Abstract
Histone lysine methylation is an important epigenetic modification with both activating and repressive roles in gene expression. Jumonji C (jmjC) domain-containing proteins have been shown to reverse histone methylation in nonplant model systems. Here, we show that plant Jumonji C proteins have both conserved and specific features compared with mammalian homologues. In particular, the rice JMJD2 family jmjC gene JMJ706 is shown to encode a heterochromatin-enriched protein. The JMJ706 protein specifically reverses di- and trimethylations of lysine 9 of histone H3 (H3K9) in vitro. Loss-of-function mutations of the gene lead to increased di- and trimethylations of H3K9 and affect the spikelet development, including altered floral morphology and organ number. Gene expression and histone modification analysis indicates that JMJ706 regulates a subset of flower development regulatory genes. Taken together, our data suggest that rice JMJ706 encodes a heterochromatin-associated H3K9 demethylase involved in the regulation of flower development in rice.
Keywords: chromatin, histone modification, demethylation
Histone lysine methylation is an important epigenetic modification with both activating and repressive roles in gene expression (1). There are six lysine residues in histone N-termini that are predominantly methylated, with the methylation of histone H3 lysine 4 (H3K4) and lysine 36 (H3K36) primarily having an activating function, whereas the methylation of histone H3 lysine 9 (H3K9) and lysine 27 (H3K27) and histone H4 lysine 20 (H4K20) is essentially associated with repressed chromatin (2). Histone lysine residues can be mono-, di- or trimethylated. Each distinct methyl state is associated with different biological functions (1). Histone methylation was considered irreversible until the recent discovery of histone demethylases. Lysine-specific demethylase 1 was the first histone demethylase to be identified in mammalian cells, and it has been shown to demethylate monomethylated and dimethylated H3K4 (H3K4me1 and H3K4me2) and H3K9 (H3K9me1 and H3K9me2) (3, 4).
Jumonji C (jmjC) domain-containing proteins have been suggested to function as histone demethylases (5). Using biochemical approaches, the first jmjC domain-containing histone demethylase, JHDM1 (jmjC domain-containing histone demethylase 1), was identified and shown to reverse H3K36 mono- and dimethylation (H3K36me1 and H3K36me2), and the jmjC domain was demonstrated to be the catalytic domain (6). Jumonji C domain-containing histone demethylases catalyze lysine demethylation through an oxidative reaction that requires iron Fe(II) and α-ketoglutarate as cofactors. Subsequently, JHDM2 was identified to reverse H3K9me2, and the JMJD2/JHDM3 subfamily, which consists of four members in mammalian cells (JMJD2A–D), was specifically found to reverse lysine trimethylation, H3K9me3, and/or H3K36me3 (7–9). More recently, JMJD3/UTX proteins were shown to be H3K27me3 demethylases (10), and members of the JARID1 subfamily were identified as H3K4me3 demethylases (11).
A phylogenetic analysis of jmjC domain-containing proteins in six nonplant model organisms was reported (12). This analysis led to the identification of seven groups of jmjC domain-containing proteins on the basis of the jmjC domain and the overall protein domain architecture. In Arabidopsis thaliana, two jmjC domain-coding genes have been studied with respect to developmental functions (13). A recent study shows that an Arabidopsis JHDM2 gene is required for genic DNA methylation (14). In this work, we show that rice (Oryza sativa) and Arabidopsis jmjC genes have both conserved and specific features compared with animal homologues. JMJ706, a rice member of the JMJD2 family of jmjC genes, is shown to encode a heterochromatin-associated protein. In vitro histone demethylation assays and analysis of T-DNA insertion mutants revealed that JMJ706 is involved in the H3K9 demethylation required for the expression of a subset of regulatory genes for rice floral development.
Results
Phylogenetic Analysis of Plant jmjC-Domain Proteins.
