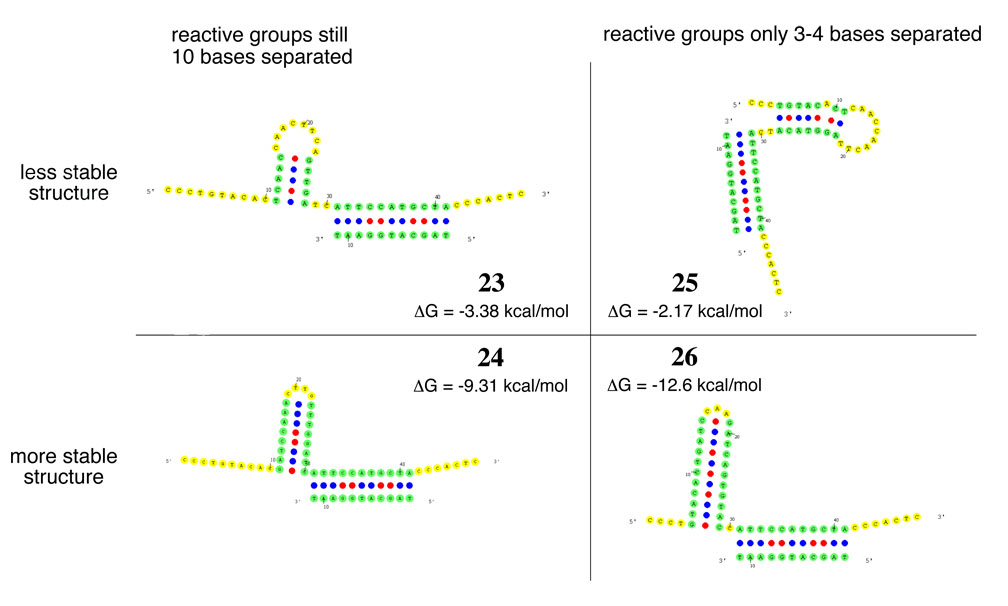Abstract
DNA-templated organic synthesis enables the translation, selection, and amplification of DNA sequences encoding synthetic small-molecule libraries. As the size of DNA-templated libraries increases, the possibility of forming intramolecularly base-paired structures within templates that impede templated reactions increases as well. In order to achieve uniform reactivity across many template sequences and to computationally predict and remove any problematic sequences from DNA-templated libraries, we have systematically examined the effects of template sequence and secondary structure on DNA-templated reactivity.
By testing a series of template sequences computationally designed to contain different degrees of internal secondary structure, we observed that high levels of predicted secondary structure involving the reagent binding site within a DNA template interfere with reagent hybridization and impair reactivity, as expected. Unexpectedly, we also discovered that templates containing virtually no predicted internal secondary structure also exhibit poor reaction efficiencies. Further studies revealed that a modest degree of internal secondary structure is required to maximize effective molarities between reactants, possibly by compacting intervening template nucleotides that separate the hybridized reactants. Therefore, ideal sequences for DNA-templated synthesis lie between two undesirable extremes of too much or too little internal secondary structure. The relationship between effective molarity and intervening nucleic acid secondary structure described in this work may also apply to nucleic acid sequences in living systems that separate interacting biological molecules.
Introduction
DNA-templated organic synthesis (DTS)1 effects the translation of a sequence of DNA into a corresponding synthetic molecule. This method does not require biosynthetic machinery and instead uses the hybridization of two oligonucleotides to increase the effective molarity of attached chemical groups, inducing reactions between sequence-programmed reaction partners. DTS has enabled new modes of chemical reactivity not accessible by conventional synthesis methods,2–4 the discovery of new chemical reactions,5,6 and the translation, selection, and amplification of DNA sequences encoding synthetic small-molecule libraries.7
To fully realize the potential of DTS to generate libraries of synthetic molecules suitable for in vitro selection requires the translation of large libraries containing many DNA sequences into corresponding small molecules. The challenge of generating codons that support efficient and sequence-specific DNA-templated synthesis grows rapidly with library size as the number of possible undesired intra- and intermolecular base pairings increases exponentially. Because the individual screening of all templates and reagents to identify problematic sequences is not practical as library sizes increase, we sought to understand principles that enable the computational design of sequences that support consistently high levels of templated reactivity.
Here we report the results of a systematic study to reveal those aspects of DNA template sequences and secondary structures that most strongly influence DNA-templated reactivity. We observed that intramolecular base pairing within the template can decrease reactivity, as expected; however we also discovered that some template secondary structure is required for efficient DNA-templated reactions. Because these key determinants of a template sequence’s ability to react can be screened computationally, the findings from this work enhance the robustness of nucleic acid-templated synthesis, especially when generating libraries of many DNA-templated products. In addition, the principles revealed in these studies may shed light on the effective molarities experienced by nucleic acid-bound biological molecules in cells.
Materials and Methods
All chemicals, unless otherwise noted, were purchased from Sigma-Aldrich. All reagents for DNA synthesis, including modified phosphoramidites and CPG resins, were purchased from Glen Research. All buffers were prepared at room temperature to match reaction conditions.
Secondary Structure Prediction
The design of specific secondary structures for the templates was preformed using the Oligonucleotide Modeling Platform (OMP; DNA Software, Inc.). All simulations to determine secondary structures were performed at 25 °C in 1.0 M NaCl with 100 nM template. Hybridization to the templates was also simulated, using a 150 nM reagent sequence with the 100 nM template sequences under the same conditions. Parallel simulations in NUPACK and MFOLD yielded similar results (see Supporting Information).
DNA Template and Reagent Synthesis
All DNA oligonucleotides were synthesized on a PerSeptive Biosystems Expedite 8090 DNA synthesizer using standard phosphoramidite protocols and purified by reverse-phase HPLC using a triethylammonium acetate (TEAA)/CH3CN gradient. The DNA sequences and structures used in this work are listed in the Supporting Information.
The template oligonucleotides (1–8, 15–22) were synthesized using 3’-(6-Fluorescein) CPG and 5’-Amino Modifier 5 Phosphoramidite (see Supporting Information for details). As UV visualization of DNA using common stains such as ethidium bromide can be affected by the amount of secondary structure in a DNA sequence, the fluorescein modification was included on the templates so that quantitation of reaction yield would be more accurate and consistent across different species.
The reagent oligonucleotides (9–14) were synthesized using 3’-Amino-Modifier C7 CPG 500. Following DNA synthesis and purification, these oligonucleotides were redissolved in 0.2 M sodium phosphate buffer, pH 7.2. Reagents for testing reductive amination were synthesized by adding 10 µL of a 20 mg/mL solution of the N-hydroxysuccinimidyl ester of p-carboxybenzaldehyde in DMF to an equal volume of the DNA reagent. After 1 h, the reaction was purified by gel filtration using Sephadex G-25 followed by reverse-phase HPLC using a TEAA/CH3CN gradient. Reagents for testing amine acylation were synthesized by first adding 0.1 volumes of a 0.1 M solution of (D)-phenylalanine in 0.2 M sodium phosphate buffer, pH 7.2 to the DNA reagent followed by the addition of 0.2 volumes of a 100 mM bis[2-(succinimidyloxycarbonyloxy)-ethyl]sulfone (BSOCOES, Pierce) solution in DMF. After 2 h, the reaction was purified by gel filtration using Sephadex G-25 followed by reverse-phase HPLC using a TEAA/CH3CN gradient. All DNA reagents were characterized by MALDI-TOF mass spectrometry.
