China; Grant 5072034 from the Beijing Municipal Science and Technology Commission; Grant 2006CB705700 from the Major State Basic Research Program of P.R. China, and Grant GM83913 from the National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0806499105/DCSupplemental.
References
- 1.Hoth M, Penner R. Depletion of intracellular calcium stores activates a calcium current in mast cells. Nature. 1992;355:353–356. doi: 10.1038/355353a0. [DOI] [PubMed] [Google Scholar]
- 2.Oh-Hora M, et al. Dual functions for the endoplasmic reticulum calcium sensors STIM1 and STIM2 in T cell activation and tolerance. Nat Immunol. 2008;9:432–443. doi: 10.1038/ni1574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gwack Y, et al. Biochemical and functional characterization of Orai proteins. J Biol Chem. 2007;282:16232–16243. doi: 10.1074/jbc.M609630200. [DOI] [PubMed] [Google Scholar]
- 4.Chang WC, et al. Local Ca2+ influx through Ca2+ release-activated Ca2+ (CRAC) channels stimulates production of an intracellular messenger and an intercellular pro-inflammatory signal. J Biol Chem. 2008;283:4622–4631. doi: 10.1074/jbc.M705002200. [DOI] [PubMed] [Google Scholar]
- 5.Baba Y, et al. Essential function for the calcium sensor STIM1 in mast cell activation and anaphylactic responses. Nat Immunol. 2008;9:81–88. doi: 10.1038/ni1546. [DOI] [PubMed] [Google Scholar]
- 6.Luik RM, Wu MM, Buchanan J, Lewis RS. The elementary unit of store-operated Ca2+ entry: Local activation of CRAC channels by STIM1 at ER-plasma membrane junctions. J Cell Biol. 2006;174:815–825. doi: 10.1083/jcb.200604015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Roos J, et al. STIM1, an essential and conserved component of store-operated Ca2+ channel function. J Cell Biol. 2005;169:435–445. doi: 10.1083/jcb.200502019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liou J, et al. STIM is a Ca2+ sensor essential for Ca2+-store-depletion-triggered Ca2+ influx. Curr Biol. 2005;15:1235–1241. doi: 10.1016/j.cub.2005.05.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zhang SL, et al. Genome-wide RNAi screen of Ca2+ influx identifies genes that regulate Ca2+ release-activated Ca2+ channel activity. Proc Natl Acad Sci USA. 2006;103:9357–9362. doi: 10.1073/pnas.0603161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vig M, et al. CRACM1 is a plasma membrane protein essential for store-operated Ca2+ entry. Science. 2006;312:1220–1223. doi: 10.1126/science.1127883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feske S, et al. A mutation in Orai1 causes immune deficiency by abrogating CRAC channel function. Nature. 2006;441:179–185. doi: 10.1038/nature04702. [DOI] [PubMed] [Google Scholar]
- 12.Varnai P, Toth B, Toth DJ, Hunyady L, Balla T. Visualization and manipulation of plasma membrane-endoplasmic reticulum contact sites indicates the presence of additional molecular components within the STIM1-Orai1 Complex. J Biol Chem. 2007;282:29678–29690. doi: 10.1074/jbc.M704339200. [DOI] [PubMed] [Google Scholar]
- 13.Xu P, et al. Aggregation of STIM1 underneath the plasma membrane induces clustering of Orai1. Biochem Biophys Res Commun. 2006;350:969–976. doi: 10.1016/j.bbrc.2006.09.134. [DOI] [PubMed] [Google Scholar]
- 14.Wu MM, Buchanan J, Luik RM, Lewis RS. Ca2+ store depletion causes STIM1 to accumulate in ER regions closely associated with the plasma membrane. J Cell Biol. 2006;174:803–813. doi: 10.1083/jcb.200604014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lioudyno MI, et al. Orai1 and STIM1 move to the immunological synapse and are up-regulated during T cell activation. Proc Natl Acad Sci USA. 2008;105:2011–2016. doi: 10.1073/pnas.0706122105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barr VA, et al. Dynamic movement of the calcium sensor stim1 and the calcium channel orai1 in activated T cells: Puncta and distal caps. Mol Biol Cell. 2008;19:2802–2817. doi: 10.1091/mbc.E08-02-0146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li Z, et al. Mapping the interacting domains of STIM1 and Orai1 in Ca2+ release-activated Ca2+ channel activation. J Biol Chem. 2007;282:29448–29456. doi: 10.1074/jbc.M703573200. [DOI] [PubMed] [Google Scholar]