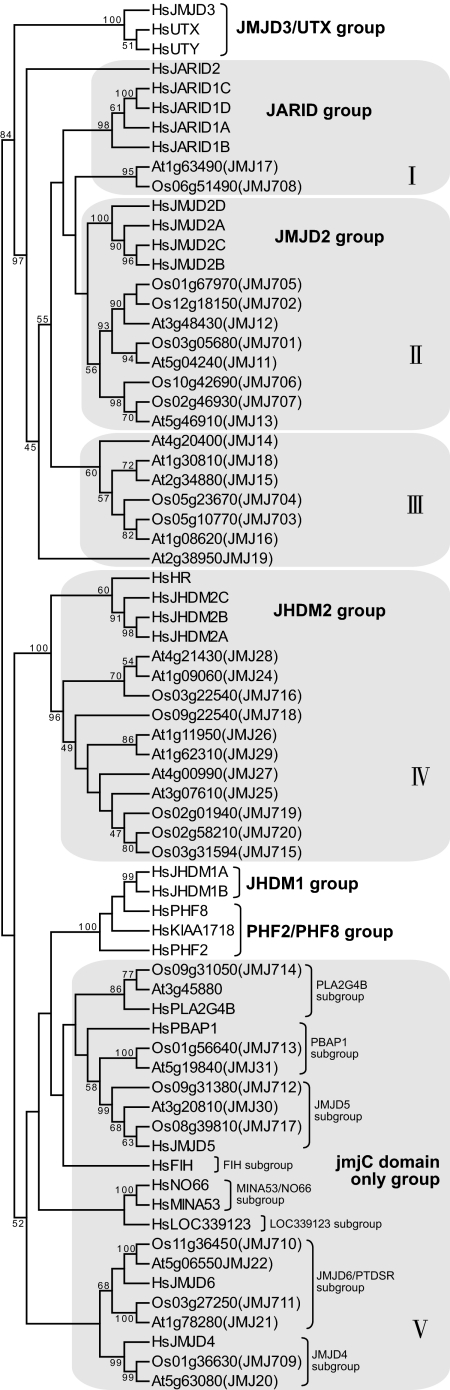
A genomic survey revealed 20 and 21 jmjC domain-containing genes in rice and Arabidopsis, respectively (Fig. 1). The sequence alignment resulted in the discrimination of five groups containing the plant proteins. Groups I, II, and IV were found to be related to the JARID, JMJD2, and JHDM2 subfamilies, respectively (12). Group V could be divided into several subgroups, each of which was found to have related members of the human “JmjC domain-only” subfamily (12). In contrast, the jmjC domains of Group III were less related to the human homologues. In addition, members of this group contained a FYRN (N-terminal FY-rich) motif and a FYRC (C-terminal FY-rich) motif that were not found in human jmjC proteins [supporting information (SI) Fig. S1A], suggesting that this group might encode proteins with different functions. Conversely, no plant proteins were found to be related to the human UTX/JMJD3, JHDM1, or PHF2/PHF8 subfamilies. Therefore, plant jmjC domain-containing proteins show both conservation and divergence with the mammalian homologues.
Fig. 1.
Phylogenetic relationship of jmjC domain-containing proteins from O. sativa (Os), A. thaliana (At), and H. sapiens (Hs). The values represent the percentages of sampled trees used in the analysis that contained the consensus partition. Names of seven evolutionarily conserved groups of Hs jmjC domain proteins previously determined by Klose et al. (12) are indicated on the right. Five groups of plant jmjC proteins are indicated by roman numerals.
JMJ706 Gene Structure, Expression Profile, and Subcellular Localization.
A more detailed analysis revealed that plant Group II members contained a C5HC2 zinc-finger (ZnF) motif in addition to the jmjC and jmjN signature domains (Fig. S1B) (12). The ZnF motif was missing from the human JMJD2 proteins but present in homologues from budding yeast (Fig. S1B). Conversely, additional modules, including the PHD (plant homeodomain) and the Tudor domain, found in human JMJD2 proteins were absent from the plant and yeast proteins.
To study the function of plant JMJD2 members, we analyzed the rice JMJ706 gene. The mRNA of the gene was found to be constitutively expressed (Fig. S1C). The transiently expressed JMJ706 protein was localized in the nucleus, and this required the ZnF and the jmjC motifs, as their deletion led to a cytoplasmic localization of the protein (Fig. 2A).
Fig. 2.
Expression pattern of rice JMJ706 and subcellular localization of the protein. (A) Subcellular localization of the rice JMJ706-GFP fusion proteins in onion cells with the constructs indicated on the left. The transfected cells expressing GFP were stained by DAPI and photographed under different light fields. (B) JMJ706 is mainly localized in heterochromatin foci. Mesophyll cell nuclei from transgenic rice expressing JMJ706-GFP were examined by GFP expression (a and d) and DAPI staining (b and e). (c and f) Overlapped images of a and b (c) and d and e (f). (C) FLAG-tagged JMJ706 expressed in jmj6–2 plants was detected by immunostaining using the anti-FLAG antibody (a). (b) Same cell stained with DAPI. (c) Overlapped image of a and b. (Scale bars: 2 μm.)
To study JMJ706 localization in rice nuclei, a construct expressing a JMJ706 fusion with the GFP was used to transform WT rice plants. Nuclei of the transgenic leaf mesophyll cells showed an enrichment of the fusion protein in spots that overlapped with 4′,6-diamidino-2-phenylindole (DAPI)–stained regions (Fig. 2B). To confirm this result, a construct expressing FLAG-tagged JMJ706 protein was used to complement JMJ706 T-DNA insertion mutant lines (see below). The FLAG-tagged protein was also observed in enriched DAPI-stained domains (Fig. 2C). These observations suggest that JMJ706 is enriched in heterochromatin domains.
In Vitro Histone Demethylase Activity of JMJ706.