Reductive Amination
DNA-templated reductive amination reactions were carried out in 0.1 M MOPS buffer, pH 7.0, 1 M NaCl, with 100 nM amine-linked DNA template and 150 nM aldehyde-linked DNA reagent. Reactions were commenced by the addition of 50 mM NaCNBH3 and reacted for 8 h at 25 °C. Reactions were then quenched by the addition of 0.1 volumes of a 1 M glycine solution, pH 7.0, and ethanol precipitated before analysis by denaturing polyacrylamide gel electrophoresis (PAGE) using Ready Gel 15% TBE-Urea Gels (BioRad).
Amine Acylation
DNA-template amine acylation reactions were carried out in 0.1 M MES buffer, pH 6.0, 1 M NaCl, with 100 nM amine-linked DNA template and 150 nM carboxylic acid-linked DNA reagent. Reactions were commenced by the addition of 24 mM sulfo-N-hydroxysuccinimide (sNHS) and 32 mM N-(3-dimethylaminopropyl)-N’-ethylcarbodiimide hydrochloride (EDC) and reacted for 8 h at 25 °C. Reactions were then quenched by the addition of 0.1 volumes of a 1 M glycine solution and ethanol precipitated before analysis by denaturing polyacrylamide gel electrophoresis (PAGE) as above.
Determination of Yield
Reaction yields were quantitated by denaturing polyacrylamide gel electrophoresis followed by CCD-based densitometry of the product and template starting material bands using the attached fluorescein label on the templates for quantitation. While the reported yields are for individual experiments, the overall yields, and in particular the reactivity trends between individual templates, were consistent in repeated trials.
Results
Design of DNA Templates to Study Secondary Structure
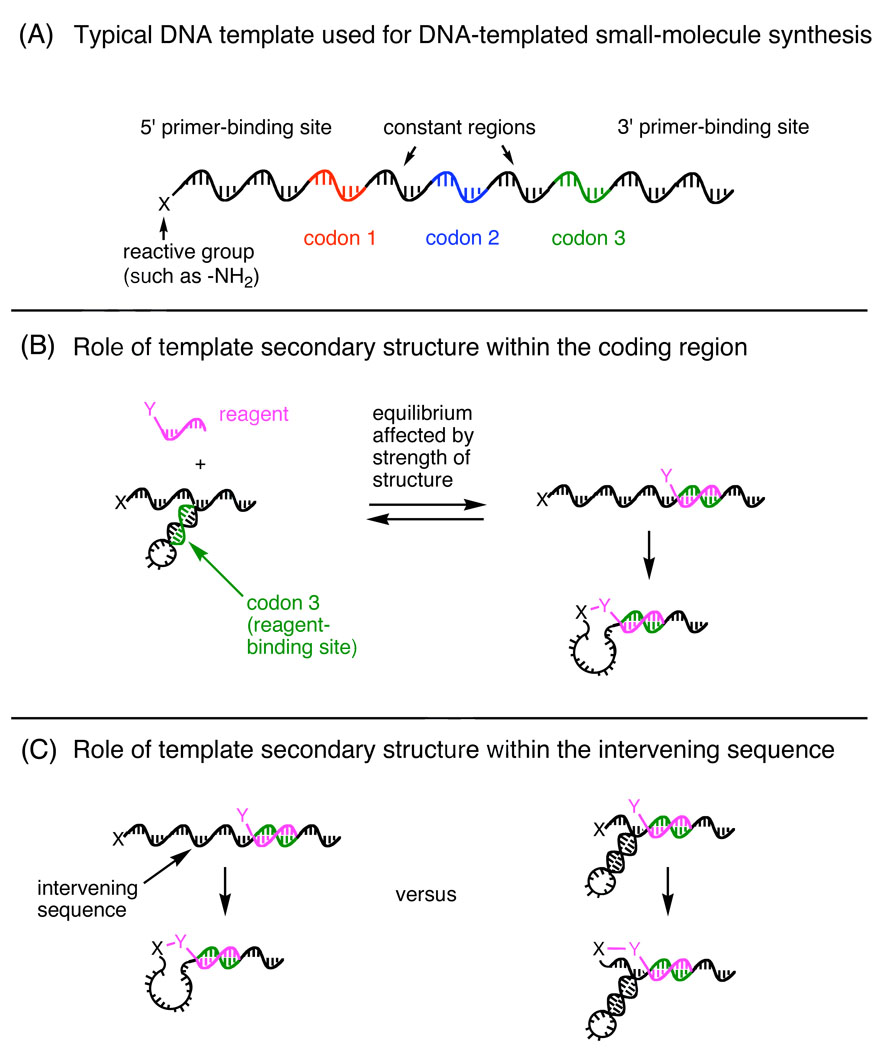
The typical design of a DNA template encoding a small-molecule library member7 is shown in Figure 1A. Each template contains three 10- to 12-base coding regions that hybridize with complementary reagent-linked oligonucleotides to effect the synthesis of the corresponding library member. Two 10-base PCR primer-binding sites flank the three coding regions. Because the starting material and subsequent intermediates are linked to the 5’ terminus of the template, each DNA-templated step requires the interaction between the 5’ end of the template and a reactant annealed approximately 10 to 30 bases away.
Figure 1.
Design of DNA templates to reveal the role of secondary structure in determining templated reactivity.
We expected that template sequences capable of forming internal secondary structure involving coding regions would impede reagent hybridization and therefore serve as poor mediators of DNA-templated synthesis (Figure 1B). To elucidate the relationship between template secondary structure and the efficiency of DNA-templated synthesis, we designed and synthesized a series of 5’-amine-linked templates (1–8) with varying internal secondary structures that are computationally predicted to span a ~10 kcal/mol range in intramolecular folding energies. The predicted secondary structures and folding energies are shown in Figure 2. All eight templates contain the same primer-binding sequences as well as the same intervening sequences between the coding regions (Figure 1A). Templates 1–8 also share the sequence for codon 3, located 30 bases away from the reactive end of the template, so that a single reagent can be used to test the reactivity of the entire series of templates. By varying the sequences used for codons 1 and 2, predicted stem-loops of different stabilities were introduced into templates 1–8 that could conceal codon 3 (Figure 2).