- 18.Vig M, et al. CRACM1 multimers form the ion-selective pore of the CRAC channel. Curr Biol. 2006;16:2073–2079. doi: 10.1016/j.cub.2006.08.085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mignen O, Thompson JL, Shuttleworth TJ. Orai1 subunit stoichiometry of the mammalian CRAC channel pore. J Physiol. 2008;586(Suppl):419–425. doi: 10.1113/jphysiol.2007.147249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liman ER, Tytgat J, Hess P. Subunit stoichiometry of a mammalian K+ channel determined by construction of multimeric cDNAs. Neuron. 1992;9:861–871. doi: 10.1016/0896-6273(92)90239-a. [DOI] [PubMed] [Google Scholar]
- 21.Soboloff J, et al. Orai1 and STIM reconstitute store-operated calcium channel function. J Biol Chem. 2006;281:20661–20665. doi: 10.1074/jbc.C600126200. [DOI] [PubMed] [Google Scholar]
- 22.Mercer JC, et al. Large store-operated calcium selective currents due to co-expression of Orai1 or Orai2 with the intracellular calcium sensor, Stim1. J Biol Chem. 2006;281:24979–24990. doi: 10.1074/jbc.M604589200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Peinelt C, et al. Amplification of CRAC current by STIM1 and CRACM1 (Orai1) Nat Cell Biol. 2006;8:771–773. doi: 10.1038/ncb1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kohout SC, Ulbrich MH, Bell SC, Isacoff EY. Subunit organization and functional transitions in Ci-VSP. Nat Struct Mol Biol. 2008;15:106–108. doi: 10.1038/nsmb1320. [DOI] [PubMed] [Google Scholar]
- 25.Ulbrich MH, Isacoff EY. Subunit counting in membrane-bound proteins. Nat Methods. 2007;4:319–321. doi: 10.1038/NMETH1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Huang GN, et al. STIM1 carboxyl-terminus activates native SOC, I(crac) and TRPC1 channels. Nat Cell Biol. 2006;8:1003–1010. doi: 10.1038/ncb1454. [DOI] [PubMed] [Google Scholar]
- 27.Brock R, Hamelers IH, Jovin TM. Comparison of fixation protocols for adherent cultured cells applied to a GFP fusion protein of the epidermal growth factor receptor. Cytometry. 1999;35:353–362. doi: 10.1002/(sici)1097-0320(19990401)35:4<353::aid-cyto8>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 28.Karasawa S, Araki T, Nagai T, Mizuno H, Miyawaki A. Cyan-emitting and orange-emitting fluorescent proteins as a donor/acceptor pair for fluorescence resonance energy transfer. Biochem J. 2004;381:307–312. doi: 10.1042/BJ20040321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hoppe A, Christensen K, Swanson JA. Fluorescence resonance energy transfer-based stoichiometry in living cells. Biophys J. 2002;83:3652–3664. doi: 10.1016/S0006-3495(02)75365-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muik M, et al. Dynamic coupling of the putative coiled-coil domain of ORAI1 with STIM1 mediates ORAI1 channel activation. J Biol Chem. 2008;283:8014–8022. doi: 10.1074/jbc.M708898200. [DOI] [PubMed] [Google Scholar]
- 31.Zhang SL, et al. Store-dependent and -independent modes regulating Ca2+ release-activated Ca2+ channel activity of human Orai1 and Orai3. J Biol Chem. 2008;283:17662–17671. doi: 10.1074/jbc.M801536200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liou J, Fivaz M, Inoue T, Meyer T. Live-cell imaging reveals sequential oligomerization and local plasma membrane targeting of stromal interaction molecule 1 after Ca2+ store depletion. Proc Natl Acad Sci USA. 2007;104:9301–9306. doi: 10.1073/pnas.0702866104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parekh AB. A CRAC current tango. Nat Cell Biol. 2006;8:655–656. doi: 10.1038/ncb0706-655. [DOI] [PubMed] [Google Scholar]
- 34.Duman JG, Chen L, Palmer AE, Hille B. Contributions of intracellular compartments to calcium dynamics: Implicating an acidic store. Traffic. 2006;7:859–872. doi: 10.1111/j.1600-0854.2006.00432.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.