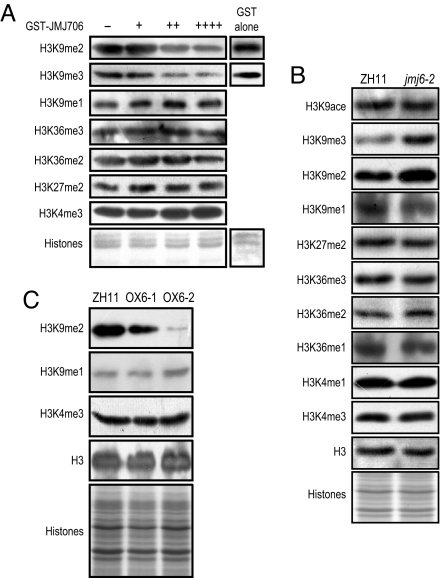
Histone demethylation activity has been shown to be jmjC domain dependent (6, 15). To study JMJ706 function, a truncated JMJ706 protein containing the jmjN and jmjC domains and an additional GST tag was purified from Escherichia coli cells and tested for in vitro histone demethylase activity. The incubated histones were analyzed by Western blot tests with antibodies specific to histone H3 modification modules. As shown in Fig. 3A, the GST-JMJ706 fusion protein reduced the level of H3K9me2 and H3K9me3 but slightly increased the level of H3K9me1. The methylation of other lysine residues was not significantly altered, except a slight decrease observed for H3K36me2. Glutathione S-transferase alone had no effect on histone modification. This suggests that JMJ706 may be mainly involved in the demethylation of H3K9me2 and H3K9me3.
Fig. 3.
JMJ706 histone demethylation activity. (A) In vitro assays of JMJ706 histone demethylation activity. Five micrograms of calf thymus histones was incubated with 0 (−), 2.5 (+), 5 (++) or 10 (++++) μg of the purified JMJ706-GST fusion protein or GST alone and was analyzed by Western blot tests using the antibodies indicated on the right. Staining of the histone proteins is shown at the bottom. (B) Western blot analysis of histones isolated from the WT and mutant jmj6–2 using the antibodies indicated on the left. Histones are shown at the bottom. One membrane was reused to test the antibodies of H3K9ace, H3K9me3, H3K27me2e, and H3K36me3, and another one was tested for the other antibodies. (C) Comparison of H3K9me2, H3K9me1, and H3K4me3 levels between WT and two JMJ706 overexpression lines by Western blot analysis.
JMJ706 Loss-of-Function Mutation Affects Floral Organogenesis.
A search of a database of a rice T-DNA insertion library (16, 17) identified two insertion lines of the JMJ706 gene (Fig. 4A). Characterization of the insertion mutants revealed similar spikelet morphology defects (Fig. 4B). The vegetative growth of both mutants appeared normal (data not shown). The phenotype cosegregated with the homozygous state of the insertions and the absence of JMJ706 mRNA accumulation (Figs. S2 and 4C). Retransformation of the jmj6-2 line with a JMJ706 cDNA under the control of the maize ubiquitin promoter in a G418 selection-based binary vector rescued the spikelet phenotype (Fig. 4B). Similarly, a FLAG-tagged JMJ706 cDNA controlled by the same promoter could also complement jmj6-2 (data not shown). In these complementation lines, JMJ706 expression levels were comparable with that in the WT plants (Fig. S3). Transgenic plants expressing an artificial micro-RNA (18) of the JMJ706 showed a phenotype that partially mimicked jmj6 (Figs. 4B and S4). Taken together, these results indicated that the spikelet phenotype of the insertion mutants was induced by JMJ706 loss of function.
Fig. 4.
JMJ706 loss-of-function mutation affects spikelet development. (A) Schematic representation of T-DNA insertion mutations jmj6–1 and jmj6–2. The primers used for genotyping and RT-PCR are indicated by arrows. (B) Comparison of panicles from WT (ZH11), jmj6–1, jmj6–2, and jmj6–2 complemented by JMJ706 cDNA and plants expressing an artificial micro (ami)-RNA of JMJ706 (artificial micro [ami]-R706). Arrowheads indicate spikelets with palea missing. (C) Reverse transcriptase polymerase chain reaction detection of JMJ706 transcripts in WT and mutant lines in homozygous (−/−) or heterozygous (+/−) states. Actin transcripts were analyzed in the presence or absence (No-RT) of reverse transcriptase as controls. (D) Spikelets with lemma and/or palea missing in the mutants. (a–c) Comparison between WT (Left), jmj6–1 (Center), and jmj6–2 (Right). (a) Both palea and lemma were missing from the mutants. (b and c) Absence of palea in the mutants. (c) Mature-stage spikelets showing seminude and deformed seeds. (E) Mutant spikelets with an extra palea or other floral organs. (a) Comparison between the WT (Left) and mutant (Center and Right) spikelets. Extra paleas are indicated by arrowheads. (b) Mutant spikelet with an additional floret (indicated by an arrowhead). (c) Mutant spikelet with an additional palea and a stamen indicated by arrowheads. (F) Mutant spikelets with increased numbers of pistils and stamens. (a) Comparison between WT (Left) and mutant carpels with the other organs removed. Pistils are indicated by arrowheads. jmj6–1 (b) and jmj6–2 (c) with seven stamens are indicated by arrowheads instead of six in the WT (data not shown). (G) Mutants with vitrified structures inside the spikelet. Comparison between the WT (Left) and mutant spikelets with the paleas and the lemmas removed. Arrowheads indicate the vitrified structures. (H) Comparison of scanning electron microscope images of palea surface from WT (a, Left) and the mutants (b and c). le, lemma; pa, palea. (Scale bars: D–G, 2 mm; H, 100 μm.)