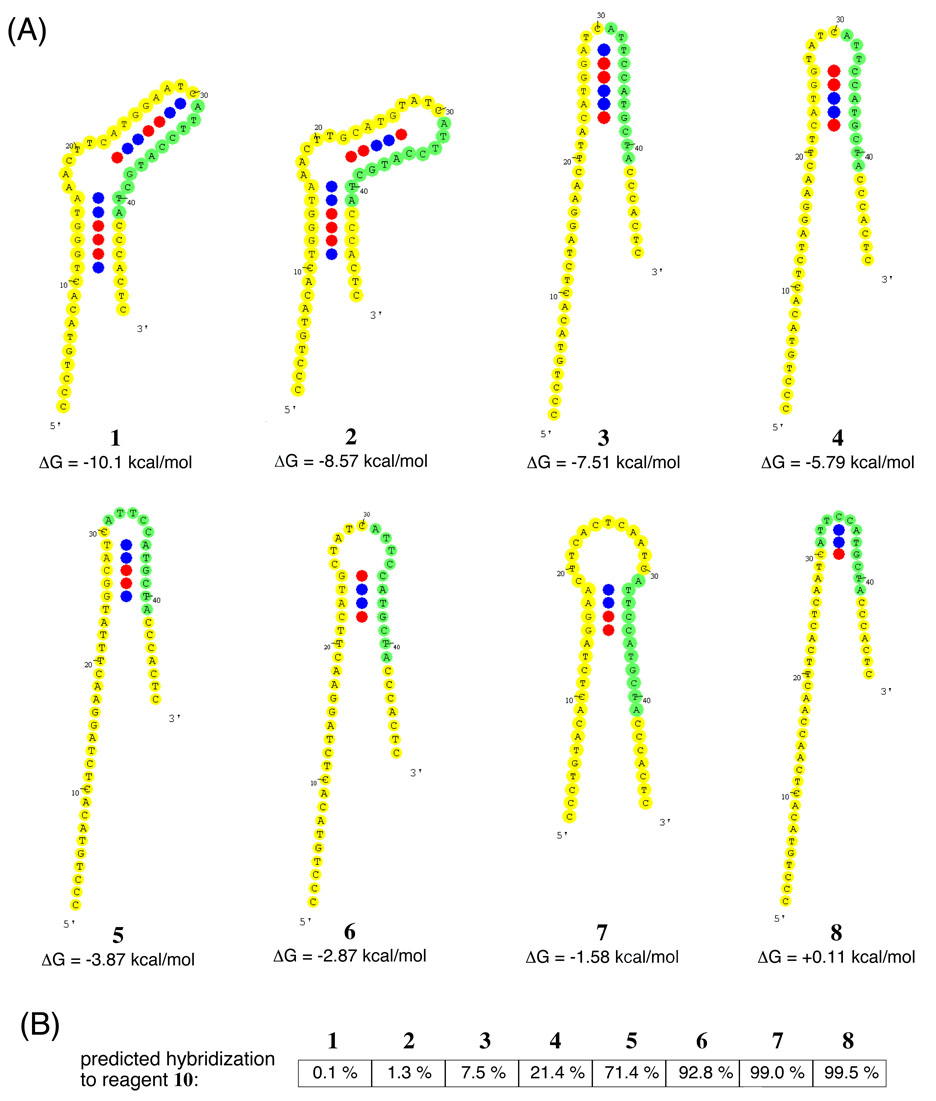
Figure 2.
Predicted folding properties of eight designed DNA templates. (A) Predicted secondary structures for each of the eight templates were generated by OMP using the conditions of 25 °C and 1 M NaCl. Each template’s predicted secondary structure contains base pairing within the binding site for reagent 10, although the energies for these structures vary over a 10 kcal/mol range. The binding site for reagent 10 is highlighted in green. In the structures, CG pairs are labeled with red circles while the less energetic AT pairs are labeled with blue circles. (B) The extent to which reagent 10 is predicted to hybridize to each of these templates was calculated using 100 nM template, 150 nM reagent 10, and 1 M NaCl at 25 °C.
The template with the highest degree of predicted internal structure is template 1, which contains a predicted stem-bulge-stem-loop structure with a folding energy of −10.1 kcal/mol. The least structured template (8) has very little predicted internal structure and has a slightly unfavorable predicted folding free energy of +0.11 kcal/mol. The remaining templates have intermediate degrees of predicted internal structure in order from 2 (−8.57 kcal/mol) to 7 (−1.58 kcal/mol) (Figure 2).
The Oligonucleotide Modeling Platform (OMP, DNA Software Inc.)8,9 was also used to model the hybridization of reagents to these templates (Figure 3A,B) under the experimental conditions (100 nM template, 150 nM reagent, 1 M NaCl, 25 °C). To support the predictions of OMP, we repeated this analysis of template secondary structure and reagent hybridization with other modeling programs, MFOLD10 and NUPACK11, and observed similar predicted structures and hybridization trends as those described below (see Supporting Information). We designed oligonucleotide reagent 9, which binds to the 5’ primer-binding site conserved in all templates and is predicted to be hybridized to > 99.5% of template molecules, to provide a benchmark for maximum reactivity when the functional groups on the template and reagent are brought very close together. We also designed oligonucleotide reagent 10, an 11-base reagent that anneals at codon 3, resulting in a 30-base separation between the reactive groups in the hybridized template and reagent. Reagent 10 must compete with any template secondary structures involving codon 3 for binding to the template (Figure 2B).
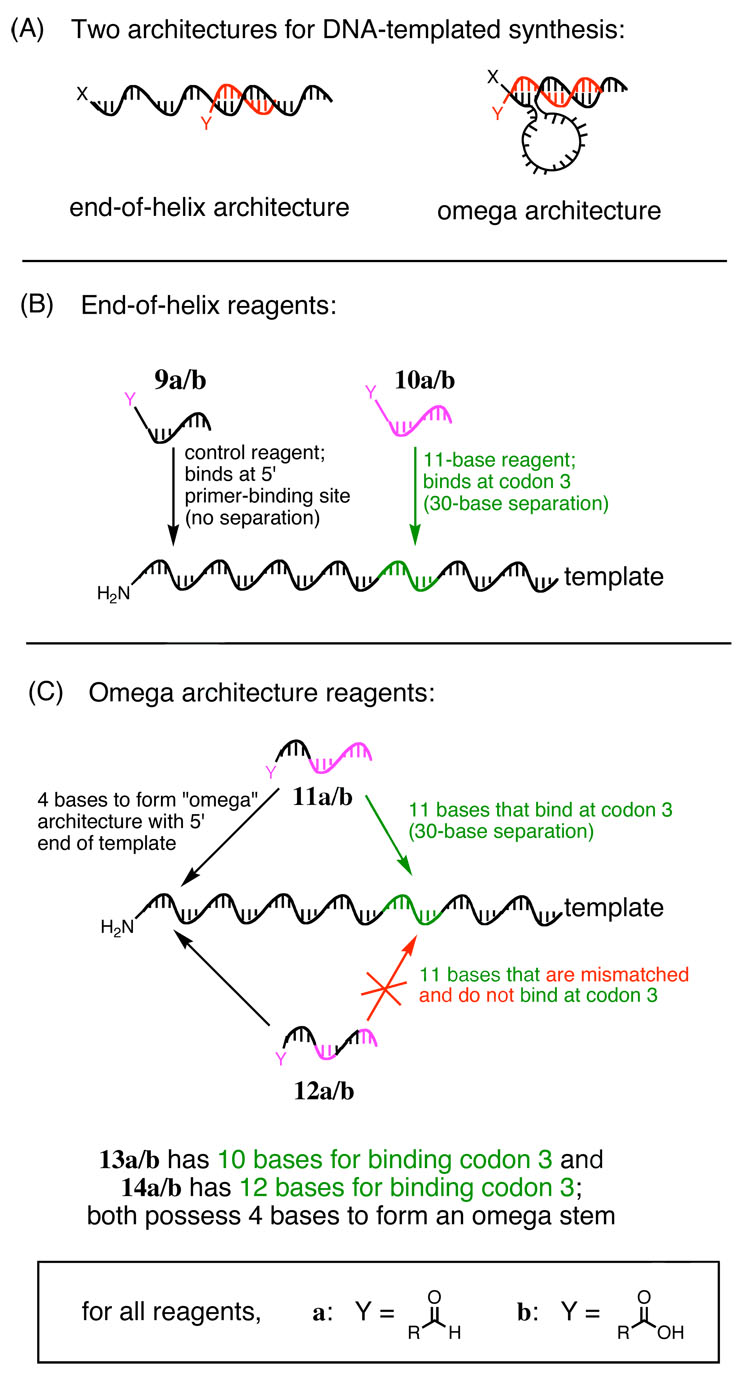
Figure 3.