The mutant spikelets showed a variety of defects, mainly on floral organ number (Fig. 4 D–H). Some spikelets were depleted of lemma and/or palea (Fig. 4D), whereas others had an additional piece of palea (Fig. 4E). Increased numbers of stamens and pistils were also observed (Fig. 4F). In addition, vitrified tissues were seen in some spikelets (Fig. 4G). Scanning microscopy revealed irregular cell size and arrangement on the mutant palea surface with increased bristle numbers (Fig. 4H). Some spikelets could still produce seeds. These mutant seeds had a deformed shape, but they germinated and produced normal seedlings (data not shown), suggesting that embryogenesis was not affected.
To study whether the mutant phenotype was associated with an altered expression of floral development genes, the transcripts of 16 rice MADS-box gene family members (19) were analyzed by RT-PCR. In the mutants, the expression of most of them, except OsMADS8 and OsMAD47, was not significantly altered (Figs. S5and 5A). OsMADS47 was repressed, but expression of OsMADS8 increased at the third stage of panicle development in the mutants (Fig. S3) (20). The jmj6 phenotype is similar to that found in the dh1 (degenerated hell1) mutants (21). DH1 encodes a putative lateral organ boundaries domain transcription factor (21). DH1 expression was repressed in the jmj6 mutants (Fig. 5A).
Fig. 5.
Effect of jmj6 mutations on the expression of relevant genes and histone methylation on the loci. (A) Expression analysis by RT-PCR of JMJ706, DH1, and 3 MADS-box genes. Actin transcripts were amplified as controls. Cycle numbers are indicated in parentheses. (B and C) ChIP analysis of H3K9 methylation on three regions (a–c) of the DH1 (B) and OsMADS47 (C) as indicated. Antibodies against histone H3, H3K9me1, H3K9me2, and H3K9me3 were used. Tests with input DNA and without antibody (Mock) were included as controls. PCR cycle numbers are indicated in parentheses.
To study whether the jmj6 mutations affected histone methylation on DH1, OsMADS47, and OsMADS8, chromatin immunoprecipitation (ChIP) assays with specific antibodies against histone H3, H3K9me1, H3K9me2, or H3K9me3 were performed. The precipitated DNA was analyzed by PCR using primer sets corresponding to three regions of the genes with three biological repeats, which produced consistent results. Results from one of the repeats are shown in Fig. 5 B and C. H3K9me2 and H3K9me3 on the promoter and 5′ regions of DH1 and OsMADS47 were more abundant in the mutants than in the WT, whereas no difference was detected in more downstream regions. Probably because of the sensitivity of the methods, there was no obvious alteration of H3K9me1 detected in the mutants. In contrast, the mutation did not seem to affect H3K9 methylations on OsMADS8 (data not shown). These results suggest that JMJ706-dependent H3K9 demethylation is required for the expression of DH1 and OsMADS47.
To characterize the function of JMJ706 further, transgenic rice plants overexpressing the gene were produced (Fig. S6A). However, the transformants did not show any aberrant phenotype (data not shown), and the expression of DH1, OsMADS47, or OsMADS8 in the overexpression lines was not affected (Fig. S6B).
Analysis of Histone Modifications in jmj6 Mutants.
To study the function of JMJ706 on histone modification, histone-enriched fractions extracted from knockout, overexpressing, or WT seedlings were analyzed by Western blot tests. An increase of H3K9me2 and H3K9me3, along with a decrease of H3K9me1, was detected in the mutants compared with the WT (Fig. 3B). In addition, a slight increase of H3K36me2 accompanied by a decrease of H3K36me1 was also observed. However, H3K36me3 was not altered by the mutation. Similarly, acetylation of H3K9 and methylation of H3K27 and H3K4 were not affected. Accordingly, overexpression of JMJ706 caused a decrease of H3K9me2 but not H3K4me3 (Fig. 3C). The decrease was more pronounced in OX6–2, probably because there was a higher level of JMJ706 expression (Fig. S4). Accordingly, an increase of H3K9me1 was detected in this line (Fig. 3C). These data confirmed the in vitro histone demethylation results (Fig. 3A) and suggested that JMJ706 mainly reverses H3K9me2 and H3K9me3 and, to a lesser extent, H3K36me2 in rice.
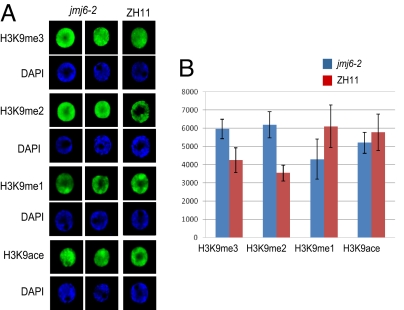
Finally, the effect of the mutations on chromatin modification was analyzed by nuclei immunostaining using antibodies against H3K9me3, H3K9me2, H3K9me1, and acetylated H3K9 (Fig. 6). Compared with WT nuclei, signals for H3K9me3 and H3K9me2 (each measured from 18 nuclei) were substantially enhanced in the mutants, whereas that for H3K9me1 in jmj6 was reduced. No significant difference was detected for H3K9 acetylation. In particular, signals for H3K9me2 were more diffused in the mutant nuclei, compared with the WT, in which H3K9me2 was detected mostly in heterochromatic regions.
Fig. 6.
Comparison of H3K9 methylation and acetylation between jmj6-2 and WT. (A) Nuclei isolated from leaves of 10-day-old seedlings of WT (ZH11) and jmj6 mutants were immunostained with antibodies indicated on the left and revealed by Alexa Fluor 488. The same cells were stained by DAPI. (B) Relative intensity of the fluorescence was measured using the software Quantity-One 4.6 (BIO-RAD). Bars represent SD from measures of 18 nuclei for each antibody. Student's t tests reveal that the differences between ZH11 and jmj6–2 for H3K9me3, H3K9me2, and H3K9me1 were significant at 5%.
Discussion
Specific Features of Plant jmjC Domain Protein Genes.
In this study, we show that plant jmjC domain-containing proteins have several distinct features compared with other model organisms. First, plant jmjC proteins can be divided into five groups, four of which contain conserved homologues in other model organisms (e.g., human). The remaining group appears to be specific to the plant lineage. In addition to jmjC, jmjN, and a C5HC2 ZnF domain, this group contains two additional motifs that are absent from human jmjC proteins (Fig. S1A). Second, there is no plant protein related to the human UTX/JMJ3, JHDM1, and PHF subfamilies. UTX/JMJ3 proteins are H3K27 demethylases (10), and the absence of UTX/JMJ3 homologues in plants means that plant H3K27 methylation is reversed by different jmjC domain-containing or distinct proteins. This may reflect the specific features of plant H3K27me3 in terms of its genomic landscape and recognition by chromatin proteins (22). Unlike in animal cells, in which H3K27me3 occupies large genomic regions, H3K27me3 is restricted to the transcribed region of a large number of genes in Arabidopsis (22). H3K27me3 is recognized and bound by LHP1 (like heterochromatin protein 1) in Arabidopsis (23, 24), whereas in Drosophila and mammals, it is bound by Polycomb, a key component of PCR1 (Polycomb repressive complex 1) (25). Third, mammalian JHDM1A and JHDM1B and their orthologues from budding yeast are demethylases of H3K36me1 and H3K36me2 (6). Again, the absence of JHDM1 homologues in plants suggests that demethylation of H3K36me1 and H3K36me2 depends on different activities. The present data suggest that rice JMJ706 may have an H3K36me2 demethylation activity. The specific features of plant jmjC proteins suggest that they may have different functions in the regulation of chromatin structure and genome activity.
Chromatin Function of Plant JMJD2 Members.
Rice JMJ706 is shown to be related to the human JMJD2 proteins. The human JMJD2 subfamily has four members, JMJD2A–D. The capacity of JMJD2A–C to reverse both H3K9me3 and H3K36me3 suggests a potential link between these two modifications. Since H3K9me3 and H3K36me3 have been implicated in transcriptional elongation and suppression of inappropriate transcription initiation within the body of the gene (1), the dual-site specificity of JMJD2 members suggests a coordinated role in regulating H3K9me3 and H3K36me3 for gene repression. This is consistent with the previous identification of JMJD2A as a transcriptional repressor in mammalian cells (26, 27). The present data showing that the rice JMJD2 homologue JMJ706 lacks this dual-site specificity for H3K9me3 and H3K36me3 demethylation infers that either dual demethylation is ensured by a different member or H3K9me3/H3K36me3 may have a different function in plant gene regulation. Although JMJ706 is not required for the demethylation of H3K36me3, the jmj6 mutations led to an increase of H3K36me2 and a decrease of H3K36me1. Therefore, JMJ706 may play a role in the conversion between H3K36me1 and H3K36me2. It has recently been shown that H3K36me1 has different function than H3K36me2/me3 in transcriptional regulation in plants (28).