Comparison of the end-of-helix architecture and omega architecture for DNA-templated reactions. (A) By introducing extra bases that complement the 5’ end of the template sequence, the omega architecture induces intervening template nucleotides to loop out, holding the reactive groups (X and Y) in close proximity and accelerating long-distance reactions. Reagents used in this study are shown in (B) and (C) for the two different architectures.
Against the strongest secondary structure in template 1, only 0.1 % of the total template is predicted by OMP to be bound by 10 at equilibrium. Instead, template 1 is predicted predominantly to engage in an intramolecular secondary structure that blocks the binding site for reagent 10. As the energy of the predicted template secondary structure decreases, reagent 10 is predicted to hybridize to the templates with increasing efficiency (Figure 2B), such that templates 6–8 are predicted to be over 90 % bound by reagent 10 at equilibrium. A simple model in which templates with the most available reagent-binding sites react the most efficiently predicts that reactivity should be highest for the least structured templates (6–8) and lowest for the most highly structured templates (1 and 2).
Reactivity of Templates Using the End-of-Helix Architecture
Two different DNA-templated reactions, amine acylation and reductive amination, were used to study reactivity. We previously showed that DNA-templated amine acylation can occur even when dozens of nucleotides separate reactive groups;4,7,12–15 in contrast, reductive amination is more distance-dependent and requires proximal hybridization of DNA-linked aldehyde and amine groups to react efficiently.14,16 Reagents 9 and 10 were therefore linked to either carboxybenzaldehyde to present an aldehyde group for reductive amination (9a and 10a) or to (D)-phenylalanine to present a carboxylic acid for amine acylation (9b and 10b).
We tested the reactivity of templates 1–8 first with a 10-base positive-control reagent (9a) complementary to the 5’ primer-binding site in the templates bearing an aldehyde group. Because this reagent should bind efficiently to each of the eight templates and because there are no intervening nucleotides separating the reactive groups in hybridized template-reagent complexes involving 9a and 1–8, this reagent establishes the maximum expected reactivity of the reagents when template secondary structure and distance are not impeding factors. Indeed, under reductive amination conditions (1–8 with 9a in 0.1 M MOPS buffer pH 7.0, 1 M NaCl, and 50 mM NaCNBH3, 25 °C for 8 h), 1–8 all reacted to 88–94 % yield (see Table 1).
Table 1.
Product yields (in %) for templates 1–8 reacting with reagents using the end-of-helix architecture and reductive amination. Reactions were performed with 150 nM reagent, 100 nM template in 0.1 M MOPS buffer, pH 7.0, 1.0 M NaCl, and 50 mM NaCNBH3 for 8 h at 25 °C. The folding energies of the eight templates are listed below the product yields.
| Template | ||||||||
|---|---|---|---|---|---|---|---|---|
| Reagent | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| 9a | 91 | 92 | 94 | 92 | 90 | 89 | 88 | 88 |
| 10a | 8 | 20 | 62 | 34 | 27 | 32 | 7 | 3 |
| ΔG (kcal/mol) |
−10.1 | −8.57 | −7.51 | −5.79 | −3.87 | −2.87 | −1.58 | +0.11 |
We then measured the reactivity of the eight templates with an 11-base reagent (10a) that anneals 30 bases away from the amine group at the 5’ end of the templates. Given the predicted hybridization of this reagent to the templates (Figure 2B), we expected reactivity to increase as the amount of internal secondary structure in the templates decreased. Indeed, for templates 1 through 3, this trend was observed. (Table 1) The most structured template (1) reacted to provide product in only 8 % yield. Template 2, with less secondary structure than 1, generated product in 20 % yield while template 3 was substantially more reactive, affording a 62 % yield of product. These results indeed were consistent with a model in which templates with the most internal secondary structure are not fully accessible for hybridization with reagents, significantly compromising reactivity. As the strength of this internal secondary structure decreases, hybridization of the reagent to the template is restored along with product yield.
Although we expected templates 4 through 8 to continue this trend, we were surprised to observe product yields falling dramatically as the total amount of secondary structure in the templates decreased. (Table 1) Templates 4 through 6 resulted in modest product yields of 27–34 %, about half of that of template 3. The least structured templates, 7 and 8, exhibited very low levels of reactivity, providing product in only 7% and 3% yield, respectively, up to 30-fold lower than the efficiency of reaction with 9a. As template 8 is predicted to be 99.5 % bound by either reagent 9a or reagent 10a, the decreased yield for this template does not likely arise from poor hybridization. Instead, we speculated that as the amount of secondary structure in the templates decreases below a certain point, the ability of the template to span its 30-base intervening distance decreases. In the extreme case of template 8, with no predicted folded structure in this intervening stretch of thirty bases, reactivity is almost completely eliminated. Collectively, these results reveal an unexpected and strong parabolic relationship between template internal secondary structure and yields of DNA-templated products encoded far from the reactive end of the template.
We performed similar experiments to study amine acylation using the end-of-helix architecture (see Supporting Information, Table S3). Just as with the reductive amination results, we observed increasing product yields for templates 1 to 3 reacting with reagent 10b (Table S3). Once again, however, as the amount of template secondary structure decreased further, product yields declined significantly, such that templates 7 and 8 were virtually unreactive. These surprising results together indicate that some template secondary structure is essential for high levels of reactivity, and that this trend is not specific to one type of chemical reaction.
Reactivity of Templates Using the Omega Template Architecture
We had previously developed the “omega” template-reagent architecture as a means of boosting reactant effective molarities and thereby augmenting reactivity when reagents are hybridized far from the reactive end of a template.14 Reagents that induce the omega architecture contain the same 10- to 12-base template-complementing sequence as the end-of-helix architecture, as well as 3–5 additional non-coding bases that exactly complement the 5’ end of the template (Figure 3A). These additional bases when paired with the template hold the 3’ end of the reagent in close proximity to the 5’ end of the template by looping out the intervening sequence. Because the reactivity of several of the templates described above was modest for reagents annealed thirty bases away from the end of the template (reagents 10a and 10b), we determined the effect of the omega architecture on the structure-reactivity trends revealed above.
We designed reagent 11 to contain the same 11-base coding sequence as reagent 10, as well as an additional four-base non-coding region that complements the 5’ end of templates 1–8 (Figure 3C). We also synthesized a mismatched reagent 12 that could not bind at codon three but still contained the four-base non-coding region as a control of sequence specificity (Figure 3C). Testing this mismatched reagent would demonstrate that any changes in reactivity were arising from changes in the ability of the template to hybridize with a reagent at codon 3, and not from the four-base non-coding region. These reagents contained either a 3’ aldehyde (11a and 12a) or a 3’ carboxylic acid (11b and 12b) for participation in reductive amination and amine acylation reactions.