Although rice JMJ706 is constitutively expressed (Fig. S1C), loss-of-function mutations resulted in only a variety of spikelet development defects (Fig. 4 D–H). The heterogeneity of the floral phenotype is reminiscent of phenotypes of mutants or transgenic plants affected in other chromatin modification activities, such as histone acetylation and DNA methylation (29–31). The jmj6 mutations led to the repression of OsMADS47 and DH1 (Fig. 5). Although the developmental function of OsMADS47 is presently unknown, the mutation of DH1 produces defects on palea and lemma formation (21), which are similar to the jmj6 phenotypes. Increased H3K9me2 and H3K9me3 on the promoter and 5′ regions of DH1 and OsMADS47 in the jmj6 mutants suggest that the genes may be targeted by JMJ706.
Two Arabidopsis members of this group (Fig. 2A), AtJMJ11/ELF6 and AtJMJ12/REF6, have been shown to be involved in the regulation of flowering time (13). Although these genes are involved in two different pathways and have opposite functions in flowering time regulation, they seem to act as transcriptional repressors of specific genes. For instance, the ref mutations induced the expression of the flowering repressor FLC and histone H4 acetylation (13). The present data show that JMJ706 is mainly involved in H3K9me2 and H3K9me3 demethylation and that H3K9 acetylation was not affected by the jmj6 mutations (Figs. 3 and 6). It is likely that rice JMJ706 and the Arabidopsis homologues target different chromatin domains. Colocalization of JMJ706-GFP and JMJ706-FLAG with enriched DAPI-stained regions in transgenic nuclei suggests that JMJ706 may have a function in heterochromatin. Similarly, in mammalian cells, JMJD2B antagonizes H3K9me3 at pericentric heterochromatin (7), whereas JMJD2A is associated with a transcriptional repressor complex (27). Therefore, members of this subfamily of H3K9 demethylases may have distinct functions in the regulation of chromatin structure and gene expression.
Plant SU(VAR)3–9 homologues of histone methyltransferases (i.e., SUVH4/KYP, SUVH5, and SUVH6 in Arabidopsis and SDG714 in rice) are involved in H3K9 methylation at transposon and/or repetitive sequences and are required for DNA methylation (32–34). The present data suggest that JMJ706 might antagonize the function of SUVH type histone methyltransferases in pericentric heterochromatin. It would be interesting to identify discriminating signals that determine whether SUVH or JMJ706 will prevail to induce or decrease H3K9 methylation at pericentric heterochromatin.
Materials and Methods
Phylogenetic Analysis.
Jumonji C domain-containing protein sequences were downloaded from the ChromDB database (www.chromdb.org). The HMMER package version 2.1 was used to search additional rice and Arabidopsis sequences. Plant jmjC proteins were named according to the ChromDB database. The SMART database (http://smart.embl-heidelberg.de) was used to retrieve the entire conserved jmjC sequences. The ClustalX program was used to generate alignments of jmjC protein sequences. The phylogenetic tree generation was performed by MEGA 3.1 using the neighbor-joining method with a Poisson correction model and a bootstrap of 1000 replicates.
For MADS-box protein tree building, databases were searched for sequences using the keywords described by Yamaguchi and Hirano (19), and analyzed with the previously mentioned methods.
Characterization of JMJ706 Insertion Mutants.
Two independent T-DNA insertion mutant alleles, jmj6–1 (03Z11FO83) and jmj6–2 (05Z11DO89), were identified in the Rice Mutant Database (http://rmd.ncpgr.cn). The insertions were confirmed by PCR using JMJ706-specific primers (Fig. 4A and Table S1) and a T-DNA specific primer, NTLB5. The primers used for RT-PCR analysis presented in Fig. 4C are listed in Table S1.
Vector Constructions and Rice Transformation.
The JMJ706 full-length cDNA clone J033044E04 obtained from the RIKEN database was used for vector construction. The binary plasmid pU1301 (35) was used to construct the JMJ706 overexpression vector. The JMJ706 artificial micro-RNA (amiR706) vector was made as described (18). The artificial micro-R706 sequence (5′-TTGACTCTAAATATGTCATAC-3′, the mismatched bases are italic) corresponding to nucleotides 2082–2102 of the JMJ706 coding region was inserted into the RS300 vector and cloned into pU1301. For JMJ706-GFP fusion protein expression, the full-length cDNA of JMJ706 was fused to the GFP coding sequence under the control of the maize ubiquitin promoter. For the mutant complementation experiments, the JMJ706 full-length cDNA was inserted downstream from the 3 × FLAG tag under the control of the maize ubiquitin promoter in a modified pU2301 vector, which contains a G418 antibiotic selection marker. Callus generated from Zhonghua11 rice cultivar (ZH11) or jmj6–2 mutant plants was used for rice transformation as described previously (35).
Histone Demethylation Assays and Western Blot Analysis.
The rice JMJ706 cDNA region encoding amino acids 1–651 was subcloned into pGEX-6P-1 to make the GST-JMJ706 fusion. The fusion protein was expressed in and affinity-purified from E. coli BL21 (DE3, RIL) cells and used for in vitro histone demethylation assays according to Whetstine et al. (9). Antibodies used to detect histone modifications are described below.