As was observed with the end-of-helix architecture, reductive amination with the matched reagent 11a and templates 1–6 resulted in increasing yields as the amount of template secondary structure decreased (Table 2). The reactivity gradually improved as the amount of structure decreased, reaching a maximum with template 6 (90 % yield), which reacted comparably to the control reagent 9a. However, the least structured templates (7 and 8) still exhibited decreased reactivity with 11a, generating only 61 % or 49 % yield. The mismatched reagent (12a) results in < 5% yield when exposed to each of these templates, indicating that reactivity still relied on coding region complementarity.
Table 2.
Product yields (in %) for templates 1–8 reacting with reagents using the omega architecture. The results for 11a under reductive amination conditions and 11b under amine acylation conditions are shown for each of the eight template sequences after 8 hours.
| Template | ||||||||
|---|---|---|---|---|---|---|---|---|
| Reagent | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 |
| 11a | 12 | 37 | 64 | 76 | 78 | 90 | 61 | 49 |
| 11b | 7 | 19 | 41 | 40 | 56 | 64 | 46 | 37 |
Similar trends were observed when these omega architecture experiments were repeated for amine acylation (Table 2). The reactivity of the most structured templates 1 and 2 was low. Reactivity increased for templates 3 through 6, reaching a maximum of 64 % yield for template 6. The least structured templates (7 and 8) once again exhibit a decrease in reactivity with 11b. Taken together, these findings indicate that the DNA-templated reactivity of the least structured templates remains impaired, even in the omega architecture.
Reagent Length as a Probe of the Relationship Between Template Structure and Reactivity
To begin to elucidate the basis of the observed parabolic relationship between template internal secondary structure and DNA-templated reactivity, we varied the length of the reagent oligonucleotides. Increasing the number of nucleotides in the reagent strand increases the number of intermolecular base pairs in the template-reagent complex and therefore shifts the equilibrium between intramolecularly paired template and intermolecular reagent-template structures to favor the latter. Conversely, shortening reagent length should shift this equilibrium to favor intramolecularly paired template. Changes in DNA-templated reactivity that arise from changes in reagent length therefore would suggest that reactivity is at least partially limited by template-reagent hybridization for a given template.
We synthesized reagents that were both one base shorter (13) and one base longer (14) than the 11-base reagent 11 (Figure 3C). These reagents contain the same 4-base omega region as 11 and still hybridize 30 bases away from the reactive end of the template. Both aldehyde-linked (13a and 14a) and carboxylic acid-linked (13b and 14b) reagents were prepared as described above.
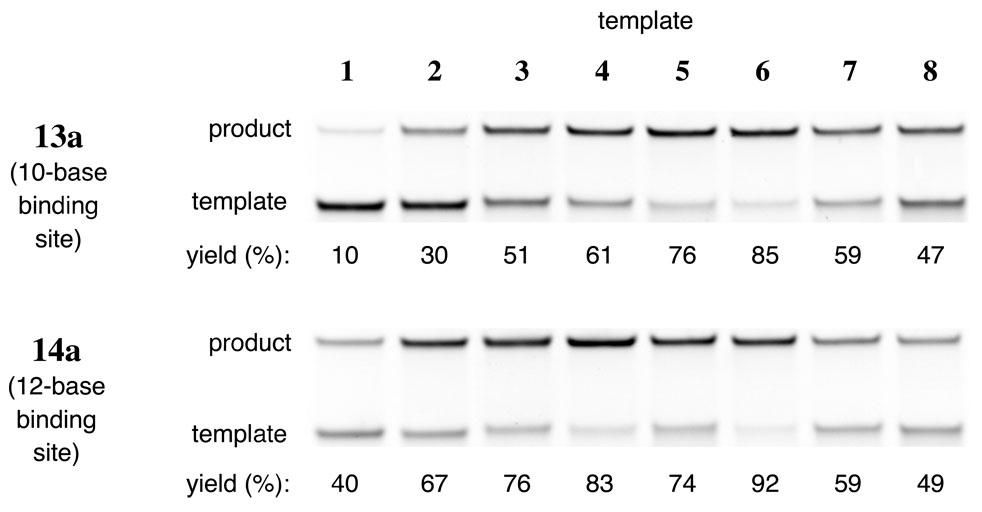
The shorter, 10-base reductive amination reagent 13a reduced product yields for highly and moderately structured templates 1–6 (Figure 4). Templates 2–4, for example, react with the 10-base reagent 13a to generate product in ~20 % lower yields than with the 11-base reagent 11a. Lengthening the reagent, conversely, increases reactivity for templates 1–6. For example, using 12-base reagent 14a, product yield with templates 1 and 2 increased by ~30 % each compared with using 11-base reagent 11a. Smaller increases were observed for templates 3–6, which already react efficiently with 11a. As expected, these results suggest that varying the length of reagent oligonucleotides can affect reactivity by altering the extent of template-reagent hybridization in the case of moderately to highly structured templates (1–6).
Figure 4.
The effect of reagent length on reductive amination yield. Denaturing PAGE analysis was performed on reactions using the templates and reagents shown. For templates 1 through 6, the increase in reagent length generally leads to an increase in reactivity. No length-dependent change in reactivity is observed, however, for templated reactions using the least structured templates, 7 and 8.
In contrast, both the shorter (13a) and longer (14a) reagents did not significantly alter the reactivity of unstructured templates 7 and 8 (Figure 4). Template 7 reacts in 59–61 % yield with reagents 11a, 13a, and 14a, while template 8 reacts in 47–49 % yield for the same three reagents. These results strongly suggest that the lower reactivity of the unstructured templates 7 and 8 is not due to inefficient formation of base paired template-reagent complexes. We instead hypothesized that the significantly impaired reactivity of templated 7 and 8 arises from the unusually low degree of secondary structure within these 30 intervening bases.
Similar experiments were performed to test the effect of reagent length on the amine acylation reaction (Supporting Information, Table S4). Just as with reductive amination, the reagent length had a strong effect on the yields with the most highly structured templates (1 and 2) with longer reagents leading to higher yields. However, neither the shorter nor the longer reagent significantly altered the reactivity of highly unstructured templates 7 and 8.
Elucidation of the Basis of Impaired Long-Distance Reactivity of Unstructured Templates
Based on the above findings, we hypothesized that highly unstructured templates do not react efficiently when a large number of bases separate the reactive groups because such templates exist in a greater number of conformational states in which the reactants are separated, compared with the case involving more structured templates. Some amount of intramolecular base pairing within the intervening sequence may favor conformations in which the intervening nucleotides are compact, thereby decreasing the average separation of the reacting groups and increasing effective molarities (Figure 1C).
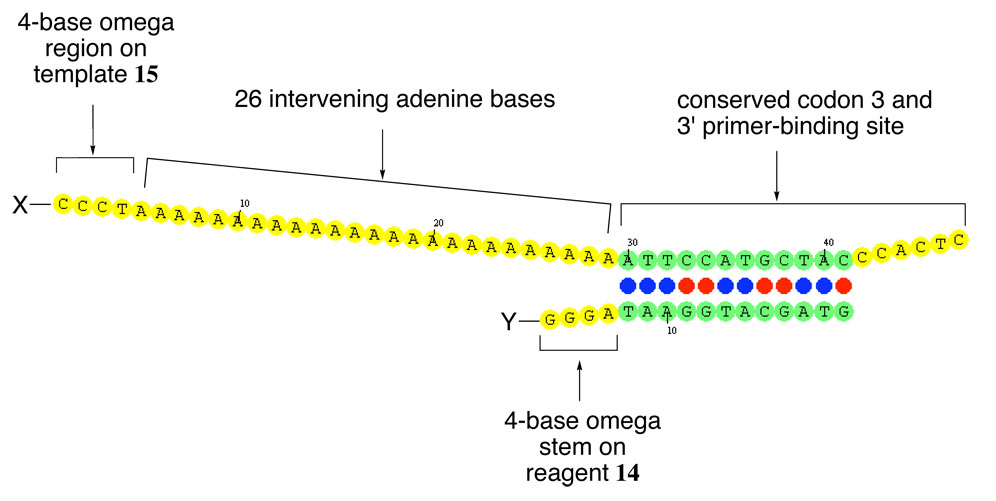
To test this model, we designed and synthesized a series of additional templates and template libraries in which we systematically varied the predicted structure of the intervening nucleotides without altering the ability of codon 3 to hybridize with the reagent. Template 15 (Figure 5) retains the four bases at the 5’ end of the template used by the omega architecture, as well as the codon 3 binding site used in the earlier templates. The other twenty-six intervening nucleotides, however, were replaced with adenosine to form a polyadenine tract separating the two functional groups. Such a stretch of sequence is not predicted to form any stable secondary structures by OMP. Prior studies17,18 that considered the optical rotatory properties and hypochromism of polyadenylic acid suggest that partially ordered structures resulting from base-stacking can occur for such a sequence, although other experiments show that the hydrodynamic properties of poly-A are consistent with a random coil model.17 Recent studies of single-stranded DNA structure in the absence of base-pairing further suggest that such a poly-A sequence could vary from the behavior of an ideal polymer due to electrostatic self-avoidance,19 and therefore might be more rigid than the classical view of a flexible random coil.20 Template 15 thus represents an extreme case of a template with a completely unstructured intervening region.
Figure 5.
Predicted structure of polyadenine-containing template 15 hybridized to reagent 14. The thirty intervening bases between the binding site for reagent 14 and the reactive 5’ end of the template contain a 4-base omega region that complements the “omega stem” in reagent 14 followed by 26 consecutive adenine bases. These thirty intervening bases are predicted to have no internal secondary structure. Template libraries 16–22 contain nucleotide mixtures of a particular composition in place of the 26 adenine bases in 15.
We reacted 15 with the end-of-helix reagents 10a or 10b under reductive amination or amine acylation conditions for 16 hours, longer than in the above reactions, and observed < 1% product yield for both reactions. Similarly, template 15 reacted with the omega architecture reagent 11a under reductive amination conditions for 8 hours to generate product in 31 % yield (compared to 49 % for highly unstructured template 8), and reacted with omega architecture reagent 11b under amine acylation conditions for 8 hours to generate product in only 24 % yield (compared to 37 % for template 8). These results collectively suggest that the presence of the highly unstructured polyadenine tract in template 15 precludes the ordering of this intervening region of the template into a conformation that allows the reactive groups in the template-reagent complex to interact. This ordering is necessary to maximize reaction efficiency in both the end-of-helix and omega architectures, and the near absence of secondary structure within the intervening region of template 15 dramatically impedes product formation.
To further test our working model behind the low reactivity of highly unstructured templates, we generated a series of template libraries in which each of the 26 intervening positions contained mixtures of nucleotides. Template libraries 16–21 contain each of the six possible mixtures of just two of the four DNA bases at all 26 intervening positions which were adenine in template 15 (Figure 5). These libraries therefore included an A/C mix (16), a G/T mix (17), a purine (A/G) mix (18), a pyrimidine (T/C) mix (19), and two mixes that contained Watson-Crick base-pairing partners: A/T (20), and C/G (21). We also synthesized a mixture that contained all four nucleotides (A/C/G/T) for library 22.
We computationally modeled the average energy distributions for these template libraries when hybridized to omega architecture reagent 14 using OMP (Table 3). The poly-A template, 15, forms no predicted secondary structure in the intervening region and has a total folding energy, including the intermolecular hybridization energy to reagent 14, of −16.4 kcal/mol. Libraries 16–19, which do not contain Watson-Crick pairing partners, have slightly more stable folding energies ranging from −17.5 to −18.8 kcal/mol with secondary structures forming exclusively to the 4-base omega stem on the reagent or to the conserved 4-base omega recognition element in the template. In contrast to 16–19, which only form secondary structures involving the 4-base omega architecture regions, 20–22 contain sequences predicted to form secondary structures throughout the intervening bases. Thus library 20, which contains an A/T mix, is predicted to hybridize intra- and intermolecularly with an average total energy of −19.7 kcal/mol, similar to the average energy of library 22 with an A/C/G/T mix. Library 21, which contains a C/G mix, has significantly more intervening region structure than the other libraries and is predicted to hybridize with an average total energy of −27.1 kcal/mol (Table 3).
Table 3.
Average folding energy and reactivity of template 15 and random libraries (16–22) with omega architecture reagents 14a and 14b. 10,000 random templates containing 26 consecutive intervening nucleotides with the composition listed were computationally generated and folded using OMP (100 nM template, 150 nM reagent 14, 1 M NaCl, 25 °C). The standard deviations for the folding energies are shown in parentheses. Product yields for reductive amination reactions with 14a and amine acylation reactions with 14b are given for each of the templates. The libraries that are not capable of forming Watson-Crick base pairs within the intervening region (15–19) are predicted to have less average structure and also exhibit lower reactivity than the libraries containing potential base pairing partners (20–22) within the intervening region.
| Library | Composition | Average Folding Energy (kcal/mol) |
Reductive Amination Yield with 14a (%) |
Amine Acylation Yield with 14b (%) |
|---|---|---|---|---|
| 15 | A only | −16.4 | 31 | 24 |
| 16 | A and C | −17.5 (1.12) | 21 | 16 |
| 17 | G and T | −18.5 (0.88) | 29 | 19 |
| 18 | C and T | −18.4 (1.69) | 16 | 27 |
| 19 | A and G | −18.8 (1.07) | 30 | 18 |
| 20 | A and T | −19.7 (1.45) | 84 | 68 |
| 21 | C and G | −27.1 (2.81) | 79 | 60 |
| 22 | A, C, G, T | −19.5 (1.97) | 74 | 64 |
We reacted these libraries with 12-base reagent 14a under reductive amination conditions (Table 3). The overall product yields for the libraries 16–19 containing mixtures of intervening nucleotides without the possibility of intramolecular Watson-Crick pairing ranged from 16–30 %, lower than that observed for template 8 and similar to the yield seen for the polyadenine template 15. In contrast, the two dimeric mixes that contain Watson-Crick pairing intervening nucleotide mixtures, 20 and 21, exhibited near maximal overall reactivity at 84 % and 79 % yield. While these libraries contain mixtures of template sequences with varying degrees of internal secondary structure, virtually all intervening template sequences within libraries 20 and 21 should be able to form some internal Watson-Crick base pairs. The library containing intervening sequences with a mixture of all four nucleotides, 22, also reacts efficiently to provide product in 74 % yield. When these experiments were repeated with the amine acylation reaction using reagent 14b, similar results were observed (Table 3).