For Western blot analysis, histone-enriched fractions were extracted from WT, mutant, or transgenic rice leaves as described previously (35). Western blot tests were performed using antibodies bought from Upstate and Abcam.
Chromatin Immunoprecipitation Assays.
Leaves of 2-week-old rice seedlings were used for chromatin immunoprecipitation assays, and the methods were performed as described (35). After extracted with phenol/chloroform/isoamyl alcohol, the immunoprecipitated DNA was washed with 75% ethanol and resuspended in 80 μl of TE (10 mM Tris/1 mM EDTA, pH 8.0) water. Polymerase chain reactions were carried out for 32 or 36 cycles, and the primers are listed in Table S2. At least three repetitions of PCR analysis were performed and obtained similar results.
Immunostaining and Electron Microscopic Analysis.
Immunostaining was performed according to Houben et al. (36). For scanning electron microscopic analysis, flowers of mutant and WT plants were fixed by glutaraldehyde (2.5%) and observed under a Hitachi S-570 microscope.
Nuclear Localization and Microscopy.
JMJ706 cDNA fragments were amplified by PCR and directionally inserted into the nuclear localization vector pU1391-GFP (35). The constructs were expressed in onion cells and observed using a confocal microscope as described by Huang et al. (35). Transgenic plants expressing the JMJ706-GFP fusion were produced as mentioned previously for in vivo localization. After being fixed in 4% PFA, mesophyll cell nuclei were stained with DAPI, and the images were visualized using a confocal microscope.
For FLAG-tagged JMJ706 immunostaining, the mouse anti-FLAG primary antibody (Sigma) and the Alexa Fluor 594–coupled goat anti-mouse second antibody (Molecular Probes) were used to detect the FLAG-tagged JMJ706.
RT-PCR and Northern Blot.
Ribonucleic acid from different tissues or organs of WT and mutant plants was used for reverse transcription. The F3 and R3 primers mentioned previously were used for JMJ706 expression analysis. The sequences of the primers used for MADS-box and DH1 gene transcript analysis are listed in Table S1. Fifteen micrograms of total RNA extracted from transgenic leaves was used for Northern blot analysis. The fragment amplified by F3 and R3 primers was 32P-labeled as a probe.
Supplementary Material
Acknowledgments.
We thank Dr. Michael Hodges for critical reading of the manuscript and Moussa Benhamed for analyzing the data presented in Fig. 6. This work was supported by grants from the National Special Key Program on Functional Genomics of Major Plants and Animals and the National Natural Science Foundation of China.
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the GenBank database [accession no. NP_001065492 (JMJ706)].
This article contains supporting information online at www.pnas.org/cgi/content/full/0805901105/DCSupplemental.
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Margueron R, Trojer P, Reinberg D. The key to development: interpreting the histone code? Curr Opin Genet Dev. 2005;15:163–176. doi: 10.1016/j.gde.2005.01.005. [DOI] [PubMed] [Google Scholar]
- 3.Lee MG, Wynder C, Cooch N, Shiekhattar R. An essential role for CoREST in nucleosomal histone 3 lysine 4 demethylation. Nature. 2005;437:432–435. doi: 10.1038/nature04021. [DOI] [PubMed] [Google Scholar]
- 4.Metzger E, et al. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature. 2005;437:436–439. doi: 10.1038/nature04020. [DOI] [PubMed] [Google Scholar]
- 5.Trewick SC, McLaughlin PJ, Allshire RC. Methylation: Lost in hydroxylation? EMBO Rep. 2005;6:315–320. doi: 10.1038/sj.embor.7400379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tsukada Y, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 7.Fodor BD, et al. Jmjd2b antagonizes H3K9 trimethylation at pericentric heterochromatin in mammalian cells. Genes Dev. 2006;20:1557–1562. doi: 10.1101/gad.388206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klose RJ, et al. The transcriptional repressor JHDM3A demethylates trimethyl histone H3 lysine 9 and lysine 36. Nature. 2006;442:312–316. doi: 10.1038/nature04853. [DOI] [PubMed] [Google Scholar]
- 9.Whetstine JR, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481. doi: 10.1016/j.cell.2006.03.028. [DOI] [PubMed] [Google Scholar]
- 10.Agger K, et al. UTX and JMJD3 are histone H3K27 demethylases involved in HOX gene regulation and development. Nature. 2007;449:731–734. doi: 10.1038/nature06145. [DOI] [PubMed] [Google Scholar]
- 11.Christensen J, et al. RBP2 belongs to a family of demethylases, specific for tri-and dimethylated lysine 4 on histone 3. Cell. 2007;128:1063–1076. doi: 10.1016/j.cell.2007.02.003. [DOI] [PubMed] [Google Scholar]
- 12.Klose RJ, Kallin EM, Zhang Y. JmjC-domain-containing proteins and histone demethylation. Nat Rev Genet. 2006;7:715–727. doi: 10.1038/nrg1945. [DOI] [PubMed] [Google Scholar]