Taken together, these results strongly support a model in which the ability of DNA-templated reactions in either the end-of-helix or omega architectures to generate product efficiently is dependent on the ability of the intervening nucleotides separating the hybridized reactive groups to participate in intramolecular base pairs. The libraries that contained the possibility for forming such structures reacted efficiently while the libraries that did not contain the possibility of forming Watson-Crick base pairing partners within this intervening region reacted poorly, despite virtually identical predicted reagent hybridization abilities.
Effects of Proximity and Strength of Intervening Sequence Secondary Structure on Reactivity
To further test our model that some degree of internal secondary structure in the intervening sequence is essential for efficient DNA-template reactivity, we explicitly designed a series of four individual templates using OMP to directly evaluate how different kinds of secondary structure in the intervening sequence can influence reactivity. These templates, 23–26, contained explicitly designed intervening region structures that varied both in their overall energy as well as in the proximity of the reactive ends of the template and reagent induced by the structure (Figure 6). Templates 23 and 24 both possess structures that bridge about 20 of the 30 intervening bases, but leave the entire primer-binding site unfolded. Template 23 has a modest folding energy while template 24 has a much stronger folding energy. Templates 25 and 26, on the other hand, possess structures that bridge all but three or four of the 30 intervening bases, placing the functional groups in much closer proximity. Template 25 has a modest folding energy similar to 23 while template 26 is predicted to form a very stable hairpin.
Figure 6.
Predicted structures of templates with designed intervening sequences annealed to 10. While the structures are shown with reagent 10 to show the proximity of the 3’ end of the 10 to the 5’ end of each template, the calculated folding energies listed reflect only the 30 intervening bases alone to provide a direct comparison of how intervening structure stability can affect reactivity. For comparison, the 30-base intervening sequence of template 8, which exhibits no significant secondary structure, is predicted to be +0.63 kcal/mole.
We compared the behavior of these four new templates with template 8, which possesses no predicted internal structure. Using the end-of-helix reagents (10a and 10b), we observed increased reactivity for the structured templates, as our model predicts (Table 4). For reductive amination with 10a, templates 25 and 26 exhibited the largest improvements in reactivity, generating product in 25 % and 55 % yield, respectively. These two templates possess the predicted structures that bring the functional groups together in closest proximity of the four templates 23–26. While template 24 has a more stable secondary structure than template 25, it does not bring the functional groups as close together and only reacted to give product in 11 % yield. For amine acylation with 10a, templates 25 and 26 again exhibited the best reactivity. Templates 23 and 24 that induced less proximity between the functional groups reacted to an intermediate degree. These results confirm that reactivity between hybridized template and reagent groups is strongly affected by intervening secondary structure and is most efficient when internal secondary structures compact intervening nucleotides, yet do not involve the reagent annealing site.
Table 4.
Reaction yields (in %) for templates containing designed intervening structures with the end-of-helix reagent 10 and omega architecture reagent 11. The results for 10a/11a under reductive amination conditions and 10b/11b under amine acylation conditions are given for each of the templates after 8 hours. Templates with intervening secondary structures that bring the reactive ends closest together (25 and 26) lead to the highest product yields for the end-of-helix reagent 10. Each of the templates with designed secondary structures (23–26) react efficiently with the omega architecture reagent 11.
| Template | |||||
|---|---|---|---|---|---|
| Reagent | 8 | 23 | 24 | 25 | 26 |
| 10a | 3 | 12 | 11 | 25 | 55 |
| 10b | 4 | 9 | 15 | 22 | 30 |
| 11a | 49 | 79 | 84 | 82 | 79 |
| 11b | 37 | 60 | 63 | 65 | 62 |
We then tested these templates containing explicitly designed intervening structures with the omega architecture reagents 11a and 11b (Table 4). For both reactions, the omega architecture resulted in very high reactivity for templates 23–26, near the maximal levels for these templates. These results indicate that templates with secondary structure within the intervening region facilitate the formation of the omega architecture to fully restore reactivity.
Discussion
Taken together, these results support a model where both very high amounts of secondary structure and very low amounts of secondary structure within DNA templates compromise DNA-templated reactivity. As expected, high amounts of secondary structure when involving the reagent-binding site can block reagent hybridization and thereby prevent reaction. Very low amounts of template structure, on the other hand, impair the natural ability of most mixed-sequence DNA strands to adopt weakly folded conformations that compact intervening nucleotides and therefore increase the effective molarities of flanking reactants. The omega architecture can restore some of this reactivity over long distances, but cannot fully restore reactivity for the most unstructured templates.
The ability of nucleic acid secondary structure to interfere with hybridization has been observed for experiments involving natural nucleic acids as well. As one example, the potency of antisense oligonucleotides to natural mRNA molecules has been shown, in both in vivo and in vitro experiments, to be inversely related to the degree of secondary structure in the target.21 In addition, siRNAs that produce unstructured guide RNAs resulted in an improved efficiency of RNA interference, suggesting that secondary structure may have been an important factor during the evolution of these sequences.22 Riboswitches provide an additional example of natural nucleic acids in which changes in secondary structure conceal a particular sequence from being recognized by macromolecular machinery.23 In response to a natural metabolite, some riboswitches will form an ordered structure that conceals the ribosome-binding site within a long stem, effectively blocking translation. Intramolecular secondary structure is therefore a common functional control element in living systems that can strongly affect the recognition of single-stranded nucleic acid sequences.
While the problematic behavior of highly structured templates was expected, the reduced reactivity of unstructured templates was surprising. It is tempting to speculate that the strongly decreased effective molarities we observed when intervening template sequences are highly unstructured may also be relevant in living systems. For example, the effective molarity of two proteins bound to the same single-stranded nucleic acid sequence may be significantly influenced by the presence or absence of secondary structure within the intervening nucleotides, even when no single intramolecular structure is obviously favored. Unstructured regions in mRNA may therefore play an important role in controlling processes such as pre-mRNA splicing or translation where multiple proteins that are bound to different sites of an RNA template must interact. It may be possible to test this hypothesis bioinformatically by integrating secondary structural predictions with the widespread availability of genome24,25 and small RNA26 sequences.
These findings have significant implications for DNA-templated library synthesis. Secondary structure involving codon sequences must be minimized to avoid impaired reactivity. Our results suggest that for typical 10–12 base coding regions, avoiding secondary structures more stable than −7 kcal/mol will be sufficient. In addition, our findings indicate that some internal secondary structure in the templates is necessary to maximize reactivity when reagents are hybridized far from the reactive end of the template. Past studies examining the behavior of DNA-templated reactions at varying distances used a single thirty-base template with a predicted folding energy of −4.38 kcal/mol.12,16 The initial eight templates studied here, particularly 7 and 8, possess much less structure in the intervening thirty bases than the sequence used in the earlier studies, indicating that different degrees of template secondary structure can influence the apparent distance dependence of a reaction. Maintaining at least ~−3 kcal/mol of predicted secondary structure in the template’s intervening region is therefore ideal to achieve long-distance reactivity at reasonable rates.
The omega template-reagent architecture promotes DNA-templated reactivity by bringing the reactive end of a reagent close to the reactive end of the template. The omega architecture induces the looping out of bases in the template and our results imply that some amount of internal structure in this looped-out intervening region is helpful to offset the entropic costs of forming the omega architecture. When a template is highly unstructured, the omega architecture cannot form as efficiently and reactivity is not completely restored.
The ideal template design for a DNA-templated library will therefore have an energy between the extremes of too much structure and too little structure. Within this regime, reagent hybridization will not be affected by competing intramolecular secondary structures in the templates, and reactivity once bound to the template will not be affected by the inability of unstructured templates to bring together distant functional groups.
Conclusion
The studies describe here have resulted in a new understanding of the relationship between DNA sequence and DNA-templated reactivity. Intramolecular base pairing involving the reagent hybridization site within a template blocks reagent binding and impairs reactivity, as expected. Surprisingly, templates devoid of internal structure also react very poorly when reactants are encoded far away from the reactive end of the template because intervening sequences that are highly unstructured keep reactants more separated than templates in which intervening regions possess some internal structure. Once hybridized, the rate of reaction is determined by how frequently the reactive ends of the template and reagent can encounter each other. Secondary structure within the intervening sequences helps to bridge long distances and improve reaction rates.
Alternate reagent architectures, such as the omega architecture, can improve reactivity significantly and also operate best when intervening sequences have the possibility to form stable structures to offset the entropic cost of looping out so many bases. Very unstructured templates react poorly even with the omega architecture, and previously distance-independent reactions such as amine acylation exhibit distance dependence with very unstructured templates. We have already begun to incorporate these principles into the design of optimized constant sequences and codon sets for DNA-templated small-molecule library synthesis, avoiding the extremes of DNA secondary structure that can compromise reactivity. These principles may also have relevance to living systems, in which the effective molarities of two molecules bound to the same strand of a nucleic acid may vary significantly depending on the degree of secondary structure within the intervening region.
Supplementary Material
DNA sequences used in this work as well as additional experimental results and complete Ref. No. 24–26. This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
This research was supported by the NIH/NIGMS (R01GM065865) and the Howard Hughes Medical Institute. T.M.S. and B.N.T. gratefully acknowledge the support of an NSF Graduate Research Fellowship. T.M.S. also acknowledges the support of an ACS Division of Organic Chemistry Fellowship sponsored by Organic Reactions, Inc.
References Cited
- 1.Li X, Liu DR. Angew Chem Int Ed Engl. 2004;43:4848–4870. doi: 10.1002/anie.200400656. [DOI] [PubMed] [Google Scholar]
- 2.Calderone CT, Puckett JW, Gartner ZJ, Liu DR. Angew Chem Int Ed Engl. 2002;41:4104–4108. doi: 10.1002/1521-3773(20021104)41:21<4104::AID-ANIE4104>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 3.Snyder TM, Liu DR. Angew Chem Int Ed Engl. 2005;44:7379–7382. doi: 10.1002/anie.200502879. [DOI] [PubMed] [Google Scholar]
- 4.Calderone CT, Liu DR. Angew Chem Int Ed Engl. 2005;44:7383–7386. doi: 10.1002/anie.200502899. [DOI] [PubMed] [Google Scholar]
- 5.Kanan MW, Rozenman MM, Sakurai K, Snyder TM, Liu DR. Nature. 2004;431:545–549. doi: 10.1038/nature02920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Momiyama N, Kanan MW, Liu DR. J Am Chem Soc. 2007;129:2230–2231. doi: 10.1021/ja068886f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gartner ZJ, Tse BN, Grubina R, Doyon JB, Snyder TM, Liu DR. Science. 2004;305:1601–1605. doi: 10.1126/science.1102629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.SantaLucia J, Jr., Hicks D. Annu Rev Biophys Biomol Struct. 2004;33:415–440. doi: 10.1146/annurev.biophys.32.110601.141800. [DOI] [PubMed] [Google Scholar]
- 9.SantaLucia J., Jr. In: Methods in Molecular Biology: PCR Primer Design. Yuryev A, editor. Totowa, New Jersey: Humana Press; 2006. [Google Scholar]
- 10.Zuker M. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dirks RM, Bois JS, Schaeffer JM, Winfree E, Pierce NA. SIAM Rev. 2007;49:65–88. [Google Scholar]
- 12.Gartner ZJ, Liu DR. J Am Chem Soc. 2001;123:6961–6963. doi: 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gartner ZJ, Kanan MW, Liu DR. J Am Chem Soc. 2002;124:10304–10306. doi: 10.1021/ja027307d. [DOI] [PubMed] [Google Scholar]
- 14.Gartner ZJ, Grubina R, Calderone CT, Liu DR. Angew Chem Int Ed Engl. 2003;42:1370–1375. doi: 10.1002/anie.200390351. [DOI] [PubMed] [Google Scholar]
- 15.Li X, Gartner ZJ, Tse BN, Liu DR. J Am Chem Soc. 2004;126:5090–5092. doi: 10.1021/ja049666+. [DOI] [PubMed] [Google Scholar]
- 16.Gartner ZJ, Kanan MW, Liu DR. Angew Chem Int Ed Engl. 2002;41:1796–1800. doi: 10.1002/1521-3773(20020517)41:10<1796::aid-anie1796>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 17.Felsenfeld G, Miles HT. Annu Rev Biochem. 1967;36:407–448. doi: 10.1146/annurev.bi.36.070167.002203. [DOI] [PubMed] [Google Scholar]
- 18.Saenger W, Riecke J, Suck D. J Mol Biol. 1975;93:529–534. doi: 10.1016/0022-2836(75)90244-2. [DOI] [PubMed] [Google Scholar]
- 19.Dessinges MN, Maier B, Zhang Y, Peliti M, Bensimon D, Croquette V. Phys Rev Lett. 2002;89:248102. doi: 10.1103/PhysRevLett.89.248102. [DOI] [PubMed] [Google Scholar]
- 20.Goddard NL, Bonnet G, Krichevsky O, Libchaber A. Phys Rev Lett. 2000;85:2400–2403. doi: 10.1103/PhysRevLett.85.2400. [DOI] [PubMed] [Google Scholar]
- 21.Vickers TA, Wyatt JR, Freier SM. Nucleic Acids Res. 2000;28:1340–1347. doi: 10.1093/nar/28.6.1340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Patzel V, Rutz S, Dietrich I, Koberle C, Scheffold A, Kaufmann SH. Nat Biotechnol. 2005;23:1440–1444. doi: 10.1038/nbt1151. [DOI] [PubMed] [Google Scholar]
- 23.Tucker BJ, Breaker RR. Curr Opin Struct Biol. 2005;15:342–348. doi: 10.1016/j.sbi.2005.05.003. [DOI] [PubMed] [Google Scholar]
- 24.Venter JC, et al. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 25.Lander ES, et al. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 26.Kapranov P, et al. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DNA sequences used in this work as well as additional experimental results and complete Ref. No. 24–26. This material is available free of charge via the Internet at http://pubs.acs.org.