- 13.Noh B, et al. Divergent roles of a pair of homologous jumonji/zinc-finger-class transcription factor proteins in the regulation of Arabidopsis flowering time. Plant Cell. 2004;16:2601–2613. doi: 10.1105/tpc.104.025353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saze H, Shiraishi A, Miura A, Kakutani T. Control of genic DNA methylation by a jmjC domain-containing protein in Arabidopsis thaliana. Science. 2008;319:462–465. doi: 10.1126/science.1150987. [DOI] [PubMed] [Google Scholar]
- 15.Chen Z, et al. Structural insights into histone demethylation by JMJD2 family members. Cell. 2006;125:691–702. doi: 10.1016/j.cell.2006.04.024. [DOI] [PubMed] [Google Scholar]
- 16.Wu C, et al. Development of enhancer trap lines for functional analysis of the rice genome. Plant J. 2003;35:418–427. doi: 10.1046/j.1365-313x.2003.01808.x. [DOI] [PubMed] [Google Scholar]
- 17.Zhang J, et al. RMD: A rice mutant database for functional analysis of the rice genome. Nucleic Acids Res. 2006;34:D745–748. doi: 10.1093/nar/gkj016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi T, Hirano HY. Function and diversification of MADS-box genes in rice. Scientific World J. 2006;6:1923–1932. doi: 10.1100/tsw.2006.320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Itoh J, et al. Rice plant development: from zygote to spikelet. Plant Cell Physiol. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- 21.Li A, et al. DH1, a LOB domain-like protein required for glume formation in rice. Plant Mol Biol. 2008;66:491–502. doi: 10.1007/s11103-007-9283-3. [DOI] [PubMed] [Google Scholar]
- 22.Zhang X, et al. Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 2007;5:e129. doi: 10.1371/journal.pbio.0050129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turck F, et al. Arabidopsis TFL2/LHP1 specifically associates with genes marked by trimethylation of histone H3 lysine 27. PLoS Genet. 2007;3:e86. doi: 10.1371/journal.pgen.0030086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang X, et al. The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat Struct Mol Biol. 2007;14:869–871. doi: 10.1038/nsmb1283. [DOI] [PubMed] [Google Scholar]
- 25.Schwartz YB, Pirrotta V. Polycomb silencing mechanisms and the management of genomic programmes. Nat Rev Genet. 2007;8:9–22. doi: 10.1038/nrg1981. [DOI] [PubMed] [Google Scholar]
- 26.Gray SG, et al. Functional characterization of JMJD2A, a histone deacetylase- and retinoblastoma-binding protein. J Biol Chem. 2005;280:28507–28518. doi: 10.1074/jbc.M413687200. [DOI] [PubMed] [Google Scholar]
- 27.Zhang D, Yoon HG, Wong J. JMJD2A is a novel N-CoR-interacting protein and is involved in repression of the human transcription factor achaete scute-like homologue 2 (ASCL2/Hash2) Mol Cell Biol. 2005;25:6404–6414. doi: 10.1128/MCB.25.15.6404-6414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu L, et al. Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol Cell Biol. 2008;28:1348–1360. doi: 10.1128/MCB.01607-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bertrand C, Bergounioux C, Domenichini S, Delarue M, Zhou DX. Arabidopsis histone acetyltransferase AtGCN5 regulates the floral meristem activity through the WUSCHEL/AGAMOUS pathway. J Biol Chem. 2003;278:28246–28251. doi: 10.1074/jbc.M302787200. [DOI] [PubMed] [Google Scholar]
- 30.Ronemus MJ, Galbiati M, Ticknor C, Chen J, Dellaporta SL. Demethylation-induced developmental pleiotropy in Arabidopsis. Science. 1996;273:654–657. doi: 10.1126/science.273.5275.654. [DOI] [PubMed] [Google Scholar]
- 31.Tian L, et al. Reversible histone acetylation and deacetylation mediate genome-wide, promoter-dependent and locus-specific changes in gene expression during plant development. Genetics. 2005;169:337–345. doi: 10.1534/genetics.104.033142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ding Y, et al. SDG714, a histone H3K9 methyltransferase, is involved in Tos17 DNA methylation and transposition in rice. Plant Cell. 2007;19:9–22. doi: 10.1105/tpc.106.048124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ebbs ML, Bender J. Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell. 2006;18:1166–1176. doi: 10.1105/tpc.106.041400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jackson JP, Lindroth AM, Cao X, Jacobsen SE. Control of CpNpG DNA methylation by the KRYPTONITE histone H3 methyltransferase. Nature. 2002;416:556–560. doi: 10.1038/nature731. [DOI] [PubMed] [Google Scholar]
- 35.Huang L, et al. Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 2007;144:1508–1519. doi: 10.1104/pp.107.099473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Houben A, et al. Methylation of histone H3 in euchromatin of plant chromosomes depends on basic nuclear DNA content. Plant J. 2003;33:967–973. doi: 10.1046/j.1365-313x.2003.01681.